Chapter summary
Conjugation is a key mechanism for horizontal gene transfer in bacteria. Some plasmids are not self-transmissible but can be mobilized by functions encoded in trans provided by other auxiliary conjugative elements. Although the transfer efficiency of mobilizable plasmids is usually lower than that of conjugative elements, mobilizable plasmidsare more frequently found in nature. In this sense, replication and mobilization can be considered as important mechanisms influencing plasmid promiscuity. Here we review the present available information on two families of small mobilizable plasmids from Gram-positive bacteria that replicate via the rolling-circle mechanism. One of these families, represented by the streptococcal plasmid pMV158, is an interesting model since it contains a specific mobilization module (MOBV) that is widely distributed among mobilizable plasmids. We discuss a mechanism in which the promiscuity of the pMV158 replicon is based on the presence of two origins of lagging strand synthesis. The current strategies to assess plasmid transfer efficiency as well as to inhibit conjugative plasmid transfer are presented. Some applications of these plasmids as biotechnological tools are also reviewed.
1. Introduction
Bacteria are everywhere simply because they can colonize and adapt to different ecological niches in a very short-term period. One important molecular mechanism underlying the abilities of bacteria to colonize new niches is the acquisition of novel traits by conjugative DNA transfer. Under these circumstances, the so-called ‘variable genome’ (as opposed to the ‘core genome’), which encodes an array of accessory functions (such as antibiotic-resistance, specific degradation pathways, symbiosis, and virulence, to name a few), is freely exchanged among bacteria (1). These newly acquired DNA pieces are represented by intra- or extra-chromosomal elements, which may or may not have self-replication and/or auto-transferable capacities. However, all of them participate in the fitness of the bacteria to colonize and to adapt to new niches; thus they contribute to create new evolutionary patterns (2). Mobile genetic elements (MGEs) constitute a reservoir of DNA that is shared among bacterial species (3) and being so, they contribute to the virulence and to the colonization of different niches by their bacterial hosts. Among MGEs, bacterial plasmids play a key role in horizontal gene transfer (HGT) and thus are important in the co-evolution and fitness of the bacterial/plasmid pair. Bacterial conjugation (described in depth elsewhere in this book) involves the unidirectional transfer of plasmid DNA from a donor to a recipient cell through physical contact (4). In the donor cell, the pre-requisite for transfer is the assembly of the plasmid-encoded relaxase and other plasmid- or host-encoded proteins on a specific cis-acting DNA plasmid region, the origin of transfer (oriT). This protein-DNA complex, the relaxosome, initiates the DNA transfer to the recipient cell by the relaxase-mediated cleavage of a phosphodiester bond at a specific dinucleotide (the nick site) within the oriT (5). It has been proposed that the relaxosome is already preformed on supercoiled DNA even before the transfer signals reach the donor-recipient cell pair (6). However, this hypothesis poses a yet unsolved question when the plasmid replicates by the rolling circle (RC) mechanism: in these RC-replicating (RCR) plasmids, initiation of replication and initiation of conjugative transfer are exerted by two different plasmid-encoded initiation proteins: Rep in the case of replication, and Mob for transfer (7). Each of these proteins recognizes a different origin on the plasmid DNA (Rep recognizes the origin of double-stranded replication, dso, whereas Mob recognizes the origin of transfer oriT); both proteins require that their DNA substrate is super coiled (7–9). If the relaxosome was already preformed, the plasmid DNA would be relaxed and the Rep-initiator could not recognize its cognate dso. As a result, the plasmid could not replicate. On the other hand, if the Rep-initiator nicks the plasmid DNA to initiate the replication, the relaxosome could not be formed and thus transfer will be hindered.
The initiation of transfer involves a Mob relaxase-mediated DNA cleavage at the oriT, the result of it being one covalent amino acyl-DNA adduct which would be actively pumped into the recipient cell by the plasmid-encoded coupling protein (CP) and the transferosome, a Type IV Secretion System (T4SS; (10–13). Thus, like almost all the nucleotidyl transferase cation-dependent enzymes, relaxases leave a free 3’-OH end while the protein remains stably bound to the 5’-phosphate product. Once in the recipient, the relaxase–DNA intermediate would restore the original circular plasmid molecule after termination of transfer by a reversion of the strand transfer reaction. Such reactions resemble the RCR termination mechanism elegantly solved in the Novick’s lab (reviewed in (14), although no indication of one modified relaxase with a short oligonucleotide covalently bound to it (as the by-product of the termination reaction) has been shown for conjugation (see (8, 14) for a detailed explanation of the termination mechanism). The last stage of transfer, the conversion of the single-stranded (ss) DNA intermediates into double-stranded (ds) plasmid molecules would take place in the recipient cell by conjugative lagging strand replication through transcription of a small primer RNA (15–18).
Since the oriT is the only cis-acting element required for DNA transfer (6), every oriT-containing plasmid should be transferred by conjugation if the protein resources (relaxase, CP, and T4SS) are available in trans from any compatible self-transferable element, a phenomenon defined as mobilization1. This interaction is therefore an important factor that can determine the frequency with which a given mobilizable element and its associated genes are involved in HGT (19). In general, self-transferable elements are transferred at higher frequencies than their mobilizable partner elements (20–21). A wide variety of mobilizable elements harbouring the oriT and the relaxase-and CP-codifying genes have been found (reviewed in (20–21). Further, many small plasmids contain a single gene cassette (oriT and relaxase-encoding gene) that allows them to be mobilized with the aid of the machinery provided by helper (auxiliary) plasmids. This is the case of many small RCR-plasmids from Gram-positive (G+) bacteria that can be mobilized by their Mob relaxase when they co-reside with an auxiliary self-transferable element (22–24). Whether the relationship between mobilizable and conjugative elements is considered as parasitic or altruistic is arguable, it seems reasonable to propose mobilization as a strategy to travel around the microbial world at low cost.
2. Nature and Diversity of Mobilizable Elements
Many MGEs share the ability to be transferred by conjugation between bacteria when they co-reside with a compatible auto-transferable element in the donor cell. Furthermore, due to their modular structure and their dynamic genetic nature, any MGE can be considered as a platform where new events of bi-directional mobilization/integration (in and out of the MGE) of other gene cassettes can occur, making it difficult to determine its original genomic location. Since the discovery of the first mobile element in 1953 (25), the diversity of the entire mobilome that one could expect in nature has been found to be very rich (21, 26). Several aspects can be considered to study the diversity of mobilizable elements, leaving aside the bacteriophages. Depending on the location of the mobile elements, they can be classified into extra- (plasmids), or intra-chromosomal elements. In the former case, they constitute the so-called plasmidome (27), whereas within the MGEs having an intra-chromosomal location two categories can be distinguished: i) Integrative and Mobilizable Elements (IMEs; (28), and ii) Mobilizable Conjugative Transposons (MCTn), which usually show non-specific integration/excision (29). The adaptive functions provided by the MGE are important because a large number of MGEs encode one or several genes involved in resistance to antibiotics, heavy metals or metabolic pathways (21, 30).
Concerning their replication autonomy, plasmids keep self-replication machinery whereas IMEs and MCTn retain the ability to integrate and excise from the host chromosome and, consequently, they do not necessarily have to be self-replicative. In the case of the extra-chromosomal MGEs, they can be generally classified considering their replication strategies, namely RC, theta, and strand displacement, although some different replication strategies can be found in linear plasmids (8, 31). Analysis of the genetic structure of the plasmid transfer-cassette shows that some plasmids contain only the oriT (plasmid pCI411), the oriT and genes encoding relaxosome components (plasmids pMV158 and pC221), or the oriT and the genes encoding the relaxase and the CP (plasmid CloDF13) (32–34). An important aspect to consider in plasmids is their host range, because replication, stability and transfer mechanisms contribute to the promiscuity of a plasmid as shown for pMV158 (7) and for IncQ plasmids (35).
3. The Mobilizable RCR-Elements: Small Plasmids Could Do It
The finding that genetic elements could be transferred by conjugation between G+ bacteria took a long while after the discovery of conjugation in Escherichia coli (36). In fact, the first reports on conjugation in G+ bacteria were over-cautious about naming ‘conjugation’ to the transfer of genetic elements encoding antibiotic-resistance determinants (the today Integrative and Conjugative Elements, ICEs) so that they were referred to as ‘DNase-resistant transfer in filter mating assays’ (37). Once accepted that the observed genetic transfer was due to genuine conjugation, it was soon apparent that small plasmids, needing helper plasmids of the Inc18 family (such as pIP501 or pAMβ1), could be also transferred by conjugation between Streptococcus pneumoniae strains (38). It was unclear which the mechanism was, since the generally believed process for transfer of these small plasmids was their co-integration with the helper, a process termed conduction (‘piggyback’). Three relevant findings at the time were: i) all RCR-plasmids studied generated large amounts of ssDNA intermediates, which were shown to be highly recombinogenic; ii) some RCR-plasmids like the staphylococcal plasmids pE194 and pT181 and, later on, pUB110 and pMV158 shared conserved sequences termed recombination site A (RSA) (22, 39); and iii) plasmid-encoded proteins, termed Pre, could be responsible for the plasmid co-integration. These findings led to the proposal that the interaction of the RSA site and the Pre protein could not only play a role in plasmid maintenance but also in the distribution of small antibiotic resistance plasmids among G+ bacteria (22, 39). These Pre proteins were claimed to contain positively charged amino acids, which were considered as probably involved in the binding of the Pre protein to the RSA site, as it is still found in some databases (http://www.uniprot.org/uniprot/P13015; http://www.ebi.ac.uk/interpro/entry/IPR001668). In spite of this, it was also clear that, at least, the ‘Pre’ protein of the streptococcal plasmid pMV158 was needed for its transfer (24), so that the term Pre/Mob to name these proteins is still retained. However, demonstration that: i) the pMV158-encoded MobM protein was able to cleave supercoiled plasmid DNA, and ii) MobM behaved like a bona fide relaxase, led to the conclusion that inter-plasmidic recombination was a by-product of the primary mobilization function (34, 40).
The only Mob proteins of RCR-plasmids that have been characterized in some detail so far are the pMV158-MobM (34, 41–42), and the mobilization proteins from plasmid pC221 (43–45). However, there are nearly 30 Mob proteins that show a high degree of similarity to MobM, including Mob proteins from well-characterized staphylococcal plasmids like pUB110, and pE194 (Table 1). Curiously enough, the staphylococcal plasmid pC194, which is closely related to pUB110, does not appear to carry a clear mobilization cassette (see below). Cross-recognition of heterologous oriTs by Mob proteins could play a role in the plasmid cassettes shuffling and spreading between bacterial species and, in fact, we have recently demonstrated the in vitro cross-recognition of oriTs of three RCR-plasmids by the pMV158-encoded MobM relaxase (41).
Table 1.
