Abstract
Site-specific gene addition can allow stable transgene expression for gene therapy. When possible, this is preferred over the use of promiscuously integrating vectors, which are sometimes associated with clonal expansion1 and oncogenesis2. Site-specific endonucleases that can induce high rates of targeted genome editing are finding increasing applications in biological discovery and gene therapy3. However, two safety concerns persist: (1) endonuclease-associated adverse effects, both on4 and off-target5,6; and (2) oncogene activation caused by promoter integration, even without nucleases7. Here, we perform recombinant adeno-associated virus (rAAV) mediated promoterless gene targeting without nucleases and demonstrate amelioration of the bleeding diathesis in haemophilia B mice. In particular, we target a promoterless human coagulation factor IX (hF9) gene to the liver-expressed albumin (Alb) locus. hF9 is targeted, along with a preceding 2A-peptide coding sequence, to be integrated just upstream to the Alb stop codon. While hF9 is fused to Alb at the DNA and RNA levels, two separate proteins are synthesized by way of ribosomal skipping. Thus, hF9 expression is linked to robust hepatic albumin expression without disrupting it. We injected an AAV8-hF9 vector into neonatal and adult mice and achieved on-target integration into ~0.5% of the albumin alleles in hepatocytes. We established that hF9 was produced only from on-target integration, and ribosomal skipping was highly efficient. Stable hF9 plasma levels at 7–20% of normal were obtained, and treated factor IX deficient mice had normal coagulation times. In conclusion, transgene integration as a 2A-fusion to a highly expressed endogenous gene may obviate the requirement for nucleases and/or vector-borne promoters. This method may allow for safe and efficacious gene targeting in both infants and adults by greatly diminishing off-target effects while still providing therapeutic levels of expression from integration.
Site-specific gene targeting is one of the fastest growing fields in gene therapy and genome engineering. The rise in popularity of gene targeting can be attributed in large part to the development of readily engineered and easy to use site-specific endonucleases (e.g. TAL- or CRISPR-based)3 that can increase rates of gene disruption, gene correction or gene addition by as much as four orders of magnitude. However, these endonucleases may have significant adverse effects including immunogenicity, uncontrolled DNA damage response, off-target cleavage and mutagenesis, induction of chromosomal aberrations, as well as off-target integration of the transgene and endonuclease vectors (if DNA-based)4–6. When a vector-borne promoter is integrated either on- or off-target, it may lead to undesired activation of nearby genes, including oncogenes. The use of endonucleases in vivo would require their vectorization, delivery and expression in a transient manner to minimize long-term side effects. It is unclear how integration of the vectored endonuclease gene could be strictly avoided.
Our promoterless, endonuclease-independent method harnesses the efficient transduction, favorable safety profile and high gene targeting rates associated with rAAV8–12, as well as the robust liver-specific expression of the Alb locus13. Different rAAV serotypes can efficiently transduce various cell types in vitro or in vivo, while other serotypes have been designed or selected for desired phenotypes14–17. rAAV is currently in use in multiple clinical trials18,19. Importantly, rAAV transduction allows high gene targeting rates in vitro8 and in vivo9,20. The increased recombination rates may be due to the viral inverted terminal repeats (ITR), the encapsidation of single-stranded DNA, or to the timing and subcellular localization of capsid uncoating.
The safety of rAAV stems from its lack of pathogenicity, as well as being devoid of viral genes. Nevertheless, non-targeted genomic integration of rAAV occurs at a low but significant rate, and there are reports of such integrations inducing hepatocellular carcinoma in mice7. Transformation was attributed to vector integration at a chromosome 12 locus coding for imprinted genes and small RNAs. Integration of rAAV-borne promoters may be the leading cause of aberrant expression, as was established for lentiviral and retroviral vector integration leading to clonal expansion and oncogenesis1,21. Vector-borne promoters are in use in many ongoing clinical trials. In contrast, the rAAV utilized in our strategy encodes no promoter, thus diminishing the chance of neighboring oncogene activation in rare off-target integrations.
As proof of concept, we targeted the hF9 gene deficient in the X-linked recessive disease haemophilia B affecting 1/30,000 males. Affected individuals suffer from serious spontaneous bleeding due to a deficiency of plasma coagulation factor IX produced from the liver. Reconstitution with as little as 1–2% clotting factor can significantly improve quality of life, while 5–20% will markedly ameliorate the bleeding diathesis. Herein, we used the liver tropic rAAV8 serotype to target hF9 for expression upon integration from the robust liver-specific Alb promoter. We postulated that: (1) the Alb promoter should allow high levels of coagulation factor production even if integration takes place in only a small fraction of hepatocytes; and (2) the high transcriptional activity at the Alb locus might make it more susceptible to transgene integration by homologous recombination.
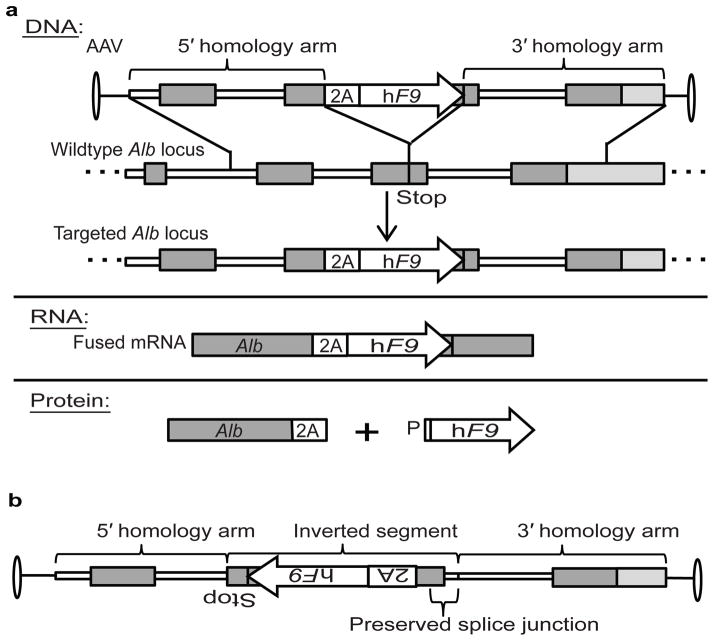
Gene targeting without nucleases should affect only a small fraction of Alb alleles in the liver. Nevertheless, we opted to minimize disruption and dysregulation of the Alb gene by targeting hF9 as a 2A-fusion at the end of the Alb reading frame (Fig. 1a). 2A-peptides, derived from plus-strand RNA viruses, allow the production of multiple proteins from a single reading frame by means of ribosomal skipping22. This process leaves the first translated protein tagged with ~20 C-terminal amino acids, and the second protein with just one additional N-terminal proline. Functionality of both proteins is typically retained, and clinical trials using 2A-peptides did not report immunogenicity23. We used single-stranded AAV to target a codon-optimized hF9 cDNA, preceded by a sequence coding for a porcine teschovirus-1 2A-peptide (P2A)22, to be integrated just 5′ of the Alb stop codon. Following integration, Alb and hF9 are co-transcribed from the strong Alb promoter and should thus be co-regulated at the levels of splicing, nuclear exit, mRNA stability, translation initiation and ER localization. Two separate proteins are translated, both containing a signal peptide, so that the ER-associated translation of Alb will be immediately followed by translation and processing of the clotting factor for secretion. Finally, in order to further reduce the chance of off-target hF9 expression, our vector has neither an ATG start codon before the hF9 signal peptide, nor a start codon in the 2A-peptide coding sequence or preceding Alb exon.
