Abstract
To visualize native or non-engineered RNA in live cells with single-molecule sensitivity, we developed multiply labeled tetravalent RNA imaging probes (MTRIPs). When delivered with streptolysin O into living human epithelial cancer cells and primary chicken fibroblasts, MTRIPs allowed the accurate imaging of native mRNAs and a non-engineered viral RNA, of RNA co-localization with known RNA-binding proteins, and of RNA dynamics and interactions with stress granules.
Currently researchers use a vast excess of probes or plasmid-derived RNA to image RNA with single-molecule sensitivity. Either both the RNA and probe are expressed from a plasmid, requiring binding of up to 48 MS2-GFP1,2 molecules, or just the RNA is expressed from a plasmid, requiring binding sites for 96 molecular beacon probes3 to achieve single-molecule sensitivity. As plasmid-derived RNA restricts usage to cell types that can be efficiently transfected and is susceptible to artifacts caused by overexpression, imaging native RNA is preferred, but requires a more sensitive probe to achieve single-molecule sensitivity with a limited number of bound probes. We designed multiply labeled tetravalent RNA imaging probes (MTRIPs) composed of a 2′-O-methyl RNA-DNA chimera nucleic acid ligand with four or five amino-modified thymidines, 5′ biotin modification and a short (5–7-base) poly(T) sequence to extend the ligands from the surface of streptavidin. We used the amino-modified thymidines to conjugate N-hydroxy-succinimide (NHS) ester–modified fluorophores to the ligand. On average, each ligand was labeled with three fluorophores, limiting self-quenching. We chose fluorophores with quantum yields above 65% and they exhibited little triplet state excitation. The multiply labeled monovalent ligands were tetramerized via their binding to streptavidin, which increased probe brightness fourfold (Fig. 1a). MTRIPs, when delivered via reversible cell membrane permeabilization4 with streptolysin O (Fig. 1b), allowed for single RNA molecule sensitivity using conventional fluorescence microscopy techniques. We identified the target RNA by the enhanced signal-to-background ratio achieved through the binding of multiple MTRIPs per RNA (two or three), via Watson-Crick base pairing3,5, or if using a single MTRIP per RNA, through the natural localization of RNA (Fig. 1c). This is analogous to the MS2-GFP binding systems, but uses native target sequences and fewer binding sites.
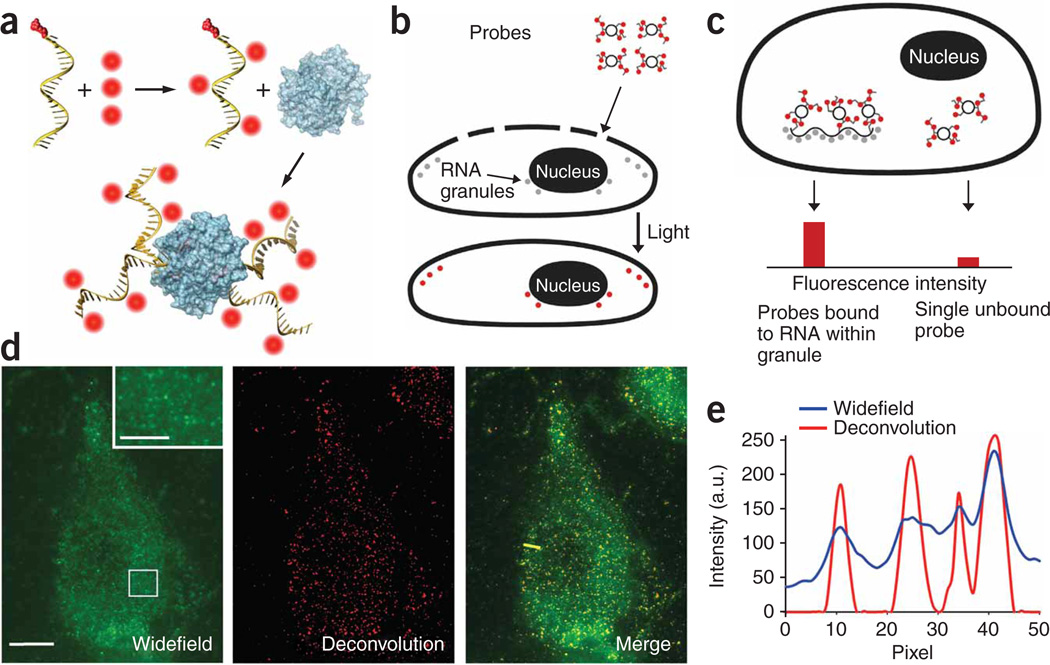
Figure 1.