RCR-plasmids from G+ bacteria belonging to the MOBV familya
| Plasmidb | Accession number | Size (bp)c | Bacterial source | Mob (aa)c | Reference |
|---|---|---|---|---|---|
| pMV158 | NC_010096 | 5540 | Streptococcus agalactiae | 494 | (47) |
| pGB2001 | NC_015973 | 4967 | Streptococcus agalactiae | 500 | (137) |
| pGB2002 | NC_015971 | 6825 | Streptococcus agalactiae | 500 | (137) |
| p5580 | NC_019370 | 4950 | Streptococcus dysgalactiae | 500 | (138) |
| pSBO2 | AB021465 | 3582 | Streptococcus equinus | 527 | (139) |
| pSMA198 | NC_016750 | 12728 | Streptococcus macedonicus | 410 | (140) |
| pRW5 | NC_010423 | 4968 | Streptococcus pyogenes | 500 | (83) |
| pGA2000 | NC_019252 | 4967 | Streptococcus pyogenes | 500 | (137) |
| pSSU1 | NC_002140 | 4975 | Streptococcus suis | 499 | (141) |
| pSMQ172 | NC_004958 | 4230 | Streptococcus thermophilus | 499 | (57) |
| pER13 | NC_002776 | 4139 | Streptococcus thermophilus | 499 | (81) |
| pE194 | NC_005908 | 3728 | Staphylococcus aureus | 403 | (142) |
| pCPS49 | NC_019142 | 5292 | Staphylococcus aureus | 409 | (143) |
| pLA106 | NC_004985 | 2862 | Lactobacillus acidophilus | 411 | (144) |
| pCD034-2 | NC_016034 | 2707 | Lactobacillus buchneri | 363 | (145) |
| pRCEID2.9 | NC_017466 | 2952 | Lactobacillus casei | 436 | (146) |
| pWCZ | NC_019669 | 3078 | Lactobacillus paracasei | 436 | Unpublished |
| pTXW | NC_013952 | 3178 | Lactobacillus paracasei | 432 | (147) |
| pMRI 5.2 | NC_019900 | 5206 | Lactobacillus plantarum | 408 | (148) |
| pLFE1 | NC_012628 | 4031 | Lactobacillus plantarum | 83 | (149) |
| pLB4 | M33531 | 3547 | Lactobacillus plantarum | 361 | (150) |
| pA1 | NC_010098 | 2820 | Lactobacillus plantarum | 103 | (55) |
| pPB1 | NC_006399 | 2899 | Lactobacillus plantarum | 355 | (58) |
| pLS55 | NC_010375 | 5031 | Lactobacillus sakei | 439 | (151) |
| pYSI8 | NC_010936 | 4973 | Lactobacillus sakei | 403 | (152) |
| pGL2 | NC_016981 | 4572 | Lactococcus garvieae | 504 | (153) |
| pBM02 | NC_004930 | 3854 | Lactococcus lactis | 304 | (154) |
| pMBLR00 | NC_019353 | 3370 | Leuconostoc mesenteroides | 361 | (155) |
| pMA67 | NC_010875 | 5030 | Paenibacillus larvae | 435 | (156) |
Data were collected by BLASTP, using as query the pMV158-initiator of replication RepB protein. We extracted 76 plasmids, and out of these, the 29 plasmids showed in this Table 1 harbour an open reading frame that could encode a relaxase.
In plasmid pSBO2 the ORF-Mob is annotated as a protein of 144 residues (corresponding to coordinates 3149–3582). Extending the analyzed DNA sequence from coordinates 1 to 1150, a larger ORF-Mob (527 residues) could be identified. Plasmids pGA2000, pGB2001, pGB2002 and p5580 share the same replicon and mobilization cassettes (137–138).
Plasmids and Mob protein sizes are indicated in number of base pairs (bp) and amino acids (aa), respectively.
Historically, mobilizable plasmids have been studied in greater detail than the intra-chromosomal MGEs. In 2004, Francia and colleagues presented the first comprehensive classification of plasmids based on the oriT-relaxase module and the homology of the relaxases identified in small mobilizable plasmids (26). Later on, this classification was updated and extended by analysing the available sequences of conjugative plasmids on the GenBank database; six MOB families were defined: MOBF, MOBH, MOBQ, MOBC, MOBP and MOBV (20–21). Some remarkable conclusions from these studies were: i) 39% of the prokaryotic plasmids could be conjugative (although it has been experimentally demonstrated only for part of them), from which 61.5% of them may be mobilizable; ii) the majority of mobilizable plasmids are small in size (mean peak of 5 kb) but some could also be larger than 1 Mb; iii) small mobilizable plasmids have a high copy number, making them more promiscuous than the conjugative elements; iv) relaxase MOB typing can be useful to classify conjugative plasmids (46), and v) MOB families are distributed differentially among bacterial phyla: for instance, MOBV is overrepresented in Firmicutes but rare in Proteobacteria and totally absent in Actinobacteria. Whereas MOBF and MOBH are almost absent, MOBV is the most frequent module in mobilizable plasmids, followed by MOBC, MOBP and MOBQ. The following sections will summarize the representative systems belonging to mobilizable RCR-plasmid families.
4. The MOBV Family
After years of research on mobilization of pMV158 and on its MobM-relaxase (24, 34), MobM is today the prototype of the MOBV family of relaxases (clade MOBV1; (20). Plasmid pMV158 (5540 bp) was originally isolated from Streptococcus agalactiae, strain MV158; historically, it was one of the first useful cloning tools since it specifies resistance to tetracycline (tetL-type determinant) which is selectable in many G+ and Gram-negative (G−) bacteria (47–50). The promiscuity of plasmid pMV158 is very high, and so far it has been established in more than 20 bacterial species. In fact, it is naturally mobilizable between G+ bacteria by functions supplied by auxiliary plasmids such as pIP501 (51), or even between G+ and G− bacteria helped by plasmids RP4 or R388 (40). In addition, its replication by the RC mode (52), and the presence of four origins, namely dso for leading strand replication, ssoA and ssoU for lagging strand synthesis, and oriTpMV158 for mobilization, has made it an ideal molecule to study macromolecular interactions between the RepB initiator of replication and the MobM initiator of transfer with their cognate origins, dso and oriTpMV158, respectively (reviewed by (7–8, 53).
During the writing of this review, we performed an in-depth search on the entire database of sequenced plasmids and bacterial genomes, and found that the family of plasmids sharing the organization of the pMV158 replicon (54) has increased up to 76 members. The search was based on homologies with the RepB-initiator of replication and with its cognate dsopMV158. Out of the 76 replicons, 29 harbour a MOB cassette that includes a putative relaxase gene or an experimentally verified relaxase (Table 1). There are a few plasmids, like pFX2, pCI411, and pXY3, which contain just an oriT but lack any recognizable relaxase gene. Another interesting exception was the case of plasmid pA1 from Lactobacillus plant arum, which appears to harbour a relaxase pseudogene that could codify a truncated Mob protein of 103 residues. However, no promoter could be identified and synthesis of the truncated protein was not detected by in vitro assays (55). Plasmid pLFE1 also seems to encode a putative truncated Mob protein (83 amino acids), which would belong to the MOBv family and harbours an oriT sequence similar to that of pMV158. Whether these pseudogenes exist as such or they are due to sequence errors remains to be determined. In the rest of the 29 plasmids, the organization of the mobilization region is similar: the oriT is placed upstream of the Mob-encoding gene. Moreover, such plasmids harbour one or two lagging-strand origins, ssos; in the latter case, the two sso are flanking the MOB cassette (Figs. 1A and 3). Out of the 29 RCR-plasmids that harbour a MOB cassette, pE194 and pMV158 are the only ones for which the nick site at the oriT has been determined in vivo (56). Both nick sites coincided with the one previously mapped for pMV158 in vitro, namely 5’-TAGTGTG↓TTA-3’, being ‘↓’ the dinucleotide cleaved by the Mob proteins (Fig. 1B; (34)). Putative nick sites for other replicons of the MOBV family, such as the S. thermophilus plasmid pSMQ172 (57) or the L. plantarum plasmid pPB1 (58) have been proposed on the basis of the pMV158 nick site. However, whereas the proposed nick site of pSMQ172 (AGTGNG↓TT) coincides with that of pMV158 (AGTGTG↓TT), the nick site of pPB1 (AGTGG↓GTT) was proposed to be located just one nucleotide upstream in the plasmid sequence. We believe that these nick sites should be experimentally determined or revised.
FIG. 1.
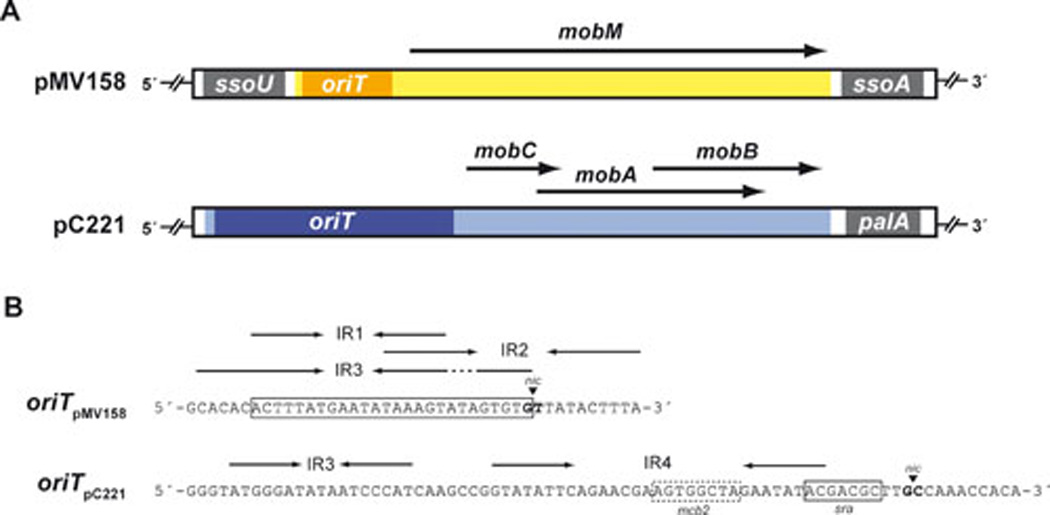
(A) The MOB modules of plasmids pMV158 and pC221. The MOB cassette of pMV158, which contains a single gene encoding the MobM relaxase, is flanked by two ssos of the types U and A, respectively (see Fig. 3). In the case of pC221, there is a single ssoA (also termed palA). Its MOB cassette contains three genes encoding the MobC, MobA, and MobB proteins. (B) Relevant features of the oriTs of plasmids pMV158 and pC221. The nucleotide sequences of the oriTpMV158 (coordinates 3564–3605) and oriTpC221 (coordinates 3084–3160) are shown. The inverted repeats (IRs, arrows) and the nick site (nic, arrowhead) are indicated. Demonstrated minimal binding site for MobM (lined box), and MobA recognition site (sra) and one of the MobC binding sites (mcb2) are also specified.
FIG. 3.