Figure 1. Vector design and experimental scheme.
a. The rAAV8 vector encodes a codon-optimized hF9 cDNA and preceding 2A-peptide coding sequence flanked by homology arms spanning the Alb stop codon. Length of the 5′ and 3′ arms are 1.3 and 1.4 Kb, respectively. Following integration by homologous recombination, Alb and hF9 are fused at the DNA and RNA levels, but two separate proteins are produced as the result of ribosomal skipping. b. With respect to the Alb homology arms, the AAV inverse control has hF9 inverted along with the 2A-peptide coding sequence, the adjacent Alb exon and the preceding splice junction. Thin white lines: Alb introns; dark gray boxes: Alb exons; white boxes: P2A; white arrows: hF9 transgene; light gray boxes: extragenic DNA; P: proline.
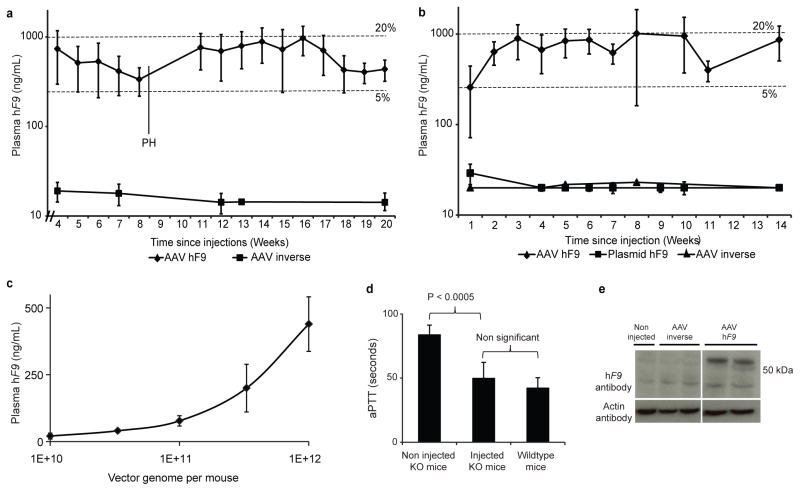
First, we performed intra-peritoneal (IP) injections of 2-day old C57BL/6 (B6) mice with 2.5e11 vector genomes (vg) per mouse (~1.25e14 per Kg) of a rAAV8 coding for the hF9 targeting cassette or a vector with an inverted cassette, controlling for off-target expression (Fig. 1b). The fragment inverted in the control with respect to the Alb homology arms includes not only the hF9 gene, but also the P2A coding sequence, the adjacent Alb exon and the preceding splice junction. The inverse control should not allow significant hF9 expression upon on-target integration, but would allow levels of off-target expression similar to that from the experimental construct. We measured plasma hF9 protein levels weekly by enzyme-linked immunosorbent assay (ELISA), starting at week 4 of life (Fig. 2a). For the experimental group, levels of plasma hF9 plateaued at 350–1000 ng/mL, which corresponds to 7–20% of normal. For the inverse control group, hF9 plasma levels were at or below the level of detection (20 ng/mL), implying that in the experimental group, hF9 expression does indeed originate from on-target integration. Importantly, hF9 retains the original plasma protein levels after a 2/3 partial hepatectomy, a surgical procedure known to reduce episomal AAV transgene expression by >90%8, further establishing stable transgene integration.
Figure 2. Human factor IX expression and activity in injected mice.
a. Plasma hF9 measured by ELISA following IP injections of 2-day-old B6 mice with 2.5e11 vg per mouse of either the hF9 experimental construct (n = 6) or inverse control (n = 3). The limit of detection was 20 ng/mL. PH = partial hepatectomy. Error bars represent standard deviation. Dashed lines denote 5% and 20% of normal F9 levels. b. Plasma hF9 measured by ELISA following tail vain injections of 9-week-old B6 mice with 1e12 vg per mouse of either the AAV hF9 experimental construct (n = 7), or inverse control (n = 3), or a hydrodynamic injection of 30 μg plasmid (3.5e12 copy number) coding for the hF9 construct in the “correct” orientation. The limit of detection was 20 ng/mL. Error bars and dashed lines as in (a). c. Plasma hF9 measured by ELISA following tail vain injections of 9-week-old B6 mice with the designated vector dose of AAV-hF9 experimental construct (n = 4 for each dose group). Error bars represent standard deviation. d. Measurement of coagulation efficiency by activated partial thromboplastin time (aPTT) 2 weeks after tail vain injections of AAV8-hF9 at 1e12 vg per mouse (n = 5). Error bars represent standard deviation. e. Western blot analysis for hF9 in liver samples from mice injected with the AAV8-hF9 construct or inverse control. The expected size of hF9 is 55-Kd.
In order to determine whether liver growth, as seen with neonates, is essential for therapeutic levels of gene targeting, we targeted hF9 to the Alb locus using the same vector in adult mice. Adult B6 mice were injected with 1e12 vg per mouse (~5e13 vg/kg) by tail vein with the rAAV8 vector, or the inverse control. A third group of control mice received hydrodynamic tail vain injections of a plasmid coding for the promoterless hF9 construct in the “correct” orientation. For the rAAV hF9 mice group, plasma hF9 levels were found to be stable at 7–20% of normal (Fig. 2b). Vector injections at lower dose led to lower plasma hF9 levels without reaching a plateau at the doses tested (Fig. 2c). For adults as well as neonates, the hF9 plasma levels of the inverse control group were at or below the limit of detection. Diminished hF9 plasma levels were also associated with mice hydrodynamically injected with plasmid (Fig. 2b). Thus, significant targeting is dependent on rAAV vectorization. Finally, we performed rAAV injections in adult F9 knockout (KO) haemophilia B mice. The functional coagulation, as determined by the activated partial thromboplastin time (aPTT) in treated KO mice, was restored to levels similar to that of wild-type mice (Fig. 2d). The hF9 biological activity correlated with plasma protein levels of 709±91 ng/mL, similar to levels in wild-type mice (Fig. 2b,c,d).