Production and imaging of MTRIPs. (a) Fluorophores (red) were conjugated to amino-modified chimera oligonucleotides and bound to streptavidin (blue). (b) Cells, permeabilized with streptolysin O, allowed probe diffusion through pores; this was followed by rapid binding of the probes to targets, which was visualized after light stimulation. (c) Single RNAs bound to multiple probes were recognized by the enhanced signal-to-background ratio. (d) Live-cell widefield, deconvolved and merged images of a single optical plane of Cy3B-labeled hRSV-targeted probes in a noninfected A549 cell. Scale bars, 10 µm (2.5 µm in inset, which is a magnification of the boxed region). (e) Intensity profile of widefield and deconvolved images through yellow line in the merge image in d.
To characterize probe sensitivity and delivery to the cytosol, we immobilized probes targeting the genomic RNA of the wild-type strain A2 of human respiratory syncytial virus (hRSV) on glass surfaces and delivered them into non-infected A549 cells using streptolysin O. After examining the images of the probe mixtures on glass, the histograms of mean probe intensity and three-dimensional plots of the intensity of individual probes on the glass surface, we concluded that the images detected single probes and not aggregates (Supplementary Table 1 and Supplementary Fig. 1 online). If the probes were aggregating, the histograms would show non-unimodal behavior and mixtures of different color probes would co-localize, which was not the case. In addition, we delivered hRSV targeting probes labeled with Cy3B (GE Healthcare) and Atto 647N (Atto-Tec GmbH) into non-infected A549 cells. From a single optical plane within the live cell, we observed individual probes to be homogenously distributed in the cytoplasm (Fig. 1d,e) and did not observe localization or accumulation of probes.
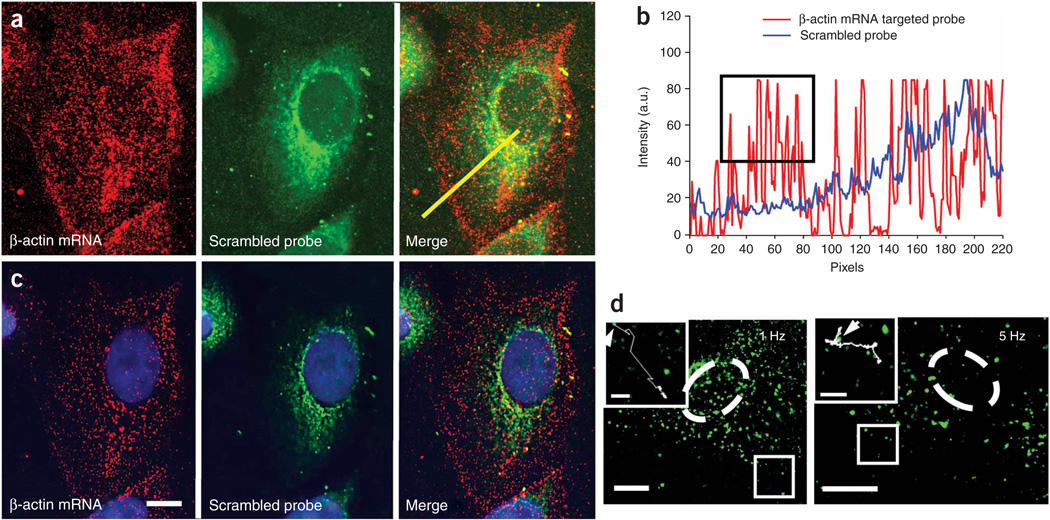
To test the ability to image single RNAs, we simultaneously delivered two Cy3B-labeled MTRIPs designed to target two regions of the human β-actin mRNA coding sequence (human β-actin mRNA probes 1 and 2; Supplementary Table 1) and an Atto 647N–labeled ‘scrambled’ probe (no target in human genome) (30 nM each) using streptolysin O into A549 cells. Twenty minutes after delivery, we fixed the cells in 4% paraformaldehyde and imaged them. We could image individual RNAs in both fixed and live cells, but we fixed the cells for quantification because of the dynamic nature of RNA granules. For β-actin, we observed individual ‘unbound’ probes as well as localized granules with twice the intensity (Fig. 2a–c). β-actin mRNA was prevalent in the perinuclear region of the cell and also localized to the leading edges, whereas the ‘scrambled’ probe produced perinuclear signals and localized not at the cell periphery but in the cytoplasm, demonstrating β-actin probe specificity. We