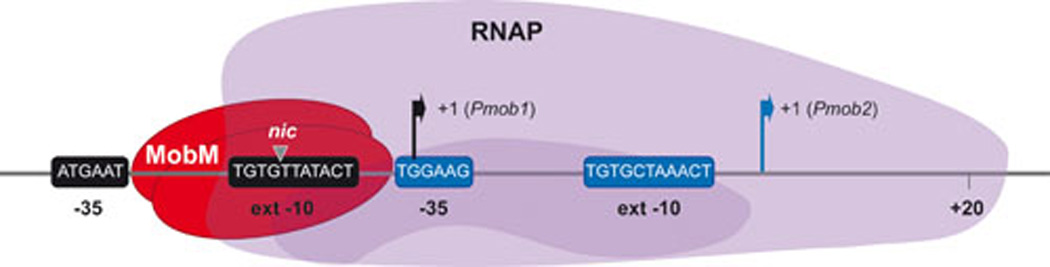
Genetic organization of the MOB cassette in pMV158. Plasmid-oriT and the mobM gene are flanked by two lagging-strand origins of replication (ssoA and ssoU). Bothssos have the possibility of generating long hairpin-loop structures exhibited as ‘ssDNA promoters’ (15, 18) where the RNAP-binding site (RSB) is recognized by the RNAP to synthesize a short RNA primer (pRNA). A consensus sequence (CS-6) located in the loop of the hairpin acts as the termination point for the pRNA synthesis. The pRNA is then used by DNA polymerase I for limited extension synthesis, followed by replication of the lagging strand by DNA Pol III.
5. The MOBP Family
The closely related plasmids pC221 and pC223 (4.6 kb) were originally isolated from Staphylococcus aureus and confer chloramphenicol resistance (59–60). They belong to the MOBP family (clade MOBP7; (20) and can be mobilized by a co-resident self-transmissible plasmid such as pGO1 (61). The transfer module of pC221/pC223 contains the oriT and three overlapping genes required for the plasmid DNA processing and mobilization. MobC and MobAreading frames overlap 16 nucleotides whereas MobA and MobBoverlap 266 nucleotides (43, 45). These genes encode the MobA relaxase and the MobB and MobC accessory proteins (Fig. 1A). Nicking of supercoiled plasmid DNA by MobA was shown to be dependent on the presence of MobC and divalent metal ions (Mg2+) both in vitro and in vivo (43, 45). Whereas the MobC proteins of pC221 and pC223 are interchangeable, the MobA proteins display specificity towards their cognate DNAs (43). The oriTs of pC221 and pC223 span a 77-bp sequence (Fig. 1B) that includes, in addition to the nick site, the MobA binding site (sra) which only differs in 4 bp between both plasmids (5’-ACGACGC-3’ and 5’-CCAGTGC-3’ in pC221 and pC223, respectively; differences in boldface letters). Remarkably, the substrate specificity of MobA could be effectively exchanged by swapping such a small region between the two plasmid oriTs (44).Little is known about the function of the MobB accessory protein, but more information is available for MobC. Protein MobC recognizes three different sites that share a consensus sequence: two sites within the oriTpC221 (mcb1 and mcb2, at 106 and 15 bp upstream of the nick site, respectively) and another site downstream of the oriTpC221 and within the mobC gene (mcb3). All the MobC binding sites are required for an efficient plasmid mobilization but only the one closest to the nick site is necessary for DNA nicking, which supports the idea that MobC works at more than one level, such as: i) assisting MobA in its nicking activity, probably by altering the substrate structure (i.e. nick site melting), and ii) positioning MobA on its recognition sequence, and mediating in a high order protein-complex structure to yield a mobilizable substrate (44).
6. Other Mobilizable RCR-Plasmids
Detailed experimental information on the features of other RCR-plasmids that can be mobilized is scarce, except for early reports on: i) transfer of staphylococcal plasmids (62–63); ii) those referring to the transfer of pUB110 from Bacillus (64), and iii) the transfer of the streptococcal plasmid pVA380-1 (65); reviewed by (9). Joint transfer of the staphylococcal plasmids pC194 and pUB110 by Tn916 from B. subtilis to B. thuringiensis has been reported (66). However, it appeared that pC194 was either not stable or non-replicative in the recipient host, which raise the question whether pC194 was conducted by the transposable element. Recently, the mobilization of pC194 (and two other plasmids lacking mobilization cassettes) by ICEBs1 was also reported, achieving higher transfer frequencies than with Tn916. These findings led the authors to propose that the initiator of replication of the plasmid could also act as a conjugative relaxase (67). Whether conduction was operating in these transfers remains to be evaluated in depth.
7. Plasmid pMV158
As stated above, the streptococcal plasmid pMV158 is the representative of the RepB-family of replicons and also of the MOBV family of relaxases. Due to its promiscuity (7) and to the numerous genetic tools based on it and developed to be used in low G+C Firmicutes (see below), we will describe here some of the main features of the pMV158 MOB-cassette (Fig. 1A).
The oriTpMV158 is located within a 41-bp region upstream of the mobM gene. The DNA sequence spanning this region exhibits a high A+T content and includes three inverted repeats (IR1, IR2, and IR3) that partially overlap (68). Such organization would permit generation of three mutually exclusive hairpin-loop structures in which the position of the dinucleotide cleaved by MobM (G/T at coordinates 3595–3596 in the pMV158 sequence; accession number NC_010096) would be different. There is a 6-bp direct repeat (5’-ACTTTA-3’), which is placed in the left arm of IR1/IR3 and in the right arm of IR2 (Fig. 1B). These features confer the oriTpMV158 a particularly complex configuration since most plasmids studied harbour only one or two IR sequences at their oriTs (69). How would MobM recognize the different IRs? This may depend on the contextual DNA topology, since, for example, extrusion of the hairpin generated by IR3, would exclude extrusion of the hairpin generated by IR2 (68). In addition, the affinity of MobM for binding to supercoiled DNA or ssDNA is much higher than for binding to linear dsDNA. Further, MobM binds to the ssDNA-encompassing IR1/3 with higher affinity (Kd ~ 60 nM) than to ssDNA-encompassing IR2 (Kd>320 nM). In fact we have shown that on ssDNA substrates the minimal oriTpMV158 sequence for an efficient MobM-binding spans 26 nucleotides that are located just upstream of the nick site (68). This minimal origin includes IR1 and eight adjacent nucleotides (Fig. 1B). Thus, differences in MobM binding could be due to the relative position of the nick site, which would be located eight nucleotides downstream of the IR1 hairpin, within the loop of the IR2 hairpin structure or just at the 3’-end of the stem of the IR3 hairpin (68).
The MobM protein (494 residues) is a dimer of identical subunits with two main domains: the N-terminal moiety harbours the DNA binding and cleaving activities (the relaxase domain), whereas the C-terminal moiety is involved in dimerization (probably through a leucine zipper motif) and, most likely, in the interaction with other proteins of the conjugative machinery (CP and association to membrane components) through coiled-coil regions (33, 41). The MobM N-terminal region has been studied in detail, and three conserved motifs have been defined: i) motif I (HxxR) of yet unknown function; ii) motif II (NYEL), which contains the proposed catalytic tyrosine, and iii) motif III (HxDExxPHuH), also known as the 3H motif of the HUH proteins, involved in the coordination of a divalent metal (70). The catalytic residue of MobM was predicted to be Tyr44 (20, 33). However, in the case of the Mob relaxase of plasmid pBBR1 (a broad-host-range theta replicating plasmid from Bordetella bronchiseptica), which belongs to the MOBV family (clade MOBV2), it was shown that none of its Tyr residues was involved in transfer (71). This finding suggested that perhaps the MOBV relaxases might present a new mechanism of conjugative DNA transfer mediated by a non-Tyr catalytic residue or by a water-mediated nucleophilic attack. This is a subject that awaits the resolution of the structure of any member of the MOBV family of relaxases.
Interestingly, MobM was able to relax super coiled DNA of other RCR-plasmids from the MOBV family (41). However, the efficiency of cleavage depended upon the homologies of the oriTs: the efficiency of MobM-mediated cleavage was similar for pMV158 and pUB110 (50% of nicking), whereas it was lower for plasmid pDL287 and pE194 (40% and 30% of nicking, respectively; (41)). Previous experiments performed with the pMV158-RepB initiator of replication protein showed that the degree of super helicity of the DNA substrate was essential for successful cleavage (72). This held true also for the RepB activity on other RCR-plasmids with origins of replication similar to the pMV158-dso (72). We postulate that, in the case of MobM, the activity of the protein would also depend on the degree of super helicity of the DNA substrate. Biochemical studies have been performed with the full length MobM protein and with two truncated derivatives that conserved the first 199 (MobMN199) or the first 243 (MobMN243) residues (41, 73). The results showed that the specificity of MobM for the DNA substrate also depends on the protein length. The three proteins relaxed super coiled DNA with the same efficiency. However, compared to MobMN199, the 44 additional residues present in MobMN243, which are predicted to form an extra α-helical region, resulted to be essential for efficient ssDNA cleavage (73). Biophysical studies showed that the thermal stability of MobMN199 increased in the presence of Mn2+ and/or DNA and that the protein retained its enzymatic activity after thermal denaturation and renaturation (68). These findings are consistent with a model in which the relaxase–DNA complex needs the protein to be unfolded to enter the recipient cell through the T4SS (74–75).
We have shown that MobM is able to negatively regulate the transcription of its own gene (76). This does not seem to be the case of several other plasmids, which require an accessory protein for transcriptional regulation of the relaxase synthesis (77). We have found that the two main protein players interacting at the oriTpMV158 region are the host RNA polymerase (RNAP) and the plasmid-encoded MobM. However, depending on the host in which the plasmid replicates, transcription of the mobM gene is controlled by different promoters (Fig. 2). Two promoters placed nearly in tandem, and termed Pmob1 and Pmob2, are used by the host RNAP: the latter is preferentially used in E. coli (76), whereas the former is used in L. lactis (51). Whereas promoter Pmob1 was mapped to reside within oriTpMV158, promoter Pmob2 was found to be adjacent to it; and yet both promoters were subjected to self-regulation by MobM in E. coli (76). Given the relative positions of RNAP and MobM proteins in the region encompassing the two promoters (Fig. 2), we could speculate that control of the mobM gene expression would be exerted by the relaxase hindering the accessibility of the RNAP to the promoters which, in turn, could also depend on the efficiency of the host RNAP to recognize one or the other promoter. Which of these two promoters is used in S. pneumoniae is a subject under investigation. Control of gene expression exerted by only a protein seems to be inefficient when a fast response to intracellular changes is required. The control role is better performed by a short-lived antisense RNA than by a relatively long-lived protein. In the case of RCR-plasmids, which are apparently devoid of any partition system, control of the synthesis of the essential Rep initiator protein is exerted at a post-transcriptional level by one or two antisense RNAs (78). However, there are instances, like replication of pMV158 or of pIP501, in which synthesis of the initiator is controlled by the joint contribution of an antisense RNA and a transcriptional regulator protein (79–80). We have no indication of the existence of an antisense RNA to control expression of the mobM gene, and we do not know yet the half-lives of the MobM-DNA and of the RNAP-Pmob1 or -Pmob2 complexes. We also lack information on the competition of the two proteins for binding to the oriTpMV158 promoter region; thus it is presently difficult to evaluate the efficiency of MobM as a transcriptional self-regulator. Several streptococcal plasmids such as pER13 (81), pSMQ172 (57), pVA380-1 (82), and pRW35 (83) maintain a similar structure at their oriT regions, with two putative tandem promoters for expression of the relaxase gene. Thus, we propose that an auto-regulation mechanism, similar to that of pMV158, would also be involved in the synthesis of their respective relaxases.