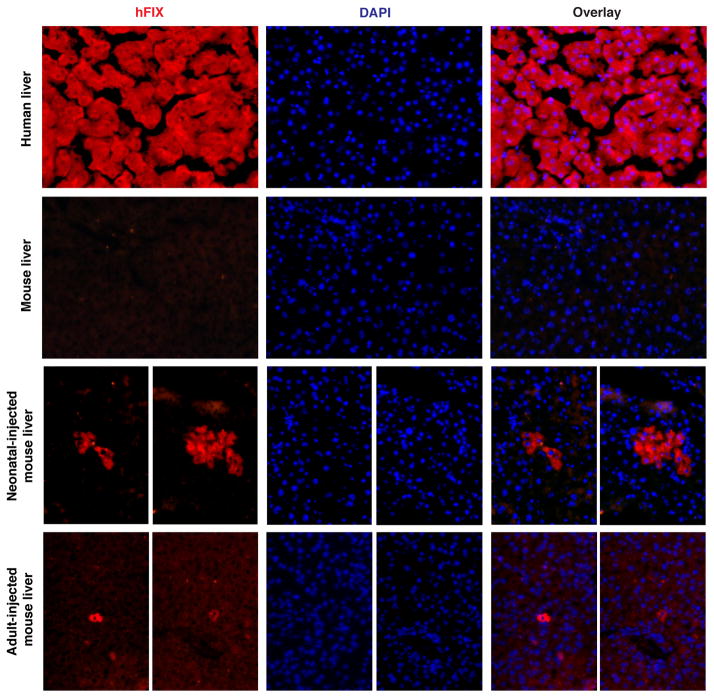
hF9 expression from the liver was confirmed by immunohistochemistry (IHC) (Extended Data Fig. 1). Western blot analysis of liver samples detected hF9 at the expected molecular weight, testifying that ribosomal skipping was efficient, and implying that both the ELISA and IHC signals correspond to accurately processed hF9 (Fig. 2d).
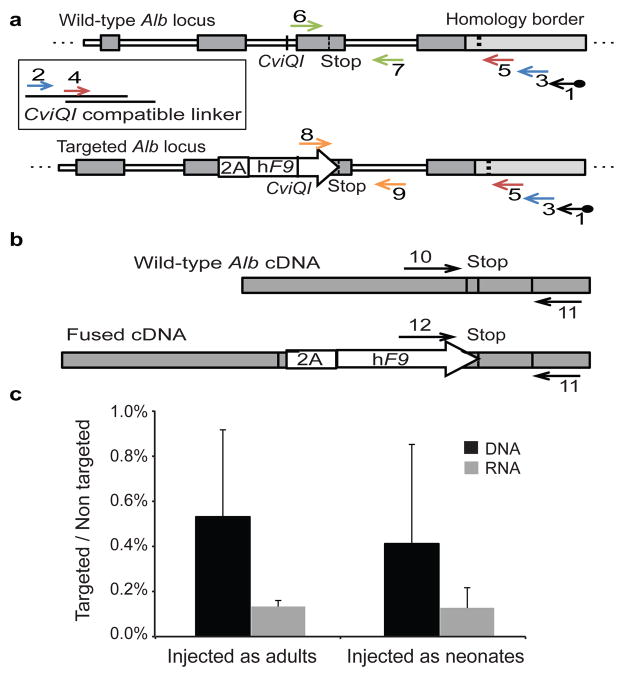
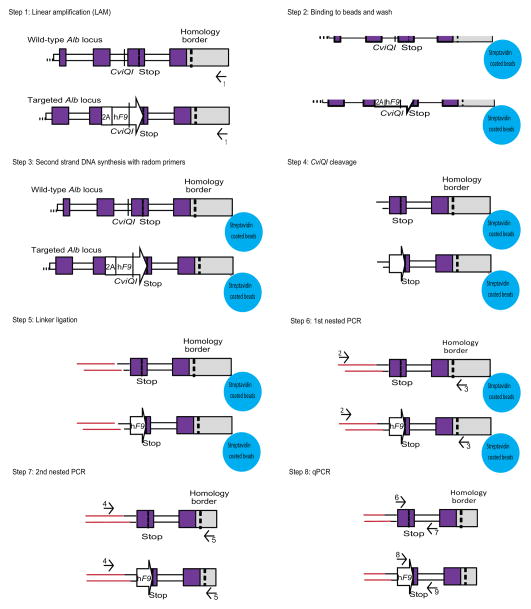
We opted to quantify targeting rates by qPCR. To avoid false signals from episomal rAAV, we first amplified a 3′ segment of the genomic Alb locus in a manner not affected by presence or absence of an integrated hF9 sequence (Fig. 3a, and Extended Data Fig. 2). The unbiased amplification was made possible by presence of a common restriction site at a roughly equal distance 3′ of the stop codon in targeted and wild-type alleles. We then used the PCR amplicon as a template for two different qPCR assays: one quantifying abundance of targeted Alb alleles, and the other quantifying abundance of untargeted wild-type alleles. In the liver, only hepatocytes are targeted by rAAV824. Therefore, we conservatively corrected for a 70% hepatocyte frequency25 and found the rate of Alb alleles targeted by hF9 to be 0.5% on average for mice injected as either neonates or adults at the highest dose (Fig. 3c and Extended Data Fig. 3). Actual rates of targeting in neonates and adults might differ because AAV distribution to the liver may vary based on the different methods used for vector infusion. We then examined the proportion of fused Alb_hF9 mRNAs to wild-type Alb mRNAs by comparing two respective qPCR assays performed on an unbiased cDNA template (Fig. 3b). The proportion was found to be 0.1% on average for mice injected as either neonates or adults (Fig. 3c). This value tended to be lower than the rate of integration at the DNA level, although the difference was not statistically significant. It is possible that the production, processing and/or stability of chimeric hF9-Alb mRNA transcripts were reduced compared to wild-type Alb mRNA. While AAV8 has been shown to target only hepatocytes in the mouse liver24, here we did not rule out the possibility that some integration occurred in non-parenchymal cells which do not express albumin. Our observed targeting rate is higher than other reports9,12,20, and is particularly noteworthy in adult mice where non-proliferating hepatocytes were expected to allow for a low rate of homologous recombination. We hypothesize that the high expression rate at the Alb locus and the associated chromatin status may contribute to the high rates of targeting. Damage-induced proliferation cannot be strictly ruled out, but no elevation in alanine Aminotransferase (ALT) levels was seen following injection (Extended Data Fig. 4).