quantified localization in an intensity profile of the confocal image (Fig. 2b). From the lower-noise, widefield-deconvolved image (Fig. 2c) we removed via thresholding the average single-probe intensities, quantified from probes on the glass surface, and counted the remaining granules using Volocity (Improvision) software. Using this approach we observed single β-actin mRNAs, containing approximately twice the single probe intensity, in the cell (Fig. 2c) and detected a total of 1,455 granules. Granule mean fluorescence intensity (calculated from the three-dimensional reconstruction) had a measured s.d. of only 25% of the mean, reflecting the uniformity of the granules when measured in three dimensions. The granule count was consistent with previous quantifications (~1,500 in serum-stimulated cells), using a similar analysis for β-actin mRNA in epithelial cells6. In addition, we imaged β-actin mRNA granules in living cells by time-lapse widefield fluorescence microscopy (Fig. 2d). We collected images with 90-ms exposure times at both 1 Hz and 5 Hz for 3 min and 30 s, respectively (Supplementary Videos 1–3 online). This demonstrated the capacity to use this method in low- and high-speed tracking experiments; similar particle trajectories have been demonstrated for plasmid-derived mRNAs1. As an additional control, we serum-starved A549 cells for 48 h and counted the β-actin mRNA granules in cells fixed after live-cell hybridization. A representative cell contained only 409 granules as compared with 1,455 granules detected in a cell grown with serum (data not shown), consistent with previous experiments6,7. We also performed single-probe imaging of clustered RNAs and co-localized them with β-actin mRNA binding protein, ZBP1 (ref. 8) in three dimensions (Supplementary Figs. 2 and 3 online), and simultaneously imaged β-actin mRNA, actin-related protein 2 homolog mRNA and ZBP1 protein, in primary chicken embryonic fibroblasts (Supplementary Fig. 4 online).
Figure 2.
Imaging of single molecules of β-actin mRNA in an A549 cell. (a) β-actin mRNA and ‘scrambled’ probe imaged with a laser scanning confocal microscope with all image planes represented. Merge images are also shown. (b) Intensity profiles along the yellow line in a. The black box highlights the large numbers of β-actin mRNA individual granules at the cell periphery detected by the targeted probe, but not by the scrambled probe. (c) Single optical plane of the same cell as in a resulting from widefield-deconvolution imaging. Scale bar, 5 µm. (d) Single optical plane of two living A549 cells (nucleus denoted with dashed line) imaged at 1 Hz for 3 min and 5 Hz for 30 s, respectively. Inset, images of boxed regions, including traces of β-actin mRNA granule trajectories for 70 s (1 Hz) and 30 s (5 Hz). Starting points are denoted by arrows. Scale bars, 10 µm (3 µm in insets).