FIG. 2.
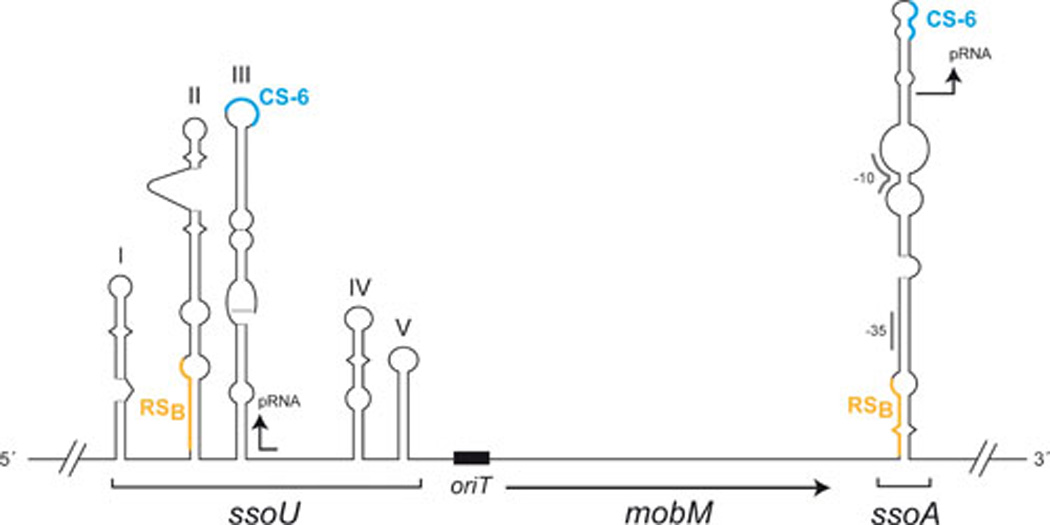
Autoregulation of mobM gene expression. The region of pMV158 that includes the Pmob1 and Pmob2 promoters of the mobM gene is shown. In E. coli, mobM transcription is mostly initiated from promoter Pmob2 (in blue) (76). Promoter Pmob1 (in black), which is located within the oriT sequence, is used in L. lactis ((51). The main sequence elements of both promoters and the nick site (nic) are indicated. Transcription start sites are also indicated with arrows. The regions recognized by MobM and E. coli RNA polymerase (RNAP) were determined by DNase I foot printing assays using linear double-stranded DNAs (42, 76). MobM is able to repress in vivo the transcription initiated from both promoters, Pmob1 and Pmob2 (76).
8. Factors contributing to the promiscuity of RCR-plasmids
The RCR-plasmids are constituted by a small number of gene modules and, in general, show an unusual promiscuity (84). Their capacity to colonize new hosts depends on their ability to: i) express their Rep initiator protein and trigger over-replication until they surpass their average copy number (around 20–30 plasmid copies per bacterial genome equivalent at the steady state); ii) control efficiently their replication once they are established in the new host and the average number of copies is reached; iii) have an efficient mechanism to convert ssDNA intermediates into dsDNA molecules, and iv) use the host machinery for their own replication and/or transfer. Thus, the philosophy of ‘travel light, not constituting a genetic load, and making a clever use of the host resources’ applies to RCR-plasmids (84). Their replication needs the Rep initiator which is provided by the plasmid, while the rest of the machinery is provided by the host: RNAP, DNA polymerases (I and III), ssDNA-binding protein, DNA gyrase, and a DNA helicase, most likely PcrA (85–87). These host proteins are synthesized in sufficient quantities so that the plasmid would not be a heavy genetic load to its host (88). Conversion of ssDNA intermediates into dsDNA forms is initiated by the host RNAP and would require the same proteins used for leading strand synthesis (15, 89).
It was soon demonstrated that small RCR-plasmids were able to replicate in several G+ bacterial hosts (49, 90), and also in G− bacteria like E. coli (50, 91–92). Some of the RCR-plasmids, like pC194, could even replicate in yeasts (93). However, some of these plasmids exhibited either structural (deletions) or segregational (loss of plasmid in the absence of selective pressure) instability. This was, most likely, associated to their replication functions (94–98), and especially to the functionality of the plasmid sso (99–100). The lagging strand origins of the RCR-plasmids are located at specific non-coding regions that can fold into imperfect hairpin structures on the ssDNA plasmid intermediates (Fig. 3). They operate in an orientation-dependent manner, indicative of the need of unpaired sequences within the hairpins that would be recognition sites of the host-encoded machinery (99, 101–102). Sequence homology analyses led to the description of various types of sso (63): ssoA (in pT181 and pC194), ssoU (in pUB110), ssoT (in pTA1060 and pBAA1), and ssoW (in pWV01 and closely related replicons; (97)). The streptococcal plasmid pMV158 harbours both ssoA and ssoU ((22, 99). Deletions affecting the ssoA functionality lead to plasmid copy number decrease, segregational instability, and accumulation of large amounts of intracellular ssDNA plasmid intermediates (22, 99, 102). Analysis of the DNA sequence and structure of the ssoA of several plasmids showed the presence of conserved unpaired regions (63) and homologies between ssoA and ssoU sequences (16, 103). One of the conserved regions encompasses part of the so-called plasmid recombination site B (Pre_RSB), which is highly conserved in nearly all the reported ssoAs. Later on, it was demonstrated that the RSB site was the binding site of the host RNAP for a class of promoters located on ssDNA (15, 18). There is another unpaired region, which was termed CS-6 (consensus sequence 6: 5’-TAGCGt/a-3’), which is a transcriptional terminator for the synthesis of a small primer RNA by the host RNAP ((15); see Fig. 3). Lagging strand synthesis is continued by DNA polymerase I, followed by DNA polymerase III and DNA gyrase to complete the plasmid replication (8, 89). An indication of a good adaptation of RCR-plasmids to their hosts is provided by the ratio of ss/ds DNA, which should be substantially low because accumulation of intracellular ssDNA can be harmful for the host bacteria (16). The MOB cassette of pMV158 (oriTpMV158 and mobM gene) is flanked by the ssoU and ssoA origins (Fig. 3). Transfer of pMV158 between S. pneumoniae and Enterococcus faecalis, allowed us to study which pMV158-sso is used in homologous and heterologous hosts (Table 2). Determination of the frequencies of transfer, plasmid copy number in the two hosts, and the efficiency of replication measured as the ratio of ss/ds DNA showed that: i) either sso supported transfer between strains of S. pneumoniae with the same efficiency as the parental pMV158; ii) only the ssoU efficiently supported transfer when it was tested between S. pneumoniae and E. faecalis, and iii) a functional ssoU is a critical plasmid region in the colonization of hosts by pMV158 (16).
Table 2.
Indicators of pMV158 promiscuity: relative efficiencies of conjugative transfer from donor S. pneumoniae to recipients S. pneumoniae and E. faecalis.
| pMV158 | Streptococcus pneumoniae | Enterococcus faecalis | ||||
|---|---|---|---|---|---|---|
| Transfera | Copy numberb | ss/dsDNAc | Transfera | Copy numberb | ss/dsDNAc | |
| WT | 1 | 30 | 1 | 10 | 20 | 1 |
| ΔssoA | 1 | 30 | 1 | 10 | 20 | 1 |
| ΔssoU | 1 | 30 | 1 | 0.01 | 5 | 15 |
| ΔssoA/ssoU | 0.01 | 10 | 50 | 0.01 | 3 | 30 |
Relative transfer in comparison to the pMV158 wild-type (WT) plasmid.
Number of plasmid molecules per chromosomal equivalent.
Ratio of ssDNA dsDNA, indicative of efficiency of conversion.
9. Inhibitors of Conjugative Transfer
Antibiotic resistance genes and virulence genes are commonly associated with MGEs that can be transferred among bacteria sharing the same niche. Many human activities related to the hospital use and to the industrial waste of antibiotics constitute a major driving force in the selection of MGEs carrying multiple antibiotic resistance genes (104). Thus, strategies for inhibiting conjugation may be crucial to preserve the effectiveness of antibiotics and to avoid the spread of resistance traits. It must be considered that many mobilizable plasmids not only are promiscuous but also harbour one or more resistance genes. Considering the molecular mechanism of conjugative transfer, several components of the relaxosome/CP/T4SS machinery could be looked at as potential targets for the development of inhibitory drugs (conjugation inhibitors, or COINS; (105)). For instance, it has been shown that unsaturated fatty acids are effective inhibitors of plasmid R388 conjugation (105). The underlying mechanism involved in this inhibition is presently unknown, but it might be related to either direct effects on the transfer machinery or to modifications of plasmatic membrane fluidity. Remarkably, whereas R388 plasmid conjugation was inhibited by unsaturated fatty acids, R388-mediated mobilization of plasmid CloDF13 was not affected, suggesting a possible effect of unsaturated fatty acids on the assembly of the R388 relaxosome. A second example of possible COINS has been provided: conjugation of plasmid F was shown to be inhibited by filamentous bacteriophages (like M13), a family of ssDNA phages that attach to the tip of the conjugative pilus (106). The proposed model contemplates that the phage coat protein gp3 occludes or reduces the assembly of the T4SS components. Therefore, these ssDNA phage coat proteins could be considered as a natural source of proteins that can compete with the conjugative T4SS, thus acting like bona fides COINS. Using again plasmid F, a third example of COINS was provided by showing that conjugative DNA relaxase was inhibited by bisphosphonates (107). However, this approach was directed against plasmids, like F, that encode multi-tyrosine relaxases and that are represented by the MOBF family (20). This may not be the general feature for all plasmid-encoded relaxases, so that bisphosphonates may be considered as limited-value COINS. A fourth approach was designed to inhibit the activity of the TrwC_R388 relaxase, and it was done by blocking its relaxation activity once the relaxase-DNA complex was within the recipient cell. This was achieved by employment of single-chain antibodies (108). All the above approaches may not be of wide usefulness, so that it would be interesting to search for COINS that target general transfer processes such as relaxosome assembly in the donor or lagging strand synthesis in the recipient bacteria.
10. Applications of the pMV158 Replicon
Due to their small size, it was soon envisaged that many of the RCR-plasmids could be used to construct useful cloning and expression vectors for G+ bacteria, especially because they were scarce or non-existent compared to those developed for G− bacteria (48). It was also apparent that generation of ssDNA intermediates, as a consequence of the RCR mechanism, could result in plasmid structural instability (94, 98, 109–111). Thus, deletions of cloned DNA happened with a certain frequency especially when the recombinant plasmids were transferred among hosts of different species (112). Nevertheless, cloning of DNA fragments up to 10 kb in size was shown to be feasible and the recombinant plasmids were structurally stable in the homologous host (48). Furthermore, employment of mobilizable RCR-plasmids to assess the HGT as well as their use as possible tools to test novel COINS has been important in the development of novel strategies to deal with the spread of antibiotic-resistance traits. To date, many different genetic tools based on RCR-plasmids have been constructed. However, small sized plasmids replicating by the theta mechanism are frequently the preferred vectors for employment in the dairy industry (113). We review below the applications of some of the plasmid vectors (mobilizable or not) that have been recently constructed using the pMV158 replicon.