Figure 3. Rate of Alb targeting at the DNA and RNA levels.
a. Assessment of on-target integration rate begins using linear amplification (LAM) with biotinylated primer 1 (black), annealing to the genomic locus but not to the vector. Linear amplicons are then bound to streptavidinylated beads and washed to exclude episomal vectors. Subsequent second-strand DNA synthesis with random primers was followed by CviQI restriction digestion. A compatible linker is then ligated, followed by two rounds of nested PCR (primers 2–3 in blue, and then primers 4–5 in red). CviQI cleaves at the same distance from the homology border in both targeted and wild-type alleles, thus allowing for unbiased amplification. The amplicons of the 2nd nested PCR then serve as a template for qPCR assays with either primers 6–7 (green) or 8–9 (orange). b. For mRNA quantification, primers 10–11 or 11–12 were used to generate a cDNA for qPCR assays. Shape and fill code as in Fig 1. c. Black bars represent the targeting rate of Alb alleles as the ratio between the abundance of the DNA template amplified by primers 6–7 to the abundance of the DNA template amplified by primers 8–9, corrected by a factor of 0.7 to account for hepatocyte frequency. Gray bars represent the expression rate of targeted Alb alleles as the ratio between the abundance of the cDNA template amplified by primers 10–11 to the abundance of the cDNA template amplified by primers 11–12. N = 3 for each group, error bars represent standard deviation.
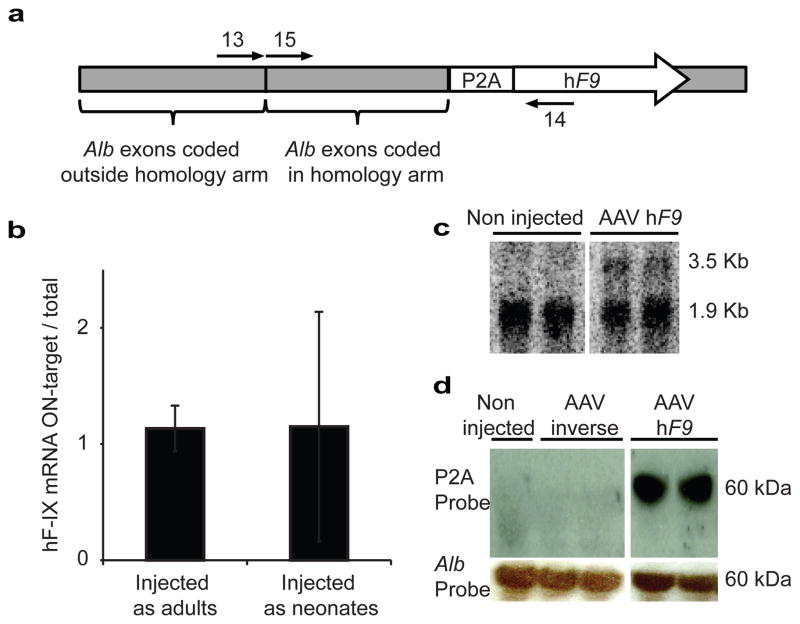
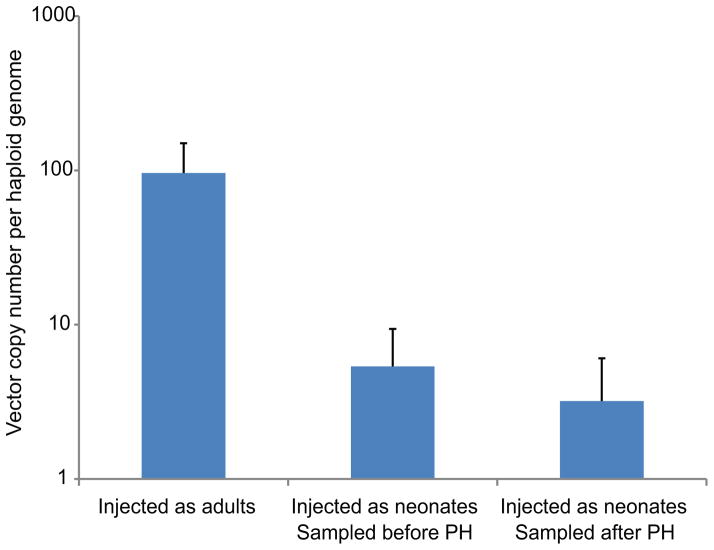
AAV genomes may be present in cells as episomes, or as on- or off-target integrants. Total vector copy number was assessed by qPCR (Extended Data Fig. 5). The minor change in vector copy number following partial hepatectomy in mice injected as neonates may imply that episomal vectors had already been greatly diluted during normal liver growth and development. In which case, vector copy number can be used as an approximate lower bound on the rate of off-target to on-target integration. However, most importantly, in the absence of a vector-borne promoter, hF9 should only be expressed from on-target integration. The reconstituted high hF9 levels following partial hepatectomy (Fig. 2a) support this assumption, as only stably integrated transgenes could rebound after such a procedure, unlike that seen with transient episomal expression26. Lack of significant hF9 plasma levels following treatment with the inverse control vector further demonstrated reduced off-target expression. We used qRT-PCR to directly assess the ratio of fused Alb_hF9 mRNAs among the total hF9 mRNA pool (Fig. 4a). The ratio was found to be 1:1 for mice injected as neonates and as adults (Fig. 4b). This implies that hF9 is expressed almost exclusively from on-target integration. Indeed, the only specific signal from a Northern blot with a P2A probe corresponded to the expected fused Alb-P2A-hF9 mRNA (Fig. 4c). Finally, a Western blot with an anti-2A-peptide antibody indicated that the 2A-peptide is associated with a single species at the expected molecular weight of Alb (Fig. 4d), as would be expected only if expression was restricted to on-target integration and followed by efficient ribosomal skipping.
Figure 4. Specificity of hF9 expression.
a. cDNA, produced from RT with a poly-dT primer, served as a template for either a qPCR assay with primers 13–14 or 14–15. b. Bars represent the rate of Alb_hF9 mRNAs to total hF9-containing mRNAs as the ratio between the abundance of the cDNA template amplified by primers 13–14 to the abundance of the cDNA template amplified by primers 14–15. N = 3 for each group, error bars represent standard deviation. c. Northern blot analysis of liver samples with a probe against P2A. The lower non-specific signal corresponds in size to 18S rRNA. d. Western blot analysis of P2A from liver samples of mice injected with the AAV-P2A-hF9 construct or inverse control. P2A is expected to be fused to Albumin (66.5-Kd).
rAAV has become a popular vector for clinical therapy. While the period of transgene expression in adults can last for several years, it is not yet known whether lifelong expression, as required for many genetic disorders, can be obtained with routine promoter-containing vectors. Episomal expression from AAV vectors is rapidly lost in dividing cells, even after just one round of cell division26. This makes it likely that diseases that induce regeneration and/or are treated in infancy while tissues continue to grow, will have limited durability. Secondary infusion of an AAV vector will be unlikely to result in a successful transduction, due to the robust humoral immunity resulting from primary vector administration27. In contrast, our approach results in vector integration that would eliminate loss of expression over time, even in dividing tissues. This relies on the choice of an appropriate AAV serotype to avoid neutralization by pre-existing immunity.
