Abstract
EGFR is the most common genetically altered oncogene in glioblastoma (GBM), but small molecule EGFR tyrosine kinase inhibitors (TKIs) have failed to yield durable clinical benefit. Here we show that in two novel model systems of acquired resistance to EGFR TKIs, elevated expression of urokinase plasminogen activator (uPA) drives signaling through the MAPK pathway, which results in suppression of the pro-apoptotic BCL2-family member protein BIM (BCL2L11). In patient-derived GBM cells and genetic GBM models uPA is shown to suppress BIM levels through ERK1/2 phosphorylation, which can be reversed by siRNA mediated knockdown of uPA. TKI-resistant GBMs are re-sensitized to EGFR TKIs by pharmacological inhibition of MEK or a BH3 mimetic drug to replace BIM function. A link between the uPA-uPAR-ERK1/2 pathway and BIM has not been previously demonstrated in GBM, and involvement of this signaling axis in resistance provides rationale for a new strategy to target EGFR TKI-resistant GBM.
Keywords: glioblastoma, EGFR, Bim, resistance, uPAR
Introduction
The epidermal growth factor receptor (EGFR) is a frequently amplified and mutated gene in glioblastoma (GBM) (1). The constitutively active deletion mutant, ΔEGFR (also known as EGFRvIII), is a therapeutic target as it confers enhanced tumorigenicity (2) and is required for maintenance of glioma growth in vivo (3). EGFR tyrosine kinase inhibitors (TKIs) are approved for treatment of certain malignancies in which kinase domain-mutated EGFR is a driver oncogene, such as non-small cell lung cancer (NSCLC), and patients with tumors that harbor these mutations are sensitive to the drugs (4, 5). By contrast, the efficacy of EGFR TKIs in GBM has been limited by inherent and acquired TKI resistance (6).
While resistance to EGFR-targeted therapy occurs in other solid tumors (7, 8), the same mechanisms do not appear to be operational in most GBMs (9–11). For example, the T790M gatekeeper mutation is a predominant mechanism of acquired resistance to EGFR inhibitors in NSCLC (12), but is not found in TKI-treated GBM tumors. Thus, there is precedence for the existence of distinct resistance mechanisms in GBM. Some GBMs demonstrate PTEN deletion, which is associated with EGFR TKI-resistance due to activation of Akt signaling (13). Activation of TKs other than EGFR also may contribute to resistance (14), as in the cases of Src- and FGFR-induced phosphorylation of PTEN at tyrosine 240 (15).
uPA receptor (uPAR) is a glycosylphosphatidylinositol-anchored membrane protein which associates with integrins and RTKs to form a potent signaling complex (16, 17). uPAR-initiated cell-signaling inhibits apoptosis, promotes release of tumor cells from dormancy (18, 19), induces stem cell-like properties in cancer cells, and promotes invasion and metastasis (20–22). We have previously shown that in GBM cells, uPAR expression is required for escape from ΔEGFR oncogene dependence in vivo (23), which suggests that uPAR-mediated signaling may serve as an EGFR TKI resistance mechanism. However, gene silencing in vivo may not model changes that occur in GBM treated with targeted drugs. Herein, we describe novel model systems of GBM acquired resistance in which EGFR TKIs were administered chronically in vitro or in vivo. Using these model systems, we demonstrate that uPA-uPAR signaling mediates repression of Bim, a pro-apoptotic protein, to promote TKI resistance in GBM. Regulating Bim levels by targeting uPAR or by modulating signaling pathways downstream of uPAR may represent a strategy to overcome resistance to EGFR-targeted therapies in GBM.
Materials and Methods
Cell culture and reagents
Ink4a/arf−/− ΔEGFR murine astrocytes were a gift from Ronald DePinho and Robert Bachoo (24). U373 ΔEGFR-dependent and escaper cells were cultured as described previously (25). U373 and murine astrocytes were recently authenticated using short tandem repeat (STR) DNA fingerprinting by ATCC (Manassas, VA, USA) and DDC (Fairfield, OH, USA), respectively. Gefitinib, erlotinib, and U0126 were obtained from LC Labs (Woburn, MA). GBM39 cells were a gift from David James, UCSF (26). ABT-737 was obtained from Selleck Chemicals and provided by the Small Molecule Development group at the Ludwig Institute for Cancer Research. GSK1120212 was kindly provided by Tona Gilmer of GlaxoSmithKline. Cisplatin was obtained from Calbiochem.
In vitro TKI resistance
Ink4a/arf−/− ΔEGFR astrocytes (24) were seeded in 0.3% agar on top of a base layer of 0.6% agar at 2×103 cells per well in a 6-well dish in 1 μM gefitinib, erlotinib, or vehicle (DMSO). A feeder layer containing drug or vehicle was replenished 2x per week. After 2 weeks, colonies greater than or equal to ~50 cells were counted in 5 random fields per well, and the drug dose was escalated to 2 μM. 3–5 weeks later, drug-resistant colonies visible to the naked eye were isolated using a blunt-ended pipette tip, dissociated, plated and maintained in 2 μM gefitinib or erlotinib.
In vivo TKI resistance
All animal procedures were approved by the IACUC at the University of California, San Diego. 1×106 ink4a/arf−/− ΔEGFR astrocytes were injected subcutaneously into the flanks of 6–8 week old female athymic mice. When average tumor volume reached ~500 mm3, mice received vehicle, 200 mg/kg gefitinib, or 150 mg/kg erlotinib daily 5x per week (M-F) by oral gavage. To generate TKI-resistance, mice continued to receive TKIs until tumors grew to 1500 mm3 or became ulcerated. Erlotinib was decreased to 75 mg/kg after 10 doses due to drug-induced skin rash. To establish cell lines, tumors were removed from mice, and mechanically dissociated in sterile PBS and cultured in media containing 1 μg/ml puromycin to ensure elimination of any contaminating mouse cells.