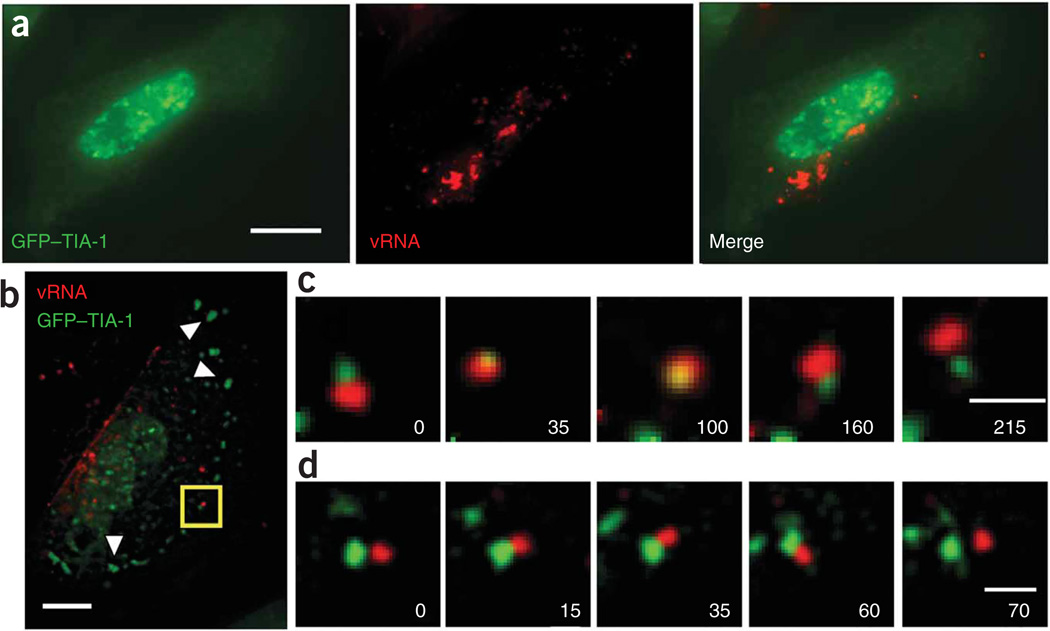
To test the utility of these probes for the study of RNA-protein colocalization in live cells, we used Cy3B-labeled MTRIPs targeted to the genomic RNA of hRSV in conjunction with a GFP–TIA-1 fusion protein in infected A549 cells, to evaluate the interaction between the stress granule protein TIA-1 and hRSV viral RNA when stress granules were induced by sodium arsenite treatment. Previous findings demonstrated that paramyxovirus RNA, which contains many possible TIA-1 or TIAR binding sites (uracil-rich regions), likely interacts with stress granules9; this interaction though, has not been characterized in living cells. In hRSV-infected cells transfected with GFP–TIA-1, stress granules had not formed 24 h after infection as identified by the lack of aggregation of GFP–TIA-1 in cells also containing viral RNA (Fig. 3a). MTRIPs were specific and did not aggregate RNA (Supplementary Figs. 5–7 and Supplementary Results online).
Figure 3.
Transient interactions between viral genomic RNA of hRSV and GFP–TIA-1. (a) Fluorescence images of a live A549 cell imaged 24 h after infection and 48 h after transfection, showing no stress granules. (b) Live-cell image of a single optical plane of GFP–TIA-1 and Cy3B-labeled hRSV-targeted MTRIPs (25 nM) in an A549 cell, 24 h after infection, 48 h after transfection and 20 min after exposure to 1 mM sodium arsenite. Arrowheads denote stress granules. (c) Time-lapse images of stress granule (green) collision, penetration and separation of a viral RNA granule (red). (d) Images of the docking of a stress and viral RNA granule, which occurred for only 45 s before separation. All granule interactions shown were imaged in the area denoted by the box in b. All times are given in seconds. Scale bars, 10 µm (a,b) and 1 µm (c,d).
To induce stress granules, we exposed the cells to 1.0 mM sodium arsenite10 and observed substantial transient interactions between stress and viral RNA granules (Fig. 3b–d). We observed a stress granule moving into a viral RNA granule and residing within it for over a minute before it was released (Fig. 3c and Supplementary Videos 4 and 5 online). Another stress granule then appeared to dock with a viral RNA granule (Fig. 3d and Supplementary Video 6 online), and appeared to be in contact for approximately 45 s. Transient interactions between RNA granules on the same time scales had been reported, supporting our observations, but previously engineered RNAs or proteins were imaged, in contrast to the non-engineered RNAs imaged in this study11–13. We also observed more stable interactions between GFP–TIA-1 and the viral RNA (Supplementary Figs. 8 and 9 and Supplementary Videos 7 and 8 online). Notably, the time-lapse results we present here cannot be obtained using probes that require high (micro-molar) probe concentrations and have lower sensitivity. The use of less-sensitive probes would result in large RNA-protein exposure time mismatches, resulting in the inability to track the interaction of individual granules accurately.