Previous work demonstrating targeting of hF9 to a chimeric locus in a transgenic mouse10 relied on the co-expression of nucleases that may be associated with immunological and genotoxic side effects. Interestingly, the same reliance on endonucleases held true even when hF8 was targeted to the Alb locus in mice and non-human primates28, probably because no homology arms were provided and integration relied instead on non-homologous end-joining. rAAV has already been used in clinical gene therapy trials to treat haemophilia B18. However, the transgene in these clinical trials was expressed from a vector-borne promoter that might induce oncogene activation, as has been reported in mice7. Measuring levels of alanine transaminases, we observed no liver toxicity with the injection of our hF9 targeting vector (Extended Data Fig. 4). However, it remains to be determined whether the transgene over-expression associated with our method will lead to toxicity when different therapeutic transgenes are targeted. 2A-induced immunogenicity could not be strictly ruled out, but notably no immune effects were reported in clinical trials when vector coding for a 2A peptide was targeted to lymphocytes23. Although we found no evidence of off-target expression and no ALT elevation, the high vector dose we used may lead to other undesired outcomes such as increased off-target integration and increased immunogenicity. In the future, this could be mitigated by the use of AAV serotypes having better tropism and/or by use of hyperactive hF9 variants. Genetic polymorphisms at the target locus in the human patient population may lead to variable therapeutic efficacy due to reduced homology. However, we found that ~95% of a 1000 genome sample of the human population have no more than just two haplotypes at the relevant Alb sequence, which may enable broad applicability (Extended Data Table 1). Our work demonstrates a therapeutic effect for in vivo gene targeting without nucleases and without a vector-borne promoter. The favorable safety profile of our promoterless and nuclease-free gene targeting strategy for rAAV makes it a prime candidate for clinical assessment in the context of haemophilia and other genetic deficiencies29. More generally, this strategy could be applied whenever the therapeutic effect is conveyed by a secreted protein (e.g. broadly neutralizing antibodies30) or when targeting confers a selective advantage12.
METHODS
Plasmid construction
A mouse genomic Alb segment (90474003-90476720 in NCBI reference sequence: NC_000071.6) was PCR-amplified and inserted between AAV2 ITRs into BsrGI and SpeI restriction sites in a modified pTRUF backbone31. The genomic segment spans 1.3 Kb upstream and 1.4 Kb downstream to the Alb stop codon. We then inserted into the Bpu10I restriction site an optimized P2A coding sequence preceded by a linker coding sequence (glycine-serine-glycine) and followed by an NheI restriction site. Finally, we inserted a codon optimized hF9 cDNA into the NheI site to get pAB269 that served in the construction of the rAAV8 vector. To construct the inverse control, we first amplified an internal segment from the BsiWI restriction site to the 3′ NheI restriction site. PCR primers used had 15 base tails to allow subsequent integration of the amplicon into a BsiWI and NheI cleaved plasmid at the inverse orientation using an In-Fusion Kit (Clontech). Primers used were: For: 5′-ATGCAAGGCACGTACGTTATGTCAGCTTGGTCTTTTCTTTGATCC-3′ and Rev: 5′-TTTAGGCTAAGCTAGCTTTACTATGTCATTGCCTATGGCTATGAAGTG-3′. Final rAAV production plasmids were generated using an EndoFree Plasmid Megaprep Kit (Qiagen).
rAAV vector production and titration
rAAV8 vectors were produced as previously described using a Ca3(PO4)2 transfection protocol followed by CsCl gradient purification31. Vectors were titered by quantitative dot blot as described31.
Mice injections and bleeding
Animal work was performed in accordance to the guidelines for animal care at both Stanford University and the University of California San Francisco. Eight-week-old wild-type C57BL/6 (B6) mice were purchased from Jackson Laboratory (Bar Harbor, ME) to serve for adult injections and as breeding pairs to produce offspring for neonatal injections. 2-day-old wild-type B6 mice were injected intraperitonealy with 2.5e11 vg per mouse of rAAV8 (hF9 or inverse) and bled weekly beginning at week 4 of life by retro-orbital bleeding for ELISA as previously described32. Adult (9-week-old) wild-type B6 mice received either tail vain injections of rAAV8 (hF9 or inverse, at the designated dose) or hydrodynamic injections of 3.5e12 plasmid copies, and were similarly bled weekly for ELISA. hF9 CD1/B6 hybrid KO mice, were injected as adults with 1e12 vg per mouse of rAAV8 hF9 and bled retro-orbitally two weeks after injection for ELISA and aPTT assays, as previously described11.
Enzyme-linked immunosorbent assay (ELISA)
ELISA for hF9 was performed as previously described32 with the following antibodies: mouse anti-hF9 IgG primary antibody at 1:1,000 (Sigma Cat#F2645), and polyclonal goat anti-hF9 peroxidase-conjugated IgG secondary antibody at 1:4,200 (Enzyme Research Cat#GAFIX-APHRP).
Partial hepatectomies
The 2/3 partial hepatectomies were performed as previously described33 with the following minor modifications: no surgical retractors were used to maintain an open abdominal cavity, 3-0 Sofsilk wax coated braided silk suture (Covidien, Cat#S-184) was used for knotting desired lobes for resection, and 6-0 Polysorb polyester suture (Covidien, Cat#GL889CV11) was used for closing the peritoneum and abdominal skin.
Activated partial thromboplastin time (aPTT) assay
The aPTT assay was carried out using a SCA2000 veterinary coagulation analyzer (Synbiotics) according to manufacturer’s instruction11.
Western blot analysis
Western blots for the detection of hF9, albumin, P2A and actin used the following antibodies: polyclonal goat anti-hF9 peroxidase-conjugated IgG primary antibody at 1:20,000 (Enzyme Research, Cat#GAFIX-APHRP), polyclonal rabbit anti-mouse serum albumin IgG primary antibody at 1:40,000 (Abcam, Cat#ab19196), donkey anti-rabbit peroxidase-conjugated IgG secondary antibody at 1:10,000 (ECL, Cat#NA-9340), polyclonal rabbit anti-2A peptide primary antibody at 1:10,000 (Millipore, Cat#ABS31), and monoclonal mouse anti-beta-actin peroxidase-conjugated IgG primary antibody at 1:50,000 (Sigma, Cat#A3854).