Cell viability assays
1×103 cells were seeded in a 96-well plate in media containing 1% FBS. 12–15 h later, drugs or vehicle were added in quadruplicate wells and incubated for 72 h. Viability was assessed with the WST-1 cell proliferation reagent (Clontech, Mountain View, CA). Drug treatments were normalized to vehicle-treated control to calculate percent cell viability.
Western blotting
15–40 μg of cell lysate was resolved on a 7.5–15% polyacrylamide gel. Proteins were transferred, blocked with 5% milk or 3% BSA followed by primary antibody incubation. Primary antibodies included: EGFR pY1068, AKT pS473, Bim, phospho-S6 ribosomal protein, S6 ribosomal protein, phospho-p42/44 MAPK, p42/44 MAPK, PARP (Cell Signaling Technology, Danvers, MA), EGFR clone 13 (BD Transduction Labs, Franklin Lakes, NJ), PTEN, Tubulin (Santa Cruz Biotechnology, Dallas, TX), and β-actin (Sigma, St. Louis, MO). Membranes were washed 3 times with TBS + 0.1% Tween-20 and incubated with anti-mouse or -rabbit-HRP antibody (Sigma). Chemiluminescence was detected using Amersham ECL Western Blotting Detection Reagent (GE Healthcare) or SuperSignal West Pico Chemiluminescent Substrate (ThermoScientific, Rockford, IL). Images were obtained using ChemiDoc MP imaging system and Image Lab 4.1 software (Bio-Rad, Hercules, CA).
Orthotopic injections and bioluminescent imaging
Cells were transduced with the pLV.FLuc.GFP lentivirus as previously described (27). 48 h following infection, in vitro luminescence was determined using the luciferase assay system (Promega, Madison, WI), or 1 × 105 cells were stereotactically injected into the brains of 6–8 week old female athymic mice, 2mm lateral and 1mm anterior to the bregma. Tumor growth was monitored by bioluminescent imaging (BLI), whereby D-luciferin was injected intraperitoneally at a dose of 150 mg/kg, and after 5 minutes luminescence was determined as measured by total flux using IVIS spectrum (Perkin Elmer). Once tumors were detectable, mice were treated 5x per week (M-F) by oral gavage with 200 mg/kg gefitinib or vehicle (0.5% Tween-80, 0.5% methylcellulose). IVIS imaging software was used to quantify total flux of luminescence in regions of interest.
Quantitative real-time PCR
RNA was isolated with the RNeasy Kit (Qiagen). cDNA was synthesized with the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed with a System 7300 (Applied Biosystems) and a one-step program: 95 °C for 10 min, 95 °C for 30 s, and 60 °C for 1 min for 40 cycles. uPA and HPRT-1 gene expression were measured using TaqMan assays (Invitrogen) in triplicate with internal duplicate determinations.
siRNA Transfection
25 nM uPA-specific siRNA (5′-CAUGUUACUGACCAGCAAC-3′) or non-targeting control (NTC) siRNA (Dharmacon) siRNAs were introduced into cells using Lipofectamine 2000 (Invitrogen) in serum-free medium (SFM) for 4 h. Cultures were allowed to recover in serum-containing medium for 12 hr and serum starved for 36 hr. The extent of gene silencing was determined by qPCR and immunoblot.
GBM 39 xenografts
1×106 GBM 39 cells in PBS were injected subcutaneously into the flanks of nude mice in a 1:1 mixture with Matrigel (BD Biosciences) in a final volume of 100 μl. Following tumor establishment as measured using calipers, mice were randomized to receive vehicle or erlotinib at a dose of 150 mg/kg delivered by oral gavage daily for 4 days.
uPA activity
Cells were serum starved in phenol-red free media overnight and conditioned media concentrated 20-fold using centricon (Millipore). Concentrated conditioned media was diluted 1:15 with 0.65 mM Val-Leu-Lys-p-nitroanilide (VLK-pNA) (Bachem, Torrance, CA) and 0.94 mM plasminogen purified from human plasma as previously described (28) in PBS. After incubating samples at 37°C for 1 hour, VLK-pNA hydrolysis was detected by measuring absorbance at 406 nm.
Immunohistochemistry
Tissue sections were cut from formalin-fixed paraffin embedded xenograft tissue. Four micron sections were stained with anti-phospho-p44/42 MAPK (Thr202/Try 204) (Cell Signaling) on a Ventana Discovery Ultra (Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval was performed using CC1 for 24 minutes at 95 °C. Primary antibody was incubated at 1:1800 for 1 hour at 37°C followed by UltraMap (Ventana) and DAB detection. Slides were dehydrated through alcohol and xylene and coverslipped. Phospho-epitope labeling was validated by eliminating staining after treating sections with lambda protein phosphatase (New England Biolabs; 2400U) for 2 h at 37°C before application of primary antibody. Prior to phosphatase treatment, tissue sections were de-paraffinized and antigen retrieval performed as described above.
TCGA analysis
TCGA glioblastoma transcriptomal profiles were downloaded from TCGA Data Portal (http://tcga-data.nci.nih.gov/tcga/). Level 3 normalized, Agilent 244K gene expression mRNA array data was downloaded and genes were median centered. For each specimen, MAPK pathway activity score was calculated using the t-score method developed by Creighton et al. (29). In brief, each gene in the MAPK signature that was over-expressed was assigned the value of +1; genes that were under-expressed were assigned −1. The normalized expression values of the signature genes in the clinical specimen were then plotted against these assigned values, and Pearson Correlation Coefficient was determined. The correlation ranged from −1 (denoting low pathway activity) to +1 (denoting high pathway activity).
Statistical analyses
All statstical analyses were performed using GraphPad Prism 6 software. The specific test used for each experiment to determine significance (p<0.05) is indicated in the text of the results and/or figure legends. Data are representative of results obtained in at least 3 independent experiments.
Results
Bim is associated with EGFR TKI sensitivity in GBM and suppressed by uPAR signaling
To define markers of EGFR TKI response in GBM, we examined the ΔEGFR-expressing patient derived xenograft GBM39, which is sensitive to EGFR blockade (30). Erlotinib caused dramatic tumor regression in mice (Fig. 1A) which coincided with upregulation of Bim (Fig. 1B), a pro-apoptotic Bcl2-family member (31). Moreover, Bim levels were, on average, higher in tumors from patients treated with the EGFR/HER2 inhibitor lapatinib (30) who were therapeutic responders than non-responders (Fig. S1A).