10.1.Construction of Green Fluorescent Bacteria using the pMV158 Replicon
Plasmid pMV158 has been employed to construct useful genetic tools for G+ bacteria in both versions, mobilizable or non-mobilizable vectors. In the latter case, a derivative of pMV158 lacking the mobM gene (plasmid pLS1) was used to construct vectors that harbour a variant of the gfp gene, which encodes a green fluorescent protein (GFP) that carries the F64L and S65T mutations (114). The F64L mutation increases GFP solubility, while the S65T mutation increases GFP fluorescence and causes a red shift in the excitation spectrum. In addition, this variant of the gfp gene contains translation initiation signals that are optimal for its expression in prokaryotes (114). In the pLS1GFP vector, the gfp gene is under the control of a pneumococcal promoter (PM) that is inducible by maltose. The MalR transcriptional repressor controls the activity of the PM promoter in S. pneumoniae (115). A second version, termed pLS1RGFP, has the malR gene placed in cis to provide an increased gene dosage of the repressor (116). Pneumococcal cells harbouring either of these plasmids would be unable to synthesize GFP by growing them in a sucrose-containing medium (repression conditions). However, when the culture medium is supplemented with maltose the MalR repressor will be inactivated and the strong PM promoter will direct synthesis of GFP (see Fig. 4).
FIG. 4.
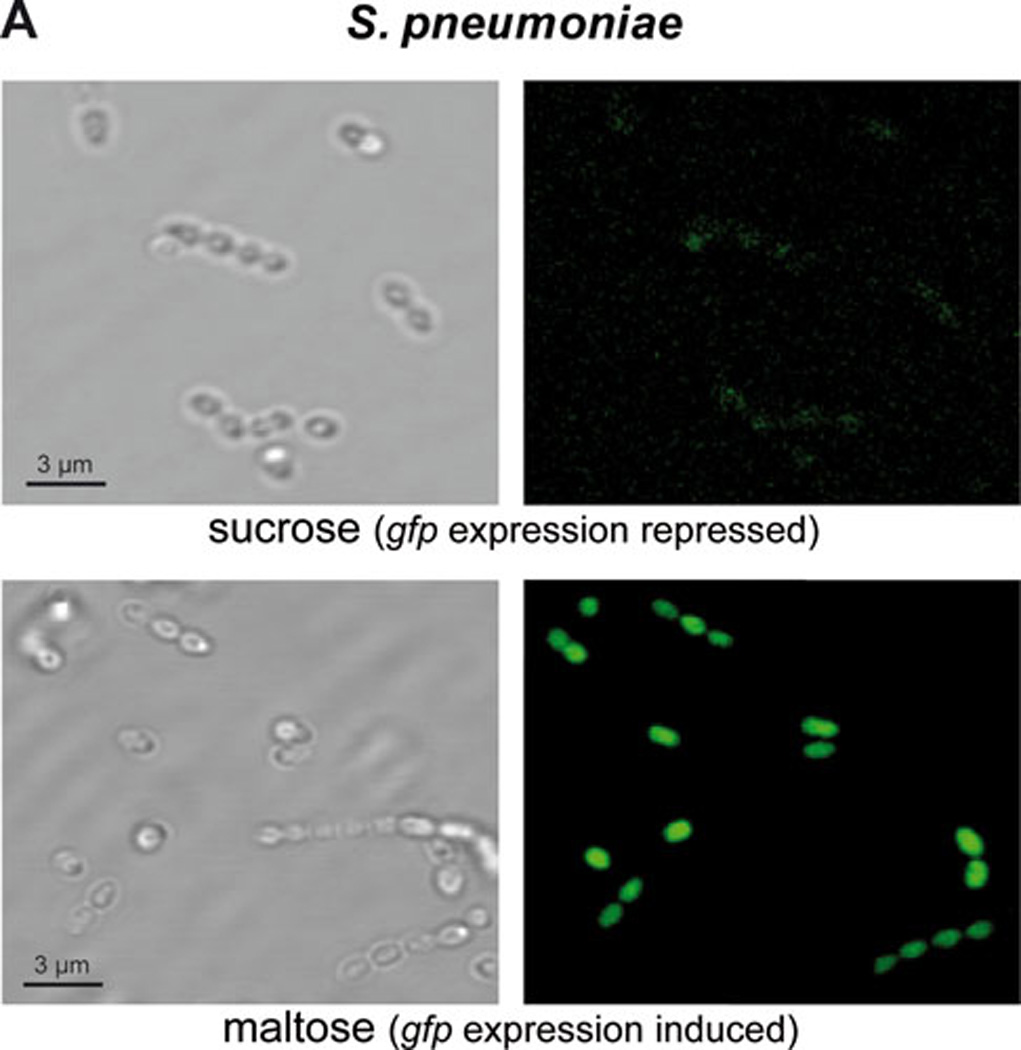
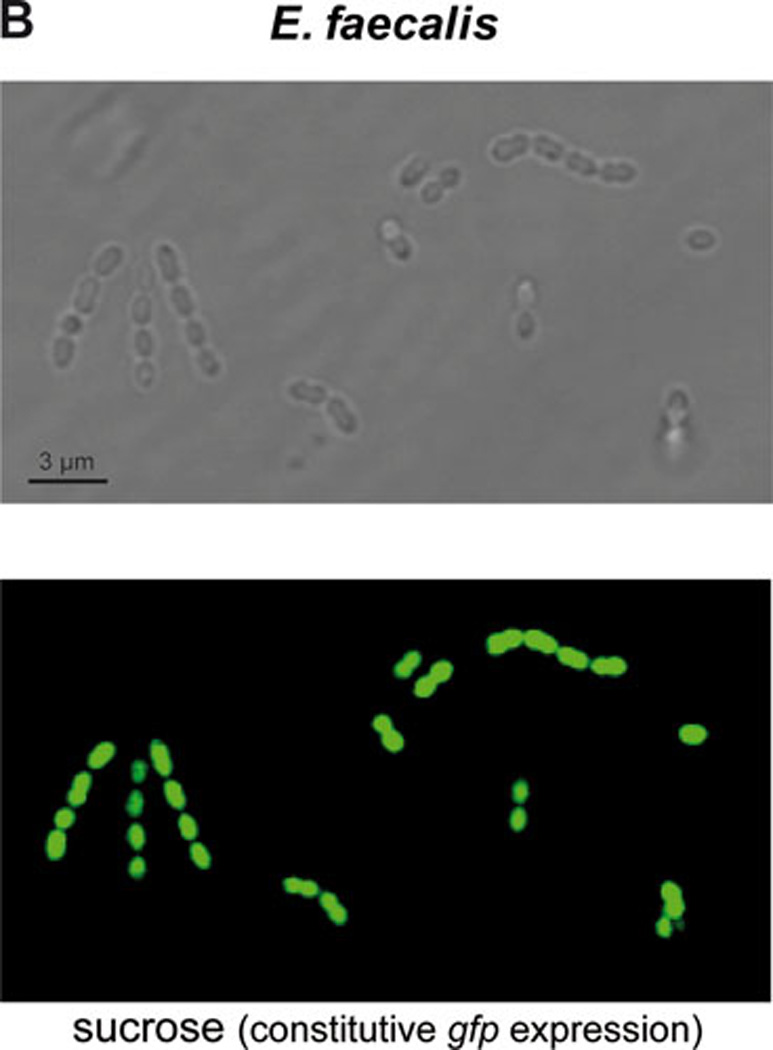
Expression of gene gfp in cells harboring plasmid pMV158GFP. Phase-contrast and fluorescence microscopy of: A) S. pneumoniae cells expressing gfp after growth in sucrose (maltose promoter PM repressed in pneumococci; top panel) or in maltose (PM induced; bottom panel), and B) E. faecalis cells expressing constitutively gfp after growth in sucrose (right panel). See text for details.
The mobilizable pMV158GFP vector is a pMV158 derivative that also carries the gfp gene fused to the maltose-inducible promoter PM (117). Thus, in pneumococcal cells carrying pMV158GFP and the chromosomal malR gene, expression of gfp depends on the carbon source added to the culture medium (repression or activation in the presence of sucrose or maltose, respectively). Nevertheless, in other G+ bacteria (like E. faecalis), the gfp gene is expressed constitutively (Fig. 4). Plasmid pMV158GFP has been mobilized not only between strains of S. pneumoniae but also from S. pneumoniae to L. lactis or E. faecalis (117), and to S. aureus (118). Plasmid pMV158GFP was shown to be a powerful tool for the construction of green fluorescent bacteria, which have been used for many different aims, such as (to name a few): i) investigation of the phagocytosis and bactericidal activity of granulocytes against live S. pneumoniae (119); ii) analysis of the intracellular localization of pneumococcal cells within murine microglial cells (120); iii) detection of L. lactis during cheese production (121); iv) visualization of the binding of E. durans to human Caco-2 cells (122); v) quantitative analysis of the intra- and inter-species conjugal transfer of pMV158 (123), and vi) quantification of adhesion of green-fluorescent-protein producing S. aureus (118). More interestingly, pMV158GFP has been used in the development of Caenorhabditis elegans as an infection model for E. faecalis. E. faecalis harbouring plasmid pMV158GFP allowed for visualization of the infection in gut of the transparent animal (Fig. 5) and can be used to quantify colonization under different conditions (D.A.G., not shown). Because the PM promoter drives constitutive expression in E. faecalis so well, it was used in the construction of an E. faecalis chromosomal integration vector to drive gfp expression in stable integrants. The expression of gfp in such a strain (SD234) was very even and stable (124).
FIG. 5.
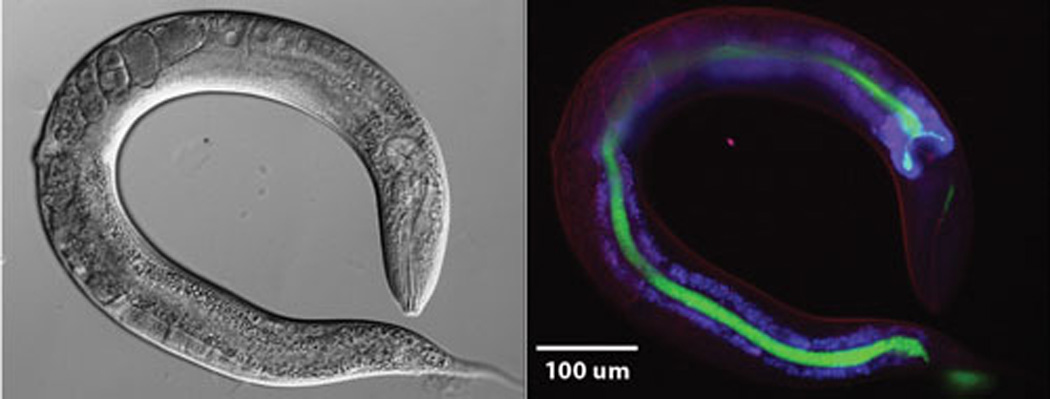
Enterococcus faecalis OG1RF expressing Green Fluorescent Protein (GFP) colonizes the intestine of C. elegans. L4 larvae were exposed for 3 hours to E. faecalis OG1RF carrying plasmid pMV158GFP. Subsequently, worms were washed with M9 medium and anesthetized with 0.25mM levamisol before imaging. DIC and fluorescence microscopy were used to visualize the worms. GFP expressing bacteria and auto-fluorescence generated by lipofuscin granules in the body of the worm were observed using FITC and DAPI filters respectively.