Northern blot analysis
Northern blots for the detection of P2A-hF9 coding mRNAs used the following P32 end-labeled anti-2A probe: 5′-GCCAGGGTTCTCTTCCACGTCGCCGGCCTGTTTCAGCAGGCTGAAATTGGTGGCGCCGCT-3′.
Assessing rate of Alb locus targeting by qPCR
Amplification of desired genomic Alb, but not undesired vector amplification, began by performing linear amplification with the following dual-biotinylated primer: 5′-/2-Biotin/GTCTCTCATTCAGAATTCTCGTAATGTTGAAG-3′, annealing outside the arm of homology. Subsequent second-strand DNA synthesis was followed by CviQI restriction digestion to produce fragments of roughly equal size and known sticky ends from both targeted and wild-type alleles. Two oligonucleotides were annealed to a CviQI-compatible linker. Oligonucleotide sequences were “Watson”: 5′-CTGAAGGCTCAGGTTACACAGGCACGCTCGTAGGAGGTGTTCCAGTTCACCACG-3′ and “Crick”: 5′-TACGTGGTGAACTGGAACACCTCCTACGAGC/3ddC/-3′. Linker ligation was followed by a primary nested PCR, using primers 2: 5′-CTGAAGGCTCAGGTTACACAGGCAC-3′ and 3: 5′-GTATTGGTTTCTAGGGTCACCACCCATAAG-3′, and a second nested PCR using primers 4: 5′-GCTCGTAGGAGGTGTTCCAGTTCACC-3′ and 5: 5′-GGGAGAGTATTAACGTTTATTTTCATTGTGTTG-3′. No nested PCR product was detected in a control without linear amplification. The PCR amplicon served as a template for two different TaqMan qPCR assays. To quantify the abundance of wild-type Alb alleles, we used a TaqMan qPCR with primers 6: 5′-TGCCTATGGCTATGAAGTGC-3′ and 7: 5′-CTGAGAAGGTTGTGGTTGTGA-3′ and TaqMan probe: 5′-TGCAAAGACGCCTTAGCCTAAACACA-3′. To quantify the abundance of targeted alleles we used a qPCR with primers 8: 5′-GATACGTGAACTGGATCAAAGAAA-3′ and 9: 5′-CAAATGGTTATCAGTCTTGATCG-3′ and TaqMan probe: 5′-CACATCACAACCACAACCTTCTCAGGT-3′. For non-injected controls, no qPCR signal was detected with primers 8–9. The abundance of the template for each amplicon was calculated using its own standard curve. We calculated the ratio between the abundance of the template for the 6–7 primer pair to the abundance of the template for the 8–9 primer pair. We then conservatively corrected this ratio by multiplying it by a factor of 0.7 to account for hepatocyte frequency in the samples25, as only hepatocytes are targeted by rAAV8 in the liver24.
Assessing rate of hF9 containing Alb mRNAs by qPCR
cDNA produced from RT with a poly dT primer served as a template for two different TaqMan qPCR assays. We quantified the abundance of wild-type Alb mRNA by a qPCR with primers 10: 5′-CTGACAAGGACACCTGCTTC-3′ and 11: 5′-TGAGTCCTGAGTCTTCATGTCTT-3′ and TaqMan probe: 5′-CCACAACCTTCTCAGGCTACCCTGA-3′. We quantified the abundance of fused mRNAs by a TaqMan qPCR with primers 12: 5′-CCAAGGTGTCCAGATACGTG-3′ and 11: 5′-TGAGTCCTGAGTCTTCATGTCTT-3′ and TaqMan probe: 5′-CCACAACCTTCTCAGGCTACCCTGA-3′. For non-injected controls, no qPCR signal was detected with primers 11–12. For the no-RT control, no signal was detected with primers 10–11 and 11–12. The rate of hF9-containing Alb mRNAs was calculated as the ratio between the abundance of template for the 10–11 primer pair to the abundance of template for the 11–12 primer pair.
Assessing specificity of hF9 expression
cDNA produced from RT with a poly-dT primer served as a template for two different TaqMan qPCR assays. We quantified the abundance of on-target hF9 expression using a qPCR with primers 13: 5′-CTTGGGCTTGTGCTTCAC-3′ and 14: 5′-AGGATCTTGTTGGCGTTCTC-3′ and TaqMan probe: 5′-CGGTACACTCGGCGCTCAGC-3′. We quantified total hF9-containing mRNA abundance using a qPCR with primers 15: 5′-GCTCTTGCTGAGCTGGTGA-3′ and 14: 5′-AGGATCTTGTTGGCGTTCTC-3′ and TaqMan probe: 5′-CGACTGAGGGTCCAAACCTTGTCA-3′. Calculations of abundance used a separate standard curve for each pair. No qPCR signal was detected for no-RT and non-injected controls. The rate of Alb_hF9 mRNAs to total hF9-containing mRNAs was determined by comparing the abundance of template for the 13–14 primer pair to abundance of template for the 14–15 primer pair.
Assessing vector copy number
DNA purified from mice liver served as a template for two different TaqMan qPCR assays. We quantified the abundance of haploid mouse genomes using a qPCR with primers “Alb copy number Ref F “: 5′-CAGAACGGTCTTTCTCGGAT-3′ and “Alb copy number Ref R”: 5′-TCTTCATCCTGCCCTAAACC-3′ and TaqMan probe: 5′-CTCAGCCCTGCAGTGCTGCA-3′. We quantified the abundance of AAV genomes (episomal or integrated) using a qPCR with primers 8: 5′-GATACGTGAACTGGATCAAAGAAA-3′ and 9: 5′-CAAATGGTTATCAGTCTTGATCG-3′ and TaqMan probe: 5′-CACATCACAACCACAACCTTCTCAGGT-3′. For non-injected controls, no qPCR signal was detected with primers 8–9. The abundance of the template for each amplicon was calculated using its own standard curve. Vector copy number per haploid genome was calculated as the ratio between the abundance of the template for the 8–9 primer pair to the abundance of the template for the “Alb copy number Ref F “and “Alb copy number Ref R” primer pair.
Statistics
Statistical analyses were conducted with Microsoft Excel. Experimental differences were evaluated by a Student one-tailed t-test assuming equal variance (except for the ALT measurement, where equal variance was not assumed).