Figure 1.
Bim is associated with EGFR inhibitor sensitivity and uPA expression. A: Volume of subcutaneous GBM 39 xenografts prior to and following treatment with vehicle (n=2) or erlotinib (n=4) for 4 days. p<0.05, unpaired t-test vehicle vs erlotinib. B: Western blot of the indicated proteins in lysates from GBM 39 tumors treated with vehicle or erlotinib. Each lane represents a different tumor. C: Western blot of the indicated proteins in U373 ΔEGFR-dependent cells −Dox or +Dox and escaper 1 and 2 cells (both +Dox) transfected with uPA siRNA (si-uPA) or control siRNA (si-Ctrl).
We previously developed a genetic model in which doxycycline (dox)-mediated suppression of ΔEGFR inhibits U373 MG xenograft tumor growth until the eventual emergence of escaper tumors that have circumvented ΔEGFR oncogene dependence. This model is analogous to sensitivity and resistance to EGFR TKIs, respectively. The Ras-MAPK pathway is thought to contribute to the escape process (3), and cell lines established from escaper tumors, which harbor sustained ΔEGFR inhibition, demonstrate increased uPA expression and dependency on uPAR signaling to maintain ERK1/2 activation (23, 32). Since Bim was associated with response to pharmacological EGFR inhibition (Fig 1A–B), we asked if there was a relationship between uPAR and Bim in this genetic model. Cells were transfected with uPA-specific or non-targeting control siRNA, and uPA gene-silencing was greater than 95% as determined by qPCR (23, 32). Interestingly, when uPA was silenced in cells lacking ΔEGFR, a very robust increase in Bim and a decrease in phospho ERK1/2 were observed (Fig. 1C, Lanes 4,6,8). These results suggest that uPAR signaling plays a key role in suppressing Bim expression when ΔEGFR is neutralized in GBM cells, possibly through the Ras-MAPK pathway.
EGFR TKI-resistance occurs in vitro and in vivo in response to chronic drug dosing
To address the hypothesis that uPAR signaling suppresses Bim and promotes cell survival during EGFR inhibition, we developed novel model systems of EGFR TKI resistance. We utilized ΔEGFR-expressing, PTEN wild-type primary ink4a/arf −/− murine astrocytes that form highly invasive intracranial tumors in mice similar to GBMs (24). Importantly, ΔEGFR and wild-type PTEN are associated with response to EGFR TKIs in GBM patients (13, 24). These cells and the tumors generated from them are hereafter referred to as “parental”. To generate resistance, gefitinib and erlotinib were administered chronically to parental cells seeded in soft agar and to mice harboring established parental xenograft tumors. As anticipated, the TKIs inhibited colony formation in soft agar (Fig. S1B) and caused dramatic regression of established tumors in vivo (Fig. S1C).
Resistance developed in vitro within 17–32 days following initial response. TKI-resistant colonies were isolated and expanded to generate clonal cell lines isolated as resistant to gefitinib or erlotonib, denoted “G−” and “E−”, respectively (Fig. S1D). Chronic TKI treatment of mice harboring xenograft tumors elicited a sustained period of dormancy for ~20 days, after which tumors became resistant and rapidly increased in size (Fig. 2A). Cell lines generated from vehicle-treated sensitive parental tumors as well as from the gefitinib or erlotinib-resistant tumors were denoted “GR−” and “ER−” respectively to distinguish them from the in vitro resistant cell lines.
Figure 2.
Models of acquired TKI resistance. A: Mean tumor volume in vehicle-treated mice and those that continued on erlotinib or gefitinib following initial 5-day treatment. n=16, vehicle and gefitinib. n=14, erlotinib. B: Viability of ΔEGFR astrocytes (Parent) or TKI-resistant cells derived from resistant colonies isolated from soft agar. G1, G5, G8, and G12 were isolated as gefitinib-resistant; E4 and E5 as erlotinib-resistant. Cells were treated with 5 μM gefitinib (top) or erlotinib (bottom) for 72 h and viability normalized to vehicle-treated control cells. *, p< 0.05 vs. parent, one-way ANOVA and Dunnett’s Multiple Comparison Test. C: Viability of cell lines generated from TKI-sensitive (parent) and –resistant (GR-1, GR-7) tumors in (A) in response to treatment with 5 μM gefitinib for 72 h. Each was normalized to vehicle-treated control. *, p< 0.05 vs. Parent, one-way ANOVA and Bonferonni Multiple Comparison test. D: Bioluminescent imaging of mice bearing intracranial tumors established from parent or G12 cells and treated with either vehicle or gefitinib for 13 days. Each image represents a different mouse at day 13. *, p<0.05, Two-way ANOVA and Bonferroni Multiple Comparison Test for parent vehicle vs. gefitinib. G12 vehicle vs. gefitinib, not significant. n=4, parent + vehicle, parent + gefitinib, G12 + gefitinib. n=3, G12 + vehicle.
TKI-resistance was confirmed in vitro in cell lines generated from soft agar (Fig. 2B, S1D) and from tumors (Fig. 2C). Parental cell viability was significantly inhibited in a dose-dependent manner by gefitinib or erlotinib, while this was not the case for the resistant cell lines (Fig. 2B–C, S2A–C). To test whether EGFR TKI resistance was maintained orthotopically in vivo, parental and G12 cells transduced with a luciferase-expressing lentivirus (Fig. S3A) were grown intracranially in mice and, once established as tumors, were treated with gefitinib or vehicle. Parental tumors decreased in size in response to gefitinib (Fig. 2D, p<0.05), but G12 tumors were unaffected (Fig. 2D). Thus, TKI resistance occurs as a result of chronic dosing of ink4a/arf −/− ΔEGFR–expressing astrocytes and xenograft tumors.