10.2. Construction of a Promoter-Probe Vector Based on the pMV158 Replicon
Promoter-probe vectors have been shown to be very useful for studies on regulation of gene expression. Although many algorithms have been developed for the prediction of promoters in bacterial genomes (125–126), definitive identification of promoter sequences requires the use of several experimental approaches, both in vivo and in vitro (127). Among these approaches, the use of promoter-probe plasmid vectors facilitates the identification of sequence elements that are essential for promoter activity. Moreover, such vectors make possible to investigate promoter activity in a variety of genetic backgrounds and under diverse environmental stimuli. Compared to E. coli, promoter-probe vectors are still scarce in many G+ bacteria. Ruiz-Cruz et al. (128) designed the pAST plasmid vector (5456 bp) (Fig. 6) that derives from a non-mobilizable pMV158 replicon (plasmid pLS1). Vector pAST contains a highly efficient transcriptional terminator signal (the tandem terminators T1 and T2 of the E. colirrnB operon) just upstream of a multiple cloning site (MCS). The MCS is followed by a promoter-less gfp reporter gene (114). The pAST vector was shown to be suitable to assess the activity of homologous and heterologous promoters in S. pneumoniae and E. faecalis. It is very likely that it can be used in numerous G+ bacteria because pMV158 replicates autonomously in streptococci, enterococci, staphylococci, bacilli and lactococci. In addition, pAST is a valuable tool for the design of novel expression vectors (see below).
FIG. 6.
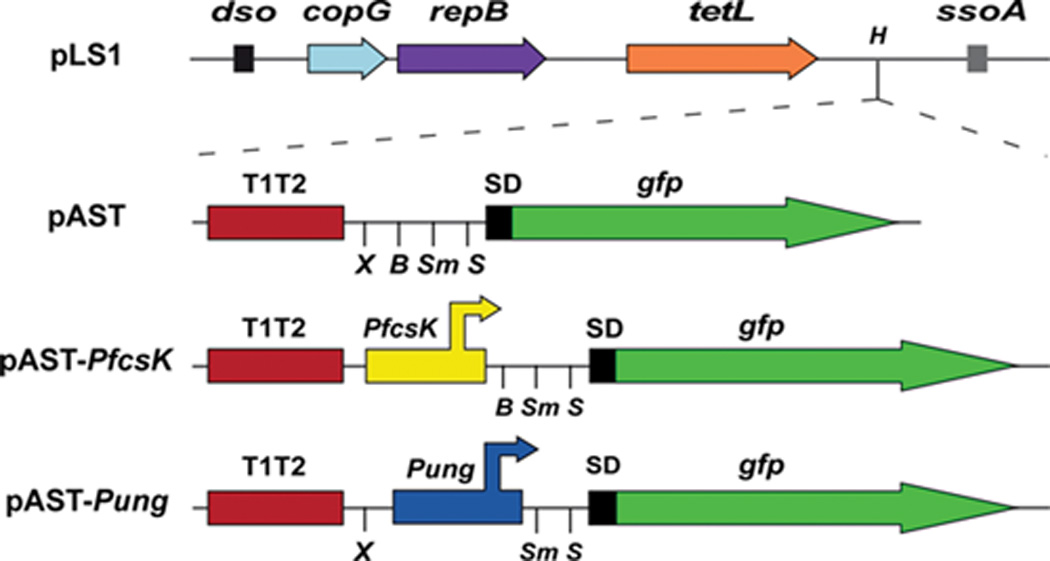
Genetic maps of plasmid pLS1 (a non-mobilizable pMV158 replicon) and its derivatives pAST, pAST-PfcsK and pAST-Pung (128). Only relevant features are indicated. copG and repB genes are involved in plasmid DNA replication. The location of the replication origins dso (double-strand origin) and ssoA (single-strand origin) is indicated. The tetL gene confers resistance to tetracycline. T1T2: tandem terminators T1 and T2 of the E. colirrnB rRNA operon. SD: translation initiation signals optimized for the expression of the gfp gene in prokaryotes (114). The position of the PfcsK and Pung promoters is indicated. H: HindIII, X: XbaI, B: BamHI, Sm: SmaI, S: SacI.
10.3. Expression Vectors Based on the pMV158 Replicon
Vector pLS1ROM (6805 bp) is a non-mobilizable pMV158 derivative that was designed for regulated gene expression in S. pneumoniae (129). It carries the malR gene under the control of a constitutive promoter and the MalR-repressed promoter PM, which is followed by a MCS. The PM promoter is activated when bacteria are grown in maltose-containing media. Under such induction conditions, pLS1ROM was shown to be structurally and segregationally stable. The functionality of this system was tested using the gfp reporter gene.
Plasmids pAST-Pung and pAST-PfcsK are a new category of plasmid pAST-derivatives (128) that can be also used as expression vectors in S. pneumoniae, and that contain useful pneumococcal promoters. Plasmid pAST-Pung (5615 bp) was constructed by cloning the promoter region (Pung) of the pneumococcal ung gene (uracil-DNA glycosylase) into the BamHI site of the pAST promoter-probe vector (Fig. 6). Promoter Pung was shown to increase 10-fold the expression of the gfp gene in pneumococcal cells (fluorescence assays). Plasmid pAST-Pung has a MCS between the Pung promoter and the gfp gene. Thus, promoter-less genes inserted into the MCS can be expressed in pneumococcus. For the construction of plasmid pAST-PfcsK (5573 bp), the PfcsK promoter of the fucose operon (130) was inserted into the XbaI site of the pAST vector (Fig. 6). Expression of gfp from the PfcsK promoter increased about 5-fold (as measured by fluorescence assays) when bacteria were grown in the presence of fucose (128). Since the putative fucose regulator gene fcsR is widely conserved in S. pneumoniaestrains (131), it can be expected that plasmid pAST-PfcsK will be valuable as inducible-expression vector in pneumococcus, and the promoter-less gene of interest can be inserted into the MCS positioned between the PfcsK promoter and the gfp gene.
11. High-throughput Assessment of the Mobility of RCR-Plasmids
Conjugation implies physical contact between the pair of donor/recipient cells, which can be established by flexible retractile pili (exemplified by F), by means of rigid pili (R388 and RP4) or, like in some G+ bacteria, by surface proteins of the sortase-type (132) or peptidoglycan degrading enzymes (133). Thus, depending on the specific conjugative system involved, cell mating can take place in liquid media or on solid/semisolid surfaces. Several high-throughput formats have been developed in order to study plasmid transfer in liquid (134), semisolid (105), or solid surfaces (123) using fluorescent as well as bioluminescent detection technologies. Such approaches permit testing plasmid transfer in single multi-well plates under the same or different conditions, reducing experimental variations and hands-on time. For instance, mobilization of plasmid pMV158GFP between different G+ bacteria has been analyzed by using microtiter plates coupled to a filtration device with sterile 0.22 µm filters (Fig. 7) to quantify: i) intra-species plasmid transfer, where the R61 pneumococcal donor strain harbouring pIP501 was mated with another pneumococcal recipient strain (carrying different selectable markers), and ii) inter-species plasmid transfer, using S. pneumoniae as donors and E. faecalis as recipients (123). Detection of transconjugants was performed by measurement of the fluorescence provided by pMV158GFP (gfp expression) and by plating suitable dilutions on selectable-marker containing plates followed by colony-forming-units counting. The frequency of transfer was high enough to be quantified: as on average, 1 out of 10,000 recipient cells received pMV158GFP. A further advantage of the employment of this format is the use of microaerophilic bacteria (like S. pneumoniae) as donors and aerobic bacteria (i.e. E. faecalis) as recipients. This improves counter-selection of donors when testing the number of transconjugants and makes the assays much easier. Therefore, this large-scale filter mating assay could be employed to analyse the parameters involved in interspecies RCR-plasmid transfers as well as to test lead compounds that could act as COINS.
FIG. 7. Strategy for high-throughput plasmid transfer detection.
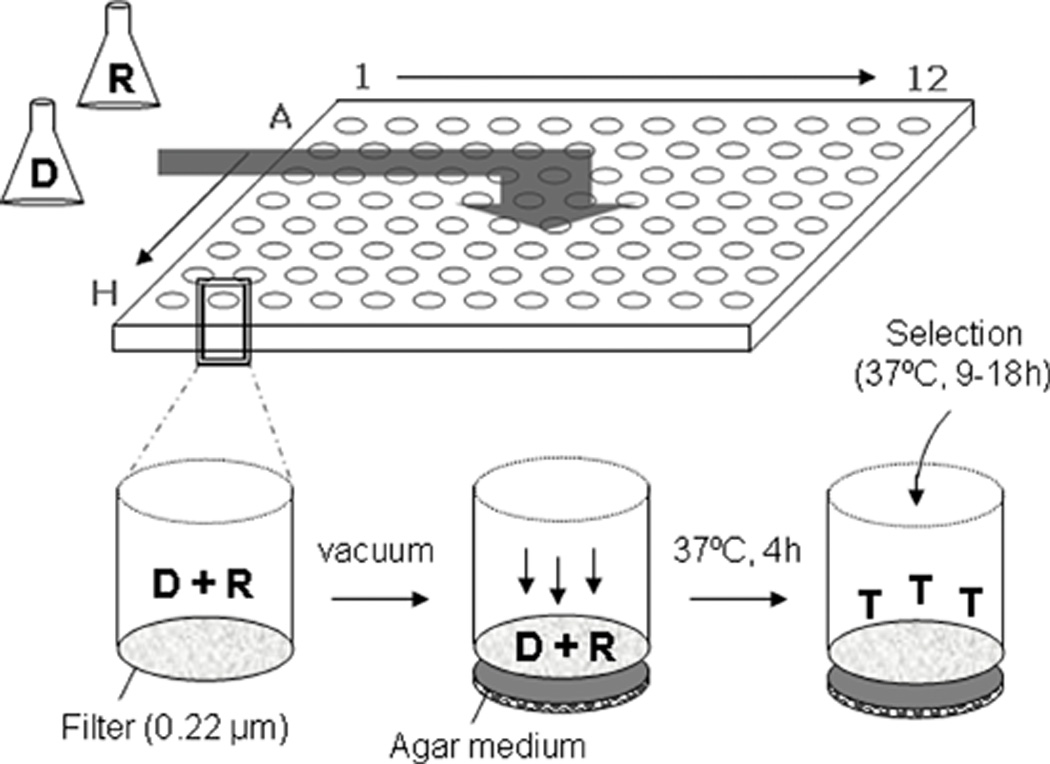
Plasmid-containing donor (D) and recipient (R) cells are mixed (it can be done at different ratios), filtered in a 96-filter-well plate (0.22 µm) and placed on a layer of solid conjugation medium (containing 5 µg/ml DNase I). After 4 h at 37°C, selection for transconjugants (T) is applied by adding antibiotic-containing medium and application of gentle vacuum. Thus, for each well the transfer frequencies can be assessed by a number of methods, such as plating on selective medium, fluorescent confocal microscopy (gfp-expressing plasmids), LacZ measurements, or quantitative PCR (Modified from (123)).