Liver immunohistochemistry
Liver lobes were harvested from mice, rinsed in ice cold dPBS, blotted dry, mounted in OCT (Tissue-Tek, Cat#4583) and frozen in cryomolds (Tissue-Tek, Cat#4557) in a liquid nitrogen cooled methylbutane bath. Cryomolds were placed at −80°C until sectioning. In all samples, a minimum of two liver sections from varying depths were cut at 5 microns and mounted onto positively charged glass slides (Thermo, Cat#6776214). Fluorescent staining was performed according to established protocols34 with minor modifications. Briefly, frozen sections were thawed to RT, fixed in acetone for 10 minutes at RT, removed and allowed to air dry. Slides were then washed 3X in dPBS for 2 min at RT, blocked with 5% donkey serum (Santa Cruz, Cat#sc-2044) for 15 min at RT in a humidified chamber, washed 1X in dPBS for 1 min at RT and then sections were circled with an ImmEdge pen (Vector Laboratories, Cat#H-4000). Slides were incubated with 200ul of goat anti-human F.IX IgG primary antibody (Affinity Biologicals, Cat#GAFIX-AP) at 1:100 in 5% donkey serum for 3 hours at RT and then washed 3X in dPBS for 2 min at RT. Slides were then incubated with 200ul of donkey anti-goat AlexaFluor 594 IgG secondary antibody (Life Technologies, Cat#A11058) at 1:400 in dPBS for 30 min at RT and then washed 3X in dPBS for 2 min at RT. Slides were rinsed with distilled water and mounted with 80ul ProLong Gold Antifade with DAPI nuclear counterstain (Life Technologies, Cat#P36935) and covered with a #1.5 coverslip (Thermo, Cat#12-544-G). Fluorescent images were taken on a Zeiss Observer.Z1 microscope equipped with a Zeiss Axiocam MRc color camera and Zeiss AxioVision software (version 4.8.2.0). Images were overlayed using Adobe Photoshop CS6 software (version 13 x64). Controls included no-primary secondary-only antibody staining, and comparison to positive control frozen human liver tissue sections (Zyagen, Cat#HF-314) and negative control frozen untreated mouse liver sections.
ALT measurements
Serum alanine aminotransferase (ALT) measurements were performed on mouse serum obtained via retro-orbital bleeding using an ALT kinetic measurement kit (Teco Diagnostics) compared with a standard curve. AAV8-U6 and H1 promoter shRNA sequences are derived from on shRNA toxicity studies performed previously35.
Assessing the distribution of Alb haplotypes in the human population
A selection of the ShapeIt2 phased haplotypes for the 1000 Genomes Phase 1 integrated variant calls, corresponding to a region 1.3 Kb upstream and 1.4 Kb downstream from the hF9 integration site at the Alb stop codon, was downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase1/analysis_results/shapeit2_phased_haplotypes/ using the 1000 Genomes Data Slicer tool (available at http://browser.1000genomes.org/Homo_sapiens/UserData/SelectSlice). Haplotypes comprised of SNPs with a substantial frequency of the alternate allele in all populations were combined (here, greater than or equal to 45%, while those excluded were less than 1%) and treated as individual strings for calculating population frequency.
Extended Data
Extended Data Figure 1. hF9 liver immunohistochemistry.
From top to bottom, panels show human factor IX staining (red) with DAPI nuclear counterstain (blue) in positive control human liver, negative control untreated mouse liver, and two sets of representative stains from mice treated as neonates or adults with AAV8-P2A-hF9.
Extended Data Figure 2. Scheme of targeting rate assessment.
Assessment of on-target integration rate begins using linear amplification (LAM) with biotinylated primer 1 (black), annealing to the genomic locus but not to the vector (step 1). Linear amplicons are then bound to streptavidinylated beads and washed to exclude episomal vectors (Step 2). Subsequent second-strand DNA synthesis with random primers (Step 3) was followed by CviQI restriction digestion (Step 4). A compatible linker is then ligated (Step 5) followed by two rounds of nested PCR amplifications (primers 2–3 in blue- Step 6, and then primers 4–5 in red- Step 7). CviQI cleaves at the same distance from the homology border in both targeted and wild-type alleles, thus allowing for unbiased amplification. The amplicons of the 2nd nested PCR then serve as a template for qPCR assays with either primers 6–7 (green) or 8–9 (orange) (Step 8).
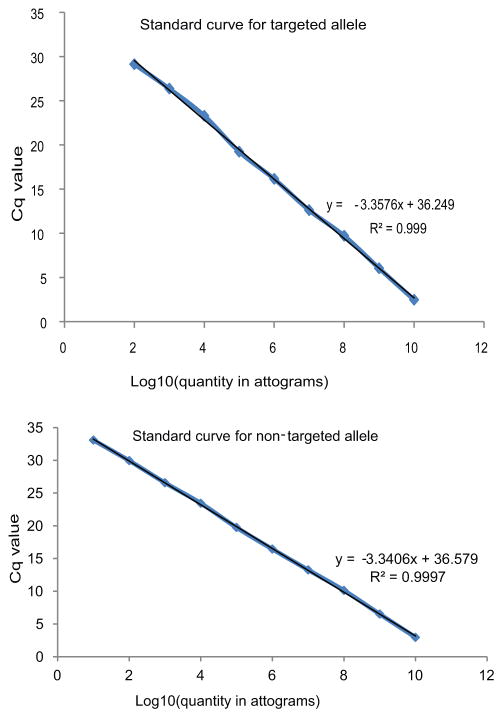
Extended Data Figure 3. Standard curves for targeting rate assessment by qPCR.
qPCR standard curves for the targeted allele (primers 8 and 9, Figure 3) and non-targeted allele (primers 6 and 7, Figure 3). Mass units used are functionally equivalent to molarity because all amplicons used were of equal length.
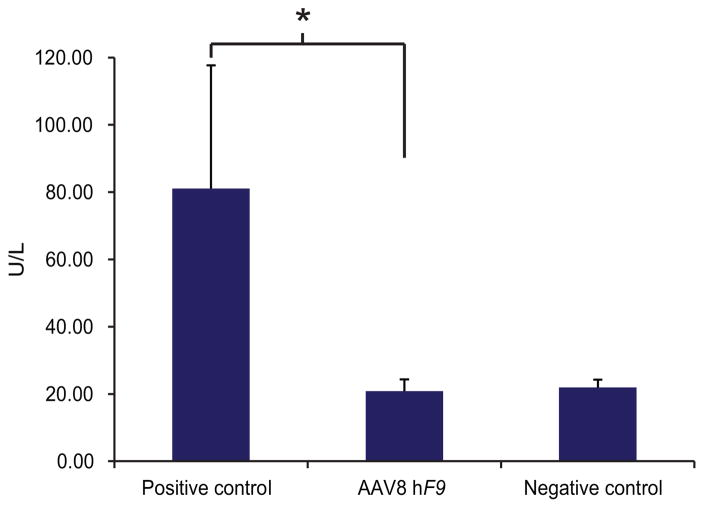
Extended Data Figure 4. Toxicity assessment by ALT measurement.