Molecular characterization of EGFR TKI resistance
We first assessed ΔEGFR expression, phosphorylation, and PI3K pathway activity as potential mechanisms contributing to EGFR TKI resistance. In resistant cells and tumors, ΔEGFR was expressed and phosphorylated upon drug removal, indicating that the cells were not resistant due to loss of the drug target (Fig. 3A, D and S3B–C). Acute treatment of parental cells with gefitinib caused a slight decrease in total EGFR, and many of the resistant cell lines exhibited similar total EGFR levels. This observation was consistent with previous reports of cells that have decreased levels of the EGFR family member ErbB2 upon treatment with afatinib, a dual EGFR/ErbB2 inhibitor (33) (Fig. 3D). Gefitinib or erlotinib inhibited EGFR phosphorylation in resistant samples (Fig. 3A, D and S3B), which indicated that the drug maintained access to and could effectively inhibit the target. These observations ruled out two candidate mechanisms of TKI resistance: efflux pump-mediated decreased intracellular drug concentration and a point mutation analogous to T790M, which decreases the ability of drug to bind to and inhibit EGFR kinase activity.
Figure 3.
Characterization of TKI resistance. A: Western blot of the indicated proteins in TKI-sensitive parent or resistant (G1, E4) cells treated for 24 h with 2 μM gefitinib or erlotinib as indicated. Resistant cells were removed from drug for 24 h before treatment. B: Western blot of the indicated proteins in parent and resistant cells isolated from tumors and treated for 24 h with 2 μM gefitinib. Resistant cells were removed from drug for 24 h before treatment. C: Western blot of PTEN in parent cells treated for 24 h with 2 μM gefitinib or erlotinib and TKI-resistant cells grown in 2 μM of gefitinib or erlotinib. D: Western blot of the indicated proteins in cell lines generated from tumors. Parent cells were treated for 24 h with 2 μM gefitinib, and resistant cell lines were maintained in 2 μM gefitinib or erlotinib as indicated.
The PI3K/AKT pathway is a canonical EGFR signaling pathway that has been implicated in resistance to EGFR-targeted drugs (13). Phosphorylation of S6, a downstream effector of PI3K, was suppressed by gefitinib or erlotinib treatment to the same extent in parental and resistant cells from the in vitro (Fig. 3A) and in vivo (Fig. 3B) model systems. Expression of PTEN, a negative regulator of the PI3K signaling pathway, was unaltered in most resistant clones with the exception of clone G5 from the in vitro system, which exhibited decreased PTEN levels compared to parent cells (Fig. 3C). As anticipated for a cell line with decreased expression of a PI3K signaling negative regulator, clone G5 had attenuated inhibition of AKT when treated with gefitinib (Fig. S3B). Decreased PTEN also occurred infrequently in the in vivo resistance system, in 2 of the 20 cell lines from resistant tumors (Fig. 3D: GR-2, GR-8). Overall, our results suggest that the predominant mechanism of acquired resistance in these model systems is not associated with modulation or function of the target itself and involves pathways other than PI3K-Akt-S6.
Increased uPA is a uniform feature of acquired resistance
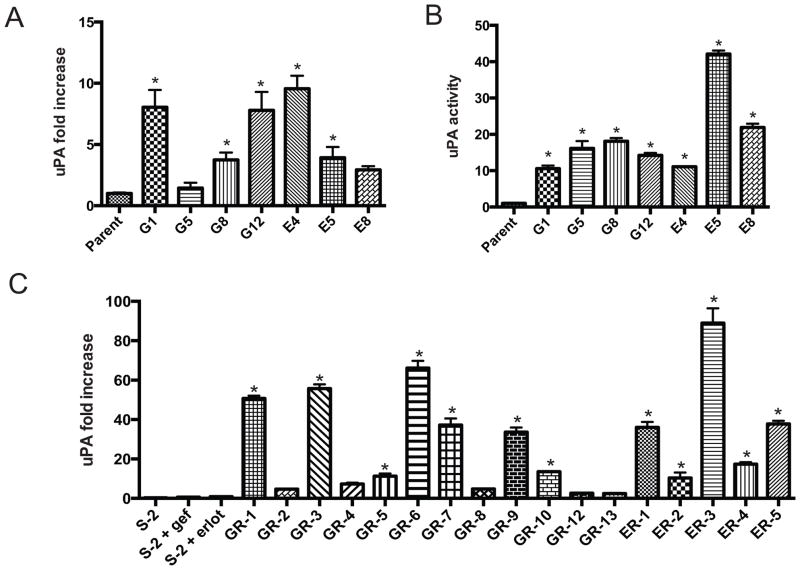
uPAR-mediated signaling was essential for escape of ΔEGFR-silenced U373 MG xenograft tumors from oncogene dependence (23). Moreover, GBM cells typically express high levels of uPAR (34), which mediates anti-apoptotic signaling (19) and induces release of cells from dormancy (18, 19). uPAR signaling is dependent upon activation by the uPAR ligand, uPA, which is often secreted by cancer cells or other cells in the tumor microenvironment (16). uPA mRNA expression was significantly increased compared to parental cells in 5/7 of the TKI-resistant cell lines from the in vitro system (Fig. 4A). uPA activity in conditioned medium was significantly increased in 7/7 resistant cell lines (Fig 4B). The slight difference in between mRNA and activity may be explained by the presence of endogenous uPA inhibitors which, if downregulated, would contribute to elevated uPA activity.
Figure 4.
uPA/uPAR activity is elevated in TKI-resistance. A: qPCR of uPA fold increase in cells. *, p<0.05 vs parent, one-way ANOVA and Dunnett’s Multiple Comparison Test. B: uPA activity assay in the indicated cells from A. Parent cells were grown in the absence of drug; G1, G5, G8, and G12 cells were grown in the presence of 2 μM gefitinib; E4, E5, E8 were grown in the presence of 2 μM erlotinib. C: qPCR of uPA fold increase as in A. Parent cells were treated for 24 h with vehicle (veh) or 2 μM gefitinib (gef) or erlotinib (erlot); GR− cells were maintained in 2 μM gefitinib; ER− cells were maintained in 2 μM erlotinib. *, p<0.05 vs parent, one-way ANOVA and Sidak’s Multiple Comparison Test.