Here we demonstrated that MTRIPs have single-molecule sensitivity and can be used to target and follow native and non-engineered RNA granules in living cells, in both a cell line and a primary fibroblast using streptolysin O delivery; streptolysin O delivery induced only minimal cell death (Supplementary Fig. 10 online) and did not change cellular morphology appreciably or induce stress granules. Owing to their brightness at multiple wavelengths, small size and ease of assembly, these probes should be broadly applicable for studying single-molecule RNA-related events in living cells.
ONLINE METHODS
MTRIPs
The 2′ O-methyl RNA-DNA chimera nucleic acid ligands were synthesized by Biosearch Technologies, Inc. Each contains a 5′ biotin modification and multiple dT-C6-NH2, modifications. The streptavidin used for the core was purchased from Pierce. Probes were assembled by first labeling the free amine groups on the ligands with either Cy3B–NHS ester (GE Healthcare) or Atto 647N–NHS ester (Atto-Tec GmbH) using manufacturers’ protocols. Free dye was removed using both Nanosep spin columns (Pall Corp.) and illustra G-25 size-exclusion columns (GE Healthcare). The purified ligands were resuspended in 1× phosphate-buffered saline (PBS; pH 7.4) and mixed at a 10:1 molar ratio with streptavidin for 1 h at room temperature (18–22 °C). Free ligands were removed using 30 kDa Nanosep spin columns, and stored at 1 µM final concentration in 1× PBS at 4 °C. When multiple probes were used, each probe was completely assembled and filtered separately, and then mixed with equimolar concentrations in streptolysin O and medium just before delivery into cells We estimated the cost of MTRIPs, based on 30 nM delivery concentration, to be $0.15 per well, for a 24-well plate.
Cells and virus
A549 lung carcinoma cells (American Type Culture Collection CCL-185) were grown in DMEM (Sigma Aldrich) with 10% fetal bovine serum (FBS; Hyclone) with 100 U ml−1 of penicillin and 100 mg ml−1 of streptomycin. Virus used was the A2 strain of hRSV (American Type Culture Collection VR-1544) at a titer of 1 × 106 50% tissue culture infectious dose (TCID50) ml−1. The titer was evaluated by serial dilution and immunostaining, 4 d after infection. Infection data shown (Fig. 3 and Supplementary Figs. 5–7) was at day 1 after infection and with a multiplicity of infection of 5. All cells were infected at greater than 80% confluence, by removing the media, washing with 1× PBS (without Ca2+ and Mg2+) and then adding virus to the cells for 30 min at 37 °C. After the 30-min incubation, complete medium was added. For the motile epithelial cell experiments, A549 cells were seeded at 5% confluence such that there were a substantial number of cells without contacts with other cells. For the motile fibroblast experiments, primary chicken embryonic fibroblasts (Charles River Laboratories) were grown in CEF growth medium (Charles River Laboratories), containing 5% FBS and seeded at 5% confluence.
Probe delivery
MTRIPs were delivered into A549 and CEF cells using a reversible permeabilization method with streptolysin O (Sigma). Cells grown in complete medium were first washed with 1× PBS and then incubated with a mixture of 0.2 U ml−1 of streptolysin O and probe (concentrations were noted in the main text) in an appropriate amount of complete growth medium for 10 min at 37 °C. The mixture of streptolysin O, probe and medium was then removed and replaced with fresh, complete growth medium or Leibovitz’s L15 medium supplemented with 10% FBS. Live cells were imaged typically 20 min immediately after delivery by epifluorescence microscopy. Using streptolysin O–based delivery, probes were delivered into A549 and CEF cells with 100% efficiency.