Alanine transaminase levels (ALT) were evaluated 7 days post-injection in mice injected with AAV8 coding for our experimental vector (1e12) or a negative control coding for a known non-toxic cassette (1e12 of H1 promoter-driven shRNA), or a positive control coding for a known toxic cassette (5e11 of U6 promoter-driven shRNA). Data represent mean of two measurements of four independent mice for each groups. The statistical significance is defined here as having p<0.05 in a one-tailed t test between samples of different variance.
Extended Data Figure 5. Vector copy number.
Vector copy number assessed by qPCR using primers 8 and 9 (Figure 3). N = 7 for mice injected as adults and N = 6 for mice injected as neonates and analyzed before or after partial hepatectomy. Error bars represent standard deviation.
Extended data Table 1.
Haplotypes in the human population at the relevant Alb locus as extracted from the 1000 genomes project: http://www.1000genomes.org
| Position | ID | REF | ALT | HAP1 | HAP2 | HAP3 | HAP4 | HAP5 | HAP6 | HAP7 |
|---|---|---|---|---|---|---|---|---|---|---|
| 74285239 | rs962004 | C | T | T | C | C | C | C | T | C |
| 74285552 | rs4076 | A | G | A | G | A | A | A | G | G |
| 74285567 | rs962005 | C | A | C | A | C | C | C | A | A |
| 74285758 | rs2236766 | G | T | G | T | T | G | G | T | T |
| 74285823 | rs2236767 | G | A | G | A | G | G | G | A | A |
| 74287403 | rs4429703 | T | C | C | T | T | C | T | T | C |
| Frequency | 50.14% | 44.69% | 2.15% | 1.51% | 1.37% | 0.09% | 0.05% |
Acknowledgments
This work was supported by a grant to M.A.K. from the National Heart Lung & Blood Institute (R01-HL064274). A.B. was supported by a fellowship from the National Center for Advancing Translational Sciences (UL1-TR001085), and a fellowship from the Child Health Research Institute of Stanford University. N.K.P. was supported by a fellowship from the National Heart Lung & Blood Institute (F32-HL119059), the Hans Popper Memorial Fellowship from the American Liver Foundation, and the Stanford Dean’s Fellowship. M.H.P. was supported by the Laurie Krauss Lacob Faculty Scholar Fund in Pediatric Translational Medicine. The funding organizations played no role in experimental design, data analysis, or manuscript preparation.
Footnotes
AUTHOR CONTRIBUTIONS
A.B., N.K.P., M.H.P., K.M.G. and M.A.K. designed the experiments. A.B., N.K.P., Y.S., Y.H., K.C., F.Z., P.N.V and L.P.S. generated reagents and performed the experiments. A.B., N.K.P. and M.A.K. wrote and edited the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Conflict Statement: A.B. and M.A.K are founders of LogicBio Therapeutics, a startup biotechnology company with interests in the technology described in the manuscript.
References
- 1.Aiuti A, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. The Journal of clinical investigation. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendel A, et al. Quantifying Genome-Editing Outcomes at Endogenous Loci with SMRT Sequencing. Cell reports. 2014;7:293–305. doi: 10.1016/j.celrep.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donsante A, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 8.Lisowski L, et al. Ribosomal DNA integrating rAAV-rDNA vectors allow for stable transgene expression. Molecular therapy : the journal of the American Society of Gene Therapy. 2012;20:1912–1923. doi: 10.1038/mt.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller DG, et al. Gene targeting in vivo by adeno-associated virus vectors. Nature biotechnology. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- 10.Li H, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Falahati R, Zhang J, Flebbe-Rehwaldt L, Gaensler KM. Role of antigen-specific regulatory CD4+CD25+ T cells in tolerance induction after neonatal IP administration of AAV-hF.IX. Gene therapy. 2013;20:987–996. doi: 10.1038/gt.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulk NK, Loza LM, Finegold MJ, Grompe M. AAV-mediated gene targeting is significantly enhanced by transient inhibition of nonhomologous end joining or the proteasome in vivo. Human gene therapy. 2012;23:658–665. doi: 10.1089/hum.2012.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Martinez R, et al. Albumin: pathophysiologic basis of its role in the treatment of cirrhosis and its complications. Hepatology. 2013;58:1836–1846. doi: 10.1002/hep.26338. [DOI] [PubMed] [Google Scholar]
- 14.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. Journal of virology. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lisowski L, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature. 2014;506:382–386. doi: 10.1038/nature12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, et al. Single amino acid modification of adeno-associated virus capsid changes transduction and humoral immune profiles. J Virol. 2012;86:7752–7759. doi: 10.1128/JVI.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalkara D, et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Science translational medicine. 2013;5:189ra176. doi: 10.1126/scitranslmed.3005708. [DOI] [PubMed] [Google Scholar]
- 18.Nathwani AC, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. The New England journal of medicine. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLaren RE, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulk NK, et al. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology. 2010;51:1200–1208. doi: 10.1002/hep.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavazza A, Moiani A, Mavilio F. Mechanisms of retroviral integration and mutagenesis. Human gene therapy. 2013;24:119–131. doi: 10.1089/hum.2012.203. [DOI] [PubMed] [Google Scholar]
- 22.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PloS one. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakai H, et al. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. Journal of virology. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Developmental cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Nakai H, et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. Journal of virology. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calcedo R, Wilson JM. Humoral Immune Response to AAV. Frontiers in immunology. 2013;4:341. doi: 10.3389/fimmu.2013.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anguela Xea. ZFN Mediated Targeting Of Albumin “Safe Harbor” Results In Therapeutic Levels Of Human Factor VIII In a Mouse Model Of Hemophilia A. Blood. 2013;122:720. [Google Scholar]
- 29.Yew NS, Cheng SH. Gene therapy for lysosomal storage disorders. Pediatric endocrinology reviews : PER. 2013;11 (Suppl 1):99–109. [PubMed] [Google Scholar]
- 30.Balazs AB, et al. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm D, Pandey K, Nakai H, Storm TA, Kay MA. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. Journal of virology. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Zhang F, Kay MA. A mini-intronic plasmid (MIP): a novel robust transgene expression vector in vivo and in vitro. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:954–963. doi: 10.1038/mt.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nature protocols. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 34.Rogers GL, Hoffman BE. Optimal Immunofluorescent Staining for Human Factor IX and Infiltrating T Cells following Gene Therapy for Hemophilia B. Journal of genetic syndrome & gene therapy. 2012;S1 doi: 10.4172/2157-7412.s1-012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]