We also examined uPA mRNA in the cell lines that acquired resistance to erlotinib or gefitinib in vivo. There was variability in the level of uPA mRNA in the different cell lines; however, in some cells, the increase in mRNA exceeded 50-fold. 12/18 cell lines demonstrated a statistically significant increase in uPA mRNA expression compared with parental cells (Fig 4C).
TKI-resistance converges on failure to undergo Bim-mediated apoptosis
Because the uPA-uPAR signaling axis is involved in survival signaling and resistance to TKIs could be caused by differences in cell death, we investigated the ability of gefitinib or erlotinib to induce apoptosis. PARP cleavage, an indicator of apoptosis, was substantial in parental cells treated with gefitinib or erlotinib but was far less pronounced in TKI-resistant cells (Fig. 5A). Furthermore, TKI treatment caused significant elevation of caspase 3/7 activity (Figure S4A) and Annexin/PI staining (Fig. S4B) in parental cells and not resistant cells (Figure S4A). Interestingly, TKI-resistant cells exhibited decreased apoptosis in response to the cytotoxic agent cisplatin (Fig. S4C). These results indicate that an anti-apoptotic TKI-resistance mechanism is induced in these model systems and suggest that it is general in nature.
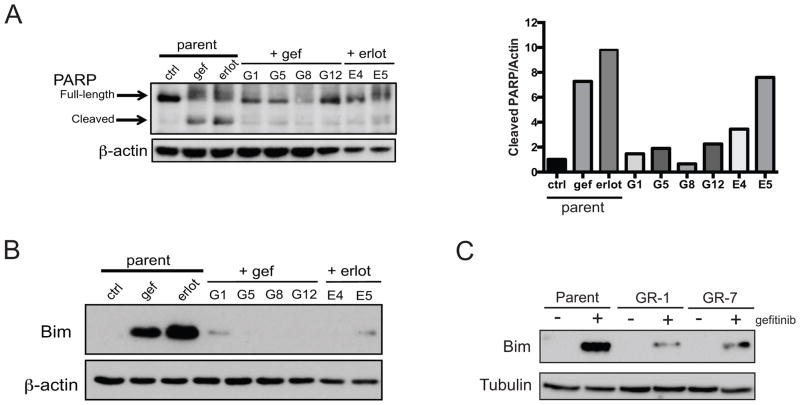
Figure 5.
Apoptotis and Bim are altered in GBM TKI-resistance. A: Left, Western blot of full-length and cleaved PARP in the indicated cell lines. Parent cells were treated with 2 μM gefitinib or erlotinib for 24 h; resistant cells were grown in 2 μM of TKI as indicated. Right, densitometry analysis of cleaved PARP normalized to β-actin. B: Western blot of Bim in the indicated cell lines treated as in (A). C: Western blot of Bim in the indicated cell lines treated with 2 μM gefitinib for 24 h. GR-1 and GR-7 were removed from maintenance dose of TKI 24 h prior to treatment.
To determine if the induction of Bim observed in erlotinib-treated GBM39 tumors (Fig. 1B) and after genetic suppression of ΔEGFR in U373 cells (Fig. 1C) is altered in our models of TKI resistance, we treated parental and resistant cells with gefitinib or elotinib. Bim was greatly induced in the TKI-sensitive cells upon treatment with either drug Fig. 5B–C). However, Bim induction was diminished in TKI-resistant cell lines generated in vitro (Fig. 5B) and in vivo (Fig. 5C). Together, these results support our findings from other GBM systems that expression of Bim is associated with TKI-sensitivity and suggest that altered apoptotic signaling and decreased expression and/or function of Bim are central features of TKI-resistance.
TKI-resistance requires Mek-ERK signaling and suppression of Bim
The Mek-ERK pathway plays an important role in mediating uPAR-dependent anti-apoptotic signaling (19). Thus, we tested whether resistance to EGFR TKIs in the astrocyte model systems reflects ERK1/2 activation. TKI-resistant cell lines and tumors possessed elevated levels of ERK1/2 phosphorylation compared with their sensitive counterparts when treated with TKIs (Fig. 6A–C).
Figure 6.
Mek inhibition or a Bim mimetic restore TKI sensitivity. A: Western blot of phospho-and total ERK1/2 in parent cells treated for 24 h with 2 μM gefitinib or erlotinib as indicated and TKI-resistant cell lines G1, G12, E4, E5, and GR-7 grown in 2 μM erlotinib or gefitinib. B: Western blot of phospho- and total ERK in TKI-sensitive and resistant xenograft tumor lysates. Parent tumors were from mice treated for 4 days with 200 mg/kg gefitinib. ‘Resistant’ indicates tumors from Figure 2A that were chronically treated with gefitinib. C: pERK immunohistochemistry in xenograft tumors. Parent tumors were from mice treated with vehicle or gefitinib for 4 days; resistant tumors were isolated from mice receiving chronic gefitinib treatment. D: Viability of TKI-sensitive (parent) and –resistant (GR-1, GR-7) cells treated with vehicle, 1 μM gefitinib, 10 μM UO126, or the combination for 24 h. E: Viability of cells from D treated with vehicle, 1 μM gefitinib, 1 μM ABT-737, or the combination for 24 h. *, p< 0.05 vs. parent, one-way ANOVA and Dunnett’s Multiple Comparison Test. F: MAPK signaling gene expression signature compared to uPA expression in GBM.
The functional importance of elevated ERK1/2 activity in promoting TKI resistance was determined by assessing whether ERK inhibition affected the resistant phenotype and TKI-induced Bim expression. In parental cells, addition of the MEK inhibitor U0126 to gefitinib produced a similar effect to gefitinib alone in suppressing cell viability and inducing Bim expression (Fig. 6D and S5A). In resistant cells, Bim induction was attenuated and cell viability was unchanged in response to treatment with gefitinib, as anticipated (Fig. 6D and S5A). Addition of a Mek inhibitor to gefitinib significantly decreased cell viability compared with either drug alone and caused increased Bim expression (Fig. 6D and S5A). Similar results were obtained with a different, allosteric Mek inhibitor, GSK1120212 (Fig. S5B). Thus, inhibition of Mek-ERK signaling conferred sensitivity to TKI-resistant GBM cells.