Counting granules and statistical analysis of RNA granule populations
For data shown in Figure 2 and Supplementary Figure 7, granules were identified and counted using Improvision’s Volocity software in three dimensions using either confocal images or from widefield fluorescence images, deconvolved using an iterative deconvolution algorithm in Volocity. For data shown in Figure 2, granules were counted using deconvolved data and identified based on the s.d. of intensities; 4 s.d. above the mean was used to locate the granules in all cases because it avoided the detection of noise or objects substantially smaller than the point spread function. Because of this, no limit on minimum granule size was necessary when counting granules. In general, the cells deconvolved using the three-dimensional interative algorithm in Volocity lacked considerable noise due to filtering in the algorithm. Notably, the s.d. in mean granule intensity for the granules in Figure 2c and in the serum-starved cell (data not shown) were only 25% and 21%, respectively, reflecting the uniformity of the granules when measured in three dimensions. For data shown in Supplementary Figure 7, the number of granules was detected using the same criteria using confocal data from tetravalent MTRIPs and a monovalent ligand (ligands not connected by streptavidin) in approximately 30 cells each. The results were compared using the Wilcoxon-Mann-Whitney (WMW) test to determine whether the two samples came from the same population. WMW is a nonparametric test is traditionally used to test equality of locations in two populations, but in its general form this test is about the equality of distributions14. The null hypothesis states that the distributions of the two samples coincide and the alternative is that the distributions differ. The P value for WMW test when comparing the number of granules detected by each probe was 0.8737, which means that the evidence for the null hypothesis is decisive; consequently, the distributions were assumed to be the same.
Plasmids and transfections
The pSRα-GFP-HA-TIA-1 plasmid15 was used to image stress granules in living cells. A549 cells plated in penicillin- and streptomycin-free medium were transfected using Fugene HD (Roche) at a ratio of Fugene HD to DNA of 2.5 µl µg−1.
Time-lapse fluorescence microscopy
Live-cell video microscopy was performed using cells grown in Bioptechs T4 plates with an objective heater. The cells were imaged in Leibovitz’s L15 medium supplemented with 10% FBS. Images were taken with a Zeiss Axiovert 200M microscope, with an ×63, numerical aperture (NA) = 1.4 Plan-Apochromat objective and Hamamatsu ORCA-ER AG camera, using Chroma 49002 ET-GFP and 49004 ET-Cy3 filter sets, controlled by Volocity software. For β-actin mRNA granule dynamics, images were taken either at 1 or 5 Hz with 90-ms exposures for 3 min and 30 s, respectively. For the stress granule-RNA experiments, images of GFP–TIA-1 and the Cy3B-labeled MTRIPS were taken at 0.2 Hz (every 5 s), with exposure times of 71 and 41 ms, respectively, for up to 8 min.
Stress granule-RNA time-lapse microscopy
A549 cells, transfected with the above plasmid for 24 h were then infected with hRSV as above. Twenty-four hours after infection, the MTRIPs were delivered at 25 nM final concentration, and the cells exposed to 1.0 mM sodium arsenite for 30 min15. During the 30-min incubation, the cells were observed periodically using epifluorescence microscopy. After 15 min of exposure, stress granules (marked by GFP–TIA-1 aggregates) were observed; an imaging plane containing both RNA granules and stress granules was chosen and time-lapse microscopy was initiated. Out-of-focus light was removed using Volocity’s 2D deconvolution algorithm.
Starvation assay
A549 cells were serum-starved in DMEM with 100 U ml−1 of penicillin and 100 mg ml−1 of streptomycin for 48 h. MTRIPS (human β-actin mRNA probes 1 and 2) were delivered with streptolysin O. Twenty minutes after delivery, the cells were fixed in 4% paraformaldehyde and stained with DAPI. The granules in a particular cell were then counted using the method described above.
Immunostaining
Twenty to thirty minutes after probe delivery, cells were fixed in 4% paraformaldehyde in 1× PBS for 10 min at room temperature. After fixation, cells were permeabilized using 0.2% Triton-X 100 for 5 min at room temperature, washed in 1× PBS, blocked for 30 min in 5% BSA (ultrapure), washed in 1× PBS, incubated with primary antibody for 30 min at 37 °C, washed 3× with 1× PBS, incubated in secondary antibody labeled with Alexa 488 for 30 min at 37 °C, washed 3× with 1× PBS, labeled with DAPI for 5 min (in the RSV experiments) and then mounted in PVA with Dabco. The monoclonal antibody for the hRSV nucleocapsid protein was from Abcam, and the polyclonal Ab for ZBP1 from the Bassell laboratory. Polyclonal ZBP1 antibodies were produced by immunizing guinea pigs with a synthetic peptide corresponding to residues 162–175 (CGPENGRRGGFGSRG; the first Cys is for conjugation) within a hinge region between two RRM and four KH domains of human ZBP1. This region is conserved completely among mouse, rat and human ZBP1 proteins, but not Imp2 or Imp3. The ZBP1 antibody does not recognize mouse Imp2 or Imp3 by western blotting (data not shown). F-actin was stained using Alexa 488–labeled phalloidin (Invitrogen).