12. Final considerations
Within the present ‘ome’ era, we are confronted with the still scarce knowledge and attention that it is given to the mobilome as a whole because the spread of the antibiotic resistance is due to the presence of mobile elements among bacteria. The discovery that many G+ bacteria encode relatively simple T4SSs (4, 13, 135), which are accompanied by surface proteins involved in the DNA traffic during conjugation or genetic competence for transformation (136), raised the concern on the massive transfer of resistance genes to many pathogens. Since these ‘minimal’ T4SSs are generally associated to MGEs (such as Tn916, ICEs, pathogenicity islands, and plasmids), the risks of explosive outbreaks of G+-mediated bacterial infectious diseases are evident. Due to their small size and their promiscuity, we cannot but be astonished that only two RCR-plasmids (pC221 and pMV158) have merited attention as to study their transfer properties. It is no a matter to be taken slightly the assumption that all mobilizable RCR-plasmids will be transferred by the same mechanism or will have only one single type of relaxase or CP, so that ‘once known one, we know them all’ is a poor scientific philosophy.
ACKNOWLEDGMENTS
Research financed by the Spanish Ministry of Economy and Competitiveness (grants CSD-2008-00013-INTERMODS to M.E. and BFU2009-11868 to A.B.), by the Spanish National Research Council (grant CSIC-PIE-201320E028 to A.B.) by the National Institute of Allergy and Infectious Disease of the National Institutes of Health under award number R01AI076406 and R56AI093699 to D.A.G., and by the 7th Framework Programme (IMBRAIN project FP7-REGPOT-2012-CT2012-31637-IMBRAIN [Capacities] to F.L.D.).
Footnotes
mobilization is used here to define the transfer of conjugative elements that do not self-transfer, and therefore they need the aid of the machinery provided by an auxiliary plasmid. It might be confused with mobile, a more general and global term to define every element that either changes from the location within a genome or moves between genomes.
REFERENCES
- 1.Osborn MA, Böltner D. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid. 2002;48:202–212. doi: 10.1016/s0147-619x(02)00117-8. [DOI] [PubMed] [Google Scholar]
- 2.Baquero F. From pieces to patterns: evolutionary engineering in bacterial pathogens. Nature Rev. Microbiol. 2004;2:510–518. doi: 10.1038/nrmicro909. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CM. The horizontal gene pool. Amsterdam: Harwood Academic Publishers; 2000. [Google Scholar]
- 4.Zechner EL, Lang S, Schildbach JF. Assembly and mechanisms of bacterial type IV secretion machines. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pansegrau W, Lanka E. Enzymology of DNA strand transfer by conjugative mechanisms. Prog. Nucl. Acid Res. Mol. Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 6.Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 7.Espinosa M. Plasmids as models to study macromolecular interactions: the pMV158 paradigm. Res. Microbiol. 2013;164:199–204. doi: 10.1016/j.resmic.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 8.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiology Reviews. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 11.Llosa M, Gomis-Rüth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 12.Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E. Biogenesis, architecture, and function of bacterial Type IV secretion systems. Annu. Rev. Microbiol. 2005;59:451–485. doi: 10.1146/annurev.micro.58.030603.123630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164:620–639. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novick RP. Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem. Sci. 1998;23:434–438. doi: 10.1016/s0968-0004(98)01302-4. [DOI] [PubMed] [Google Scholar]
- 15.Kramer MG, Khan SA, Espinosa M. Plasmid rolling circle replication: identification of the RNA polymerase-directed primer RNA and requirement of DNA polymerase I for lagging strand initiation. EMBO J. 1997;16:5784–5795. doi: 10.1093/emboj/16.18.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzo-Díaz F, Espinosa M. Lagging strand DNA replication origins are required for conjugal transfer of the promiscuous plasmid pMV158. J. Bacteriol. 2009;191:720–727. doi: 10.1128/JB.01257-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker C, Meyer R. Mechanisms of strand replacement synthesis for plasmid DNA transferred by conjugation. J. Bacteriol. 2005;187:3400–3406. doi: 10.1128/JB.187.10.3400-3406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masai H, Arai K. Frpo: a novel single-stranded DNA promoter for transcription and for primer RNA synthesis of DNA replication. Cell. 1997;89:897–907. doi: 10.1016/s0092-8674(00)80275-5. [DOI] [PubMed] [Google Scholar]
- 19.Thomas CM, Nielsen KM. Mechanisms of barriers to, Horizontal Gene Transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 20.Garcillán-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Lelie D, Bron S, Venema G, Oskam L. Similarity of minus origins of replication and flanking open reading frames of plasmids pUB110, pTB913 and pMV158. Nucleic Acids Res. 1989;17:7283–7294. doi: 10.1093/nar/17.18.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abajy MY, Kopeć J, Schiwon K, Burzynski M, Döring M, Bohn C, Grohmann E. A Type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J. Bacteriol. 2007;189:2487–2496. doi: 10.1128/JB.01491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Priebe SD, Lacks SA. Region of the streptococcal plasmid pMV158 required for conjugative mobilization. J. Bacteriol. 1989;171:4778–4784. doi: 10.1128/jb.171.9.4778-4784.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lederberg J, Tatum E. Sex in bacteria; genetic studies, 1945–1952. Science. 1953;118:169–175. doi: 10.1126/science.118.3059.169. [DOI] [PubMed] [Google Scholar]
- 26.Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Walker A. Welcome to the plasmidome. Nature Rev. Microbiol. 2012;10:379. doi: 10.1038/nrmicro2804. [DOI] [PubMed] [Google Scholar]
- 28.Waldor MK. Mobilizable genomic islands: going mobile with oriT mimicry. Mol. Microbiol. 2010;78:537–540. doi: 10.1111/j.1365-2958.2010.07365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 30.Garcillán-Barcia MP, Alvarado A, de la Cruz F. Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol. Rev. 2011;35:936–956. doi: 10.1111/j.1574-6976.2011.00291.x. [DOI] [PubMed] [Google Scholar]
- 31.Espinosa M, Cohen S, Couturier M, del Solar G, Diaz-Orejas R, Giraldo R, Jannière L, Miller C, Osborn M, Thomas CM. Plasmid replication and copy number control. In: Thomas CM, editor. The horizontal gene pool. Amsterdam: Harwood Academic publishers; 2000. pp. 1–47. [Google Scholar]
- 32.Cabezon E, Sastre JI, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 33.de Antonio C, Farias ME, de Lacoba MG, Espinosa M. Features of the plasmid pMV158-encoded MobM, a protein involved in its mobilization. J. Mol. Biol. 2004;335:733–743. doi: 10.1016/j.jmb.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 34.Guzmán L, Espinosa M. The mobilization protein, MobM, of the streptococcal plasmid pMV158 specifically cleaves supercoiled DNA at the plasmid oriT. J. Mol. Biol. 1997;266:688–702. doi: 10.1006/jmbi.1996.0824. [DOI] [PubMed] [Google Scholar]
- 35.Meyer R. Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid. 2009;62:57–70. doi: 10.1016/j.plasmid.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lederberg J, Tatum E. Gene recombination in E. coli. Nature. 1946;158:558. doi: 10.1038/158558a0. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker NB, Smith MD, Guild WR. DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in Pneumococcus. Plasmid. 1980;3:80–87. doi: 10.1016/s0147-619x(80)90036-0. [DOI] [PubMed] [Google Scholar]
- 38.Smith MD, Shoemaker NB, Burdett V, Guild WR. Transfer of plasmids by conjugation in Streptococcus pneumoniae. Plasmid. 1980;3:70–79. doi: 10.1016/s0147-619x(80)90035-9. [DOI] [PubMed] [Google Scholar]
- 39.Gennaro ML, Kornblum J, Novick RP. A site-specific recombination function in Staphylococcus aureus plasmids. J. Bacteriol. 1987;169:2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farías ME, Espinosa M. Conjugal transfer of plasmid pMV158: uncoupling of the pMV158 origin of transfer from the mobilization gene mobM and modulation of pMV158 transfer in Escherichia coli mediated by IncP plasmids. Microbiology. 2000;146:2259–2265. doi: 10.1099/00221287-146-9-2259. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-López C, Lorenzo-Díaz F, Pérez-Luque R, Rodríguez-González L, Boer R, Lurz R, Bravo A, Coll M, Espinosa M. Nicking activity of the pMV158 MobM relaxase on cognate and heterologous origins of transfer. Plasmid. 2013:120–130. doi: 10.1016/j.plasmid.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Grohmann E, Guzmán LM, Espinosa M. Mobilisation of the streptococcal plasmid pMV158: interactions of MobM protein with its cognate oriT DNA region. Mol. Gen. Genet. 1999;261:707–715. doi: 10.1007/s004380050014. [DOI] [PubMed] [Google Scholar]
- 43.Caryl JA, Smith MCA, Thomas CD. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J. Bacteriol. 2004;186:3374–3383. doi: 10.1128/JB.186.11.3374-3383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caryl JA, Thomas CD. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol. Microbiol. 2006;60:1302–1318. doi: 10.1111/j.1365-2958.2006.05188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith MCA, Thomas CD. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J. Bacteriol. 2004;186:3363–3373. doi: 10.1128/JB.186.11.3363-3373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarado A, Garcillán-Barcia MP, de la Cruz F. A degenerate primer MOB typing (DPMT) method to classify Gamma-Proteobacterial plasmids in clinical and environmental settings. PLoS ONE. 2102;7:e40438. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burdett V. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B) Antimicrob. Agents Chemother. 1980;18:753–760. doi: 10.1128/aac.18.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stassi DL, Lopez P, Espinosa M, Lacks SA. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA. 1981;78:7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espinosa M, López P, Pérez-Ureña MT, Lacks SA. Interspecific plasmid transfer between Streptococcus pneumoniae and Bacillus subtilis. Mol. Gen. Genet. 1982;188:195–201. doi: 10.1007/BF00332675. [DOI] [PubMed] [Google Scholar]
- 50.Lacks SA, López P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J. Mol. Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 51.Farias ME, Grohmann E, Espinosa M. Expression of the mobM gene of the streptococcal plasmid pMV158 in Lactococcus lactis subsp lactis. FEMS Microbiol. Lett. 1999;176:403–410. doi: 10.1111/j.1574-6968.1999.tb13690.x. [DOI] [PubMed] [Google Scholar]