To directly test the role of Bim as a downstream mediator of ERK1/2 in controlling GBM cell viability, we utilized ABT-737, a BH-3 mimetic compound which functions by inhibiting Bcl-2, Bcl-w, and Bcl-xL (35). TKI-sensitive and –resistant cells were treated with ABT-737 and gefitinib, alone or in combination. In TKI-sensitive parental cells, treatment with ABT-737, gefitinib, or the combination significantly decreased cell viability (p<0.05) (Fig. 6E). In TKI-resistant cells, cell viability was decreased only upon treatment with the combination of gefitinib and ABT-737 (p<0.05) (Fig. 6E).
Finally, to determine if levels of uPA expression are directly correlative with MAPK signaling in clinical samples, glioblastoma transcriptional profiles from the TCGA Data Portal were analyzed. There was a highly statistical correlation (p<0.0001) between a gene signature indicative of MAPK activity (29) and uPA expression levels (Fig 6F), indicating that uPA-mediated Bim repression through MAPK activity, which we detected in two models of EGFR TKI acquired resistance and in a genetic system of ΔEGFR-independent glioma growth, is an accurate reflection of activity of this signaling pathway in patients. Together, these data implicate an ERK-Bim signaling axis in GBM EGFR TKI resistance and support a model in which restoration of Bim function and TKI sensitivity can be achieved either by blocking MEK activity or with a Bim mimetic.
Discussion
We have identified a novel function of the uPA-uPAR-ERK1/2 signaling axis in mediating Bim repression during resistance to EGFR inhibition in glioblastoma. Bim has not been previously implicated in resistance or response of GBM to EGFR TKIs, but is known to be required for EGFR TKI-induced apoptosis in other solid tumors (36, 37). In our model systems of resistance, elevated uPA caused an increase in signaling through uPAR to Mek-ERK1/2. To our knowledge, this is the first study that directly implicates the uPA/uPAR signaling axis as playing a critical function in a pharmacological model of resistance to EGFR targeted therapy. uPA levels are increased in primary glioblastoma cell lines resistant to cisplatin as a result of chronic drug dosing (38). Interestingly, the TKI-resistant cell lines we generated were also resistant to cisplatin-induced apoptosis. We hypothesized that uPA/uPAR is functionally important in EGFR TKI resistance based on our previous findings detailing the role of this signaling system in escape from ΔEGFR oncogene dependence (23). The in vitro and in vivo pharmacological models that we generated here independently support this genetic model and confirm a role for uPA/uPAR signaling in EGFR TKI resistance.
The inherent molecular heterogeneity of GBM makes it likely that several resistance mechanisms are acting simultaneously within one tumor. Like other oncogenes, EGFR amplification and/or mutation can be sub-clonal (39). Expression of EGFR itself often varies in response to TKI treatment, and tumors could overcome EGFR oncogene dependence in multiple ways. We observed a minority of resistance tumors exhibiting decreased EGFR expression, and some others that appeared to have slightly decreased PTEN. The current study demonstrates the emergence of a uPA/uPAR signaling axis, but does not preclude that other, non-redundant mechanisms may exist. In fact, it is possible that Bim represents a central, downstream, resistance node and targeting this node would have the benefit of bypassing the heterogeneity of upstream resistance mechanisms.
Bim expression is induced transcriptionally by factors such as FoxO3a (40), and its function is suppressed post-translationally by Mek and ERK1/2-mediated phosphorylation at serine 55 and 69, respectively (41) (42). Bim interacts with dynein light chains (DLCs) in non-apoptotic cells, which sequester the protein in the cytosol (43). JNK-mediated phosphorylation of Bim within the DLC binding motif prevents this interaction, allowing Bim to bind to Bcl-2 family members and activate Bax and Bak at the mitochondria. In our model systems of resistance, Bim was induced by TKI treatment in sensitive cells and tumors, but expression was suppressed during TKI resistance. We were able to achieve Bim induction in resistant cells by using a combination of gefitinib and Mek inhibitor. Although we cannot rule out a transcriptional component of Bim control, we believe that the primary avenue by which Bim is suppressed in our models of resistant GBM cells and tumors is likely via a Mek-mediated mechanism of phosphorylation and subsequent degradation (42). In this study, we utilized the BH3 mimetic compound ABT-737 to restore Bim function, which conferred TKI-sensitivity upon resistant cells. ABT-737 is selective for binding to Bcl-2, Bcl-w, and Bcl-xl and has no activity against Mcl-1 (44). Interestingly some tumors become resistant to ABT-737 by up-regulating expression of Mcl-1(44), which is a possible scenario in the setting of combined chronic treatment with ABT-737 and EGFR TKI. Thus, it is likely that a more suitable BH3 mimetic would be one that also targets Mcl-1, such as TW-37(44).
To avoid apoptosis, tumor cells may up-regulate any number of pathways known to suppress Bim. Signaling through PI3K/AKT or JNK (45) causes attenuation of Bim expression or pro-apoptotic function, and TKI-resistant tumors in which PTEN has been lost or mutated typically harbor a higher sustained level of PI3K/Akt activity. Mechanistically, Akt-induced phosphorylation and inactivation of FoxO3a prevents transcriptional up-regulation of Bim (46). In support of this notion, it has been shown that in the context of HER2-amplified breast cancer with mutated PIK3CA, combination of lapatinib with a PI3K/mTOR inhibitor increases apoptosis significantly more than either agent alone (47). Our results suggest that the combination of EGFR TKIs with Mek inhibitors, explored for EGFR-mutant NSCLC resistant cell lines (48), might be an effective therapeutic strategy for TKI-resistant GBM. An alternative strategy is to interfere with the upstream mechanism responsible for driving Mek signaling and Bim suppression, which in our model system is the uPA-uPAR signaling axis. While uPAR signaling has been associated with Bim down-regulation in macrophages (49), a direct link between uPA/uPAR and Bim has not previously been shown in tumor cells.