Fluorescence imaging
Immobilized Cy3B and Atto 647N probes on the glass surface were imaged using a Zeiss Axiovert 200M microscope with an ×63, NA = 1.4 Plan-Apochromat objective, using Chroma 49004 ET-Cy3 and 49006 ET-Cy5 filter sets, with 500-ms exposures. An EXFO excite 120 light source with a ND (neutral density) = 0.4 (40% transmission) was used for fluorescence excitation, and a Hamamatsu ORCA-ER AG for taking digital images. Live-cell images of single probes within A549 cells were taken with 350 ms exposures under the same illumination conditions. Z-dimension stacks were taken in both cases, in 200-nm steps, and deconvolved using Volocity iterative deconvolution algorithm. Cells used in the human β-actin mRNA-scrambled probe experiments were fixed after live-cell hybridization and imaged similarly to the immobilized probes, but with 200-ms exposures and deconvolved in Volocity. Time-lapse live cell images were taken as discussed above, and were processed with Volocity’s 2D or fast deconvolution algorithm. In the stress granule control experiments, live cell images were taken similarly to the single probe images but with 50-ms exposures. hRSV after-delivery, fixed-cell control experiments were imaged with a ZeissLSM 510 Meta using an ×63, NA = 1.4, Plan-Apochromat objective. All images were taken using multi-track scanning for each fluorophore to prevent bleed-through. Z-dimension stacks were taken in 0.5-µm increments; the 543 nm laser (Cy3B probe) was set at 25% power, the 488 nm laser (for N protein immunostaining) was set at 37%, and the pinholes were set to an airy unit of 1 (equal to airy disk). β-actin mRNA, actin-related protein 2 homolog mRNA and ZBP1, in the chicken embryonic fibroblasts, were imaged under similar conditions to the human β-actin mRNA experiments.
Image analysis
Images were analyzed using Volocity software and NIH ImageJ. Volocity was used to deconvolve the widefield images (2D and full-iterative), reconstruct the images in three dimensions, identify individual probes based on intensity and measure the mean intensity within granules of all sizes. It was also used to perform the three-dimensional co-localization calculations of Manders coefficient. The Color Profiler tool in ImageJ was used to obtain profile data for the intensity plots from the merged images.
Live-dead assay
To asses the effects of streptolysin O, and the Invitrogen L-3224 Live/Dead Viability/Cytotoxicity kit for mammalian cells was used as per the manufacturers instructions. The images were taken on a Zeiss 200M widefield epi-fluorescence microscope with an LD Plan-Neofluar ×20, NA = 0.4 objective, Hamamatsu ORCA ER-AG camera, and appropriate filter sets described above.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Georgia Institute of Technology administration (P.J.S.).
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
P.J.S. designed and performed experiments, analyzed data and wrote manuscript, A.W.L. performed experiments and analyzed data, P.C. performed experiments, Y.S. and G.J.B. provided novel antibody, M.E.L. and J.E.C. provided virus and antibody and edited manuscript.
References
- 1.Fusco D, et al. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shav-Tal Y, et al. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Proc. Natl. Acad. Sci. USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santangelo PJ, Bao G. Nucleic Acids Res. 2007;35:3602–3611. doi: 10.1093/nar/gkm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Utley TJ, et al. Proc. Natl. Acad. Sci. USA. 2008;105:10209–10214. doi: 10.1073/pnas.0712144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Femino AM, Fay FS, Fogarty K, Singer RH. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 7.Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. J. Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Mol. Cell. Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iseni F, et al. EMBO J. 2002;21:5141–5150. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedersha N, et al. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitelhofer M, et al. J. Neurosci. 2008;28:7555–7562. doi: 10.1523/JNEUROSCI.0104-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kedersha N, et al. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mollet S, et al. Mol. Biol. Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conover WJ. Practical Nonparametric Statistics 3rd edn. New York: Wiley; 1999. [Google Scholar]
- 15.Kedersha N, et al. J. Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.