- 52.Puyet A, del Solar G, Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucleic Acids Res. 1988;16:115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.del Solar G, Hernández-Arriaga AM, Gomis-Rüth FX, Coll M, Espinosa M. A genetically economical family of plasmid-encoded transcriptional repressors in control of plasmid copy number. J. Bacteriol. 2002;184:4943–4951. doi: 10.1128/JB.184.18.4943-4951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.del Solar G, Moscoso M, Espinosa M. Rolling circle-replicating plasmids from Gram-positive and Gram-negative bacteria: a wall falls. Mol. Microbiol. 1993;8:789–796. doi: 10.1111/j.1365-2958.1993.tb01625.x. [DOI] [PubMed] [Google Scholar]
- 55.Vujcic M, Topisirovic L. Molecular analysis of the rolling-circle replicating plasmid pA1 of Lactobacillus plantarum A112. Appl. Environm. Microbiol. 1993:274–280. doi: 10.1128/aem.59.1.274-280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grohmann E, Zechner EL, Espinosa M. Determination of specific DNA strand discontinuities with nucleotide resolution in exponentially growing bacteria harbouring rolling circle-replicating plasmids. FEMS Microbiol. Lett. 1997;152:363–369. doi: 10.1111/j.1574-6968.1997.tb10453.x. [DOI] [PubMed] [Google Scholar]
- 57.Turgeon N, Moineau S. Isolation and characterization of a Streptococcus thermophilus plasmid closely related to the pMV158 family. Plasmid. 2001;45:171–183. doi: 10.1006/plas.2001.1517. [DOI] [PubMed] [Google Scholar]
- 58.de las Rivas B, Marcobal A, Muñoz R. Complete nucleotide sequence and structural organization of pPB1, a small Lactobacillus plantarum cryptic plasmid that originated by modular exchange. Plasmid. 2004;52:203–211. doi: 10.1016/j.plasmid.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Iordanescu S, Surdeanu M. Interactions between small plasmids in Staphylococcus aureus. Arch. Roum. Pathol. Exp. Microbiol. 1978;37:155–160. [PubMed] [Google Scholar]
- 60.Projan SJ, Novick RP. Comparative analysis of five related staphylococcal plasmids. Plasmid. 1988;19:203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- 61.Projan SJ, Archer GL. Mobilization of the Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novick RP. Plasmid incompatibility. Microbiol. Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novick RP. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- 64.Oskam L, Hillenga DJ, Venema G, Bron S. The large Bacillus plasmid pTB19 contains two integrated rolling-circle plasmids carrying mobilization functions. Plasmid. 1991;26:30–39. doi: 10.1016/0147-619x(91)90034-t. [DOI] [PubMed] [Google Scholar]
- 65.Le Blanc DJ, Chen Y-Y, Lee LN. Identification and characterization of a mobilization gene in the straptococcal plasmid pVA380-1. Plasmid. 1993;30:296–302. doi: 10.1006/plas.1993.1063. [DOI] [PubMed] [Google Scholar]
- 66.Naglich JG, Andrews REJ. Tn916-dependent conjugal transfer of pC194 and pUB110 from Bacillus subtilis into Bacillus thuringiensis subsp. israelensis. Plasmid. 1988;20:113–126. doi: 10.1016/0147-619x(88)90014-5. [DOI] [PubMed] [Google Scholar]
- 67.Lee CA, Thomas J, Grossman AD. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 2012;194:3165–3172. doi: 10.1128/JB.00301-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lorenzo-Díaz F, Dostál L, Coll M, Schildbach JF, Menendez M, Espinosa M. The MobM-relaxase domain of plasmid pMV158: thermal stability and activity upon Mn2+-and DNA specific-binding. Nucl. Acids Res. 2011;39:4315–4329. doi: 10.1093/nar/gkr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.González-Pérez B, Carballeira JD, Moncalián G, de la Cruz F. Changing the recognition site of a conjugative relaxase by rational design. Biotechnology Journal. 2009;4:554–557. doi: 10.1002/biot.200800184. [DOI] [PubMed] [Google Scholar]
- 70.Chandler M, de la Cruz F, Dyda F, Hickman AB, Moncalian G, Ton-Hoang B. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nature Rev. Microbiol. 2013;11:625–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Szpirer CY, Faelen M, Couturier M. Mobilization function of the pBHR1 plasmid, a derivative of the broad-host-range plasmid pBBR1. J. Bacteriol. 2001;183:2101–2110. doi: 10.1128/JB.183.6.2101-2110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moscoso M, del Solar G, Espinosa M. In vitro recognition of the replication origin of pLS1 and of plasmids of the pLS1 family by the RepB initiator protein. J. Bacteriol. 1995;177:7041–7049. doi: 10.1128/jb.177.24.7041-7049.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernández-López C, Pluta R, Pérez-Luque R, L R-G, Espinosa M, Coll M, Lorenzo-Díaz F, Boer R. Functional properties and structural requirements of the plasmid pMV158-encoded MobM relaxase domain. J. Bacteriol. 2013;195:3000–3008. doi: 10.1128/JB.02264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. PNAS. 2005;102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonzalez-Prieto C, Agundez L, Linden RM, Llosa M. HUH site-specific recombinases for targeted modification of the human genome. Trends Biotech. 2013;31:305–312. doi: 10.1016/j.tibtech.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 76.Lorenzo-Díaz F, Solano-Collado V, Lurz R, Bravo A, Espinosa M. Autoregulation of the synthesis of the MobM relaxase encoded by the promiscuous plasmid pMV158. J. Bacteriol. 2012;194:1789–1799. doi: 10.1128/JB.06827-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varsaki A, Moncalián G, Garcillán-Barcia MdP, Drainas C, de la Cruz F. Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J. Bacteriol. 2009;191:1446–1455. doi: 10.1128/JB.01342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kumar C, Novick RP. Plasmid pT181 replication is regulated by two counter transcripts. Proc. Natl. Acad. Sci. USA. 1985;82:638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.del Solar G, Espinosa M. The copy number of plasmid pLS1 is regulated by two trans-acting plasmid products: the antisense RNA II and the repressor protein, RepA. Mol. Microbiol. 1992;6:83–94. doi: 10.1111/j.1365-2958.1992.tb00840.x. [DOI] [PubMed] [Google Scholar]
- 80.Brantl S, Wagner EGH. An unusually long-lived antisense RNA in plasmid copy number control: in vivo RNAs encoded by the streptococcal plasmid pIP501. J. Mol. Biol. 1996;255:275–288. doi: 10.1006/jmbi.1996.0023. [DOI] [PubMed] [Google Scholar]
- 81.Somkuti GA, Steinberg DH. Molecular organization of plasmid pER13 in Streptococcus thermophilus. Biotechnol. Lett. 2007;29:1991–1999. doi: 10.1007/s10529-007-9542-z. [DOI] [PubMed] [Google Scholar]
- 82.LeBlanc DJ, Chen YYM, Lee LN. Identification and characterization of a mobilization gene in the streptococcal plasmid pVA380-1. Plasmid. 1993;30:296–302. doi: 10.1006/plas.1993.1063. [DOI] [PubMed] [Google Scholar]
- 83.Woodbury RL, Klammer KA, Xiong Y, Bailiff T, Glennen A, Bartkus JM, Lynfield R, Van Beneden C, Beall BW for the Active Bacterial Core Surveillance T. Plasmid-Borne erm(T) from invasive, Macrolide-Resistant Streptococcus pyogenes strains. Antimicrob. Agents Chemother. 2008;52:1140–1143. doi: 10.1128/AAC.01352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.del Solar G, Alonso JC, Espinosa M, Díaz-Orejas R. Broad host range plasmid replication: an open question. Mol. Microbiol. 1996;21:661–666. doi: 10.1046/j.1365-2958.1996.6611376.x. [DOI] [PubMed] [Google Scholar]
- 85.Anand SP, Khan SA. Structure-specific DNA binding and bipolar helicase activities of PcrA. Nucl. Acids. Res. 2004;32:3190–3197. doi: 10.1093/nar/gkh641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anand SP, Mitra P, Naqvi A, Khan SA. Bacillus anthracis and Bacillus cereus PcrA helicases can support DNA unwinding and in vitro rolling-circle replication of plasmid pT181 of Staphylococcus aureus. J. Bacteriol. 2004;186:2195–2199. doi: 10.1128/JB.186.7.2195-2199.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz-Masó JA, Anand SP, Espinosa M, Khan SA, del Solar G. Genetic and biochemical characterization of the Streptococcus pneumoniae PcrA helicase and its role in plasmid rolling circle replication. J. Bacteriol. 2006;188:7416–7425. doi: 10.1128/JB.01010-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hernández-Arriaga AM, Espinosa M, Del Solar G. Fitness of the pMV158 replicon in Streptococcus pneumoniae. Plasmid. 2012;67:162–166. doi: 10.1016/j.plasmid.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 89.Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 90.Gruss AD, Ehrlich SD. The family of highly interrelated single-stranded deoxyribonucleic avid plasmids. Microbiol. Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.del Solar G, Díaz R, Espinosa M. Replication of the streptococcal plasmid pMV158 and derivatives in cell-free extracts of Escherichia coli. Mol. Gen. Genet. 1987;206:428–435. doi: 10.1007/BF00428882. [DOI] [PubMed] [Google Scholar]
- 92.Goze A, Ehrlich SD. Replication of plasmids from Staphylococcus aureus in Escherichia coli. Proc Natl Acad Sci U S A. 1980;77:7333–7337. doi: 10.1073/pnas.77.12.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Goursot R, Goze A, Niaudet B, Ehrlich SD. Plasmids from Staphylococcus aureus replicate in yeast Saccharomyces cerevisiae. Nature. 1982;298:488–490. doi: 10.1038/298488a0. [DOI] [PubMed] [Google Scholar]
- 94.Bron S, Luxen E, Swart P. Instability of recombinant pUB110 plasmids in Bacillus subtilis: plasmid-encoded stability function and effects of DNA inserts. Plasmid. 1988;19:231–241. doi: 10.1016/0147-619x(88)90041-8. [DOI] [PubMed] [Google Scholar]
- 95.Meijer WJJ, van der Lelie D, Venema G, Bron S. Effects of the generation of single-stranded DNA on the maintenance of plasmid pMV158 and derivatives in Lactococcus lactis. Plasmid. 1995;33:91–99. doi: 10.1006/plas.1995.1011. [DOI] [PubMed] [Google Scholar]
- 96.Meijer WJJ, van der Lelie D, Venema G, Bron S. Effects of the generation of single-stranded DNA on the maintenance of plasmid pMV158 and derivatives in different Bacillus subtilis strains. Plasmid. 1995;33:79–89. doi: 10.1006/plas.1995.1010. [DOI] [PubMed] [Google Scholar]
- 97.Seegers JFML, Zhao AC, Meijer WJJ, Khan SA, Venema G, Bron S. Structural and functional analysis of the single-strand origin of replication from the lactococcal plasmid pWV01. Mol. Gen. Genet. 1995;249:43–50. doi: 10.1007/BF00290234. [DOI] [PubMed] [Google Scholar]
- 98.Ballester S, Lopez P, Espinosa M, Alonso JC, Lacks SA. Plasmid structural instability associated with pC194 replication functions. J. Bacteriol. 1989;171:2271–2277. doi: 10.1128/jb.171.5.2271-2277.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.del Solar G, Puyet A, Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987;15:5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hernández-Arriaga AM, Espinosa