One potential approach to targeting uPA-uPAR would be the use of uPA-neutralizing antibodies (50), or a peptide that interferes with uPA function (51). An alternative therapeutic approach for combating resistance that is supported by our findings ignores the upstream mechanisms of Bim suppression and involves the use of BH-3 mimetics to restore Bim function in combination with EGFR TKIs. In fact, the killing effect of EGFR TKIs against NSCLC cell lines is enhanced by BH-3 mimetics (36), and these drugs are being tested clinically as chemo-sensitizers for several different tumor types (44).
In conclusion, we have found that EGFR TKI-resistant GBM cells and tumors have up-regulated uPA to achieve sustained signaling through Mek-ERK1/2 such that Bim expression is maintained at low levels, rendering them incapable of efficiently inducing apoptosis. Through the generation and characterization of multiple model systems, we have identified a novel role for the uPA/uPAR signaling axis in resistance of GBM to EGFR-targeted therapy and have provided the rationale for therapeutic strategies targeting uPA/uPAR or downstream signaling mediators to prevent or reverse resistance.
Supplementary Material
Acknowledgments
Financial support:
This work was supported by grants from the NIH (F32-NS066519) and the American Brain Tumor Association (to J.W.), support from the Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society (to W.K.C., P.S.M., F.B.F.), R01-CA169096 (to S.L.G.), R01-NS080939 and the James S. McDonnell Foundation (to F.B.F). W.K.C. is a fellow of the National Foundation for Cancer Research.
Footnotes
Conflicts of Interest: The authors declare no conflicts of interest.
References
- 1.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes & development. 2012;26:756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci U S A. 1994;91:7727–31. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukasa A, Wykosky J, Ligon KL, Chin L, Cavenee WK, Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Wykosky J, Fenton T, Furnari F, Cavenee WK. Therapeutic targeting of epidermal growth factor receptor in human cancer: successes and limitations. Chinese journal of cancer. 2011;30:5–12. doi: 10.5732/cjc.010.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelman JA, Janne PA. Mechanisms of Acquired Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small Cell Lung Cancer. Clin Cancer Res. 2008;14:2895–9. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 8.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras Mutations and Benefit from Cetuximab in Advanced Colorectal Cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 10.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–7. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, et al. Molecular Determinants of the Response of Glioblastomas to EGFR Kinase Inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 14.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of Receptor Tyrosine Kinases Affects the Response of Tumor Cells to Targeted Therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 15.Fenton TR, Nathanson D, Ponte de Albuquerque C, Kuga D, Iwanami A, Dang J, et al. Resistance to EGF receptor inhibitors in glioblastoma mediated by phosphorylation of the PTEN tumor suppressor at tyrosine 240. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14164–9. doi: 10.1073/pnas.1211962109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–43. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 17.Jo M, Thomas KS, Marozkina N, Amin TJ, Silva CM, Parsons SJ, et al. Dynamic Assembly of the Urokinase-type Plasminogen Activator Signaling Receptor Complex Determines the Mitogenic Activity of Urokinase-type Plasminogen Activator. Journal of Biological Chemistry. 2005;280:17449–57. doi: 10.1074/jbc.M413141200. [DOI] [PubMed] [Google Scholar]
- 18.Aguirre-Ghiso JA, Liu D, Mignatti A, Kovalski K, Ossowski L. Urokinase Receptor and Fibronectin Regulate the ERKMAPK to p38MAPK Activity Ratios That Determine Carcinoma Cell Proliferation or Dormancy In Vivo. Molecular Biology of the Cell. 2001;12:863–79. doi: 10.1091/mbc.12.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Z, Webb DJ, Jo M, Gonias SL. Endogenously produced urokinase-type plasminogen activator is a major determinant of the basal level of activated ERK/MAP kinase and prevents apoptosis in MDA-MB-231 breast cancer cells. J Cell Sci. 2001;114:3387–96. doi: 10.1242/jcs.114.18.3387. [DOI] [PubMed] [Google Scholar]
- 20.Smith HW, Marra P, Marshall CJ. uPAR promotes formation of the p130Cas–Crk complex to activate Rac through DOCK180. The Journal of Cell Biology. 2008;182:777–90. doi: 10.1083/jcb.200712050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adachi Y, Chandrasekar N, Kin Y, Lakka SS, Mohanam S, Yanamandra N, et al. Suppression of glioma invasion and growth by adenovirus-mediated delivery of a bicistronic construct containing antisense uPAR and sense p16 gene sequences. Oncogene. 2002;21:87–95. doi: 10.1038/sj.onc.1204999. [DOI] [PubMed] [Google Scholar]
- 22.Jo M, Eastman BM, Webb DL, Stoletov K, Klemke R, Gonias SL. Cell Signaling by Urokinase-type Plasminogen Activator Receptor Induces Stem Cell–like Properties in Breast Cancer Cells. Cancer Research. 2010;70:8948–58. doi: 10.1158/0008-5472.CAN-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, Jo M, Cavenee WK, Furnari F, VandenBerg SR, Gonias SL. Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc Natl Acad Sci U S A. 2011;108:15984–9. doi: 10.1073/pnas.1113416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, et al. Epidermal growth factor receptor and Ink4a/Arf: convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 25.Mukasa A, Wykosky J, Ligon KL, Chin L, Cavenee WK, Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci U S A. 2010;107:2616–21. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–71. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 27.Shah K, Hingtgen S, Kasmieh R, Figueiredo JL, Garcia-Garcia E, Martinez-Serrano A, et al. Bimodal Viral Vectors and In Vivo Imaging Reveal the Fate of Human Neural Stem Cells in Experimental Glioma Model. J Neurosci. 2008;28:4406–13. doi: 10.1523/JNEUROSCI.0296-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutsch DG, Mertz ET. Plasminogen: purification from human plasma by affinity chromatography. Science. 1970;170:1095–6. doi: 10.1126/science.170.3962.1095. [DOI] [PubMed] [Google Scholar]
- 29.Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D. Activation of Mitogen-Activated Protein Kinase in Estrogen Receptor α–Positive Breast Cancer Cells In vitro Induces an In vivo Molecular Phenotype of Estrogen Receptor α–Negative Human Breast Tumors. Cancer Research. 2006;66:3903–11. doi: 10.1158/0008-5472.CAN-05-4363. [DOI] [PubMed] [Google Scholar]
- 30.Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, et al. Targeted Therapy Resistance Mediated by Dynamic Regulation of Extrachromosomal Mutant EGFR DNA. Science. 2014;343:72–6. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–37. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Muller KA, Furnari F, Cavenee WK, VandenBerg SR, Gonias SL. Targeting the EGF receptor in glioblastoma cells stimulates cell migration by activating uPAR-initiated cell signaling. Oncogene. 2014 doi: 10.1038/onc.2014.336. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janjigian YY, Viola-Villegas N, Holland JP, Divilov V, Carlin SD, Gomes-DaGama EM, et al. Monitoring Afatinib Treatment in HER2-Positive Gastric Cancer with 18F-FDG and 89Zr-Trastuzumab PET. Journal of Nuclear Medicine. 2013;54:936–43. doi: 10.2967/jnumed.112.110239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salajegheh M, Rudnicki A, Smith TW. Expression of urokinase-type plasminogen activator receptor (uPAR) in primary central nervous system neoplasms. Applied immunohistochemistry & molecular morphology: AIMM/official publication of the Society for Applied Immunohistochemistry. 2005;13:184–9. doi: 10.1097/01.pai.0000138448.85231.da. [DOI] [PubMed] [Google Scholar]
- 35.Chonghaile TN, Letai A. Mimicking the BH3 domain to kill cancer cells. Oncogene. 2009;27:S149–S57. doi: 10.1038/onc.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cragg MS, Kuroda J, Puthalakath H, Huang DC, Strasser A. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires BIM and can be enhanced by BH3 mimetics. PLoS medicine. 2007;4:1681–89. doi: 10.1371/journal.pmed.0040316. discussion 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS medicine. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osmak M, Vrhovec I, Skrk J. Cisplatin resistant glioblastoma cells may have increased concentration of urokinase plasminogen activator and plasminogen activator inhibitor type 1. Journal of neuro-oncology. 1999;42:95–102. doi: 10.1023/a:1006125629887. [DOI] [PubMed] [Google Scholar]
- 39.Francis JM, Zhang C-Z, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, et al. EGFR Variant Heterogeneity in Glioblastoma Resolved through Single-Nucleus Sequencing. Cancer Discovery. 2014;4:956–71. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunters A, Fernández de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a Transcriptional Regulation of Bim Controls Apoptosis in Paclitaxel-treated Breast Cancer Cell Lines. The Journal of biological chemistry. 2003;278:49795–805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 41.Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci U S A. 2004;101:15313–7. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 Signaling Pathway Promotes Phosphorylation and Proteasome-dependent Degradation of the BH3-only Protein, Bim. The Journal of biological chemistry. 2003;278:18811–6. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 43.Puthalakath H, Huang DCS, O’Reilly LA, King SM, Strasser A. The Proapoptotic Activity of the Bcl-2 Family Member Bim Is Regulated by Interaction with the Dynein Motor Complex. Mol Cell. 1999;3:287–96. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 44.Billard C. BH3 Mimetics: Status of the Field and New Developments. Molecular Cancer Therapeutics. 2013;12:1691–700. doi: 10.1158/1535-7163.MCT-13-0058. [DOI] [PubMed] [Google Scholar]
- 45.Yuan Z, Wang F, Zhao Z, Zhao X, Qiu J, Nie C, et al. BIM-mediated AKT phosphorylation is a key modulator of arsenic trioxide-induced apoptosis in cisplatin-sensitive and -resistant ovarian cancer cells. PLoS One. 2011;6:e20586. doi: 10.1371/journal.pone.0020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt Promotes Cell Survival by Phosphorylating and Inhibiting a Forkhead Transcription Factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 47.Tanizaki J, Okamoto I, Fumita S, Okamoto W, Nishio K, Nakagawa K. Roles of BIM induction and survivin downregulation in lapatinib-induced apoptosis in breast cancer cells with HER2 amplification. Oncogene. 2011;30:4097–106. doi: 10.1038/onc.2011.111. [DOI] [PubMed] [Google Scholar]
- 48.Huang M-H, Lee J-H, Chang Y-J, Tsai H-H, Lin Y-L, Lin AM-Y, et al. MEK inhibitors reverse resistance in epidermal growth factor receptor mutation lung cancer cells with acquired resistance to gefitinib. Molecular Oncology. 2013;7:112–20. doi: 10.1016/j.molonc.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paland N, Aharoni S, Fuhrman B. Urokinase-type plasminogen activator (uPA) modulates monocyte-to-macrophage differentiation and prevents Ox-LDL-induced macrophage apoptosis. Atherosclerosis. 2013;231:29–38. doi: 10.1016/j.atherosclerosis.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 50.Lund IK, Jögi A, Rønø B, Rasch MG, Lund LR, Almholt K, et al. Antibody-mediated Targeting of the Urokinase-type Plasminogen Activator Proteolytic Function Neutralizes Fibrinolysis in Vivo. The Journal of biological chemistry. 2008;283:32506–15. doi: 10.1074/jbc.M801924200. [DOI] [PubMed] [Google Scholar]
- 51.Ghamande SA, Silverman MH, Huh W, Behbakht K, Ball G, Cuasay L, et al. A phase 2, randomized, double-blind, placebo-controlled trial of clinical activity and safety of subcutaneous Å6 in women with asymptomatic CA125 progression after first-line chemotherapy of epithelial ovarian cancer. Gynecologic Oncology. 2008;111:89–94. doi: 10.1016/j.ygyno.2008.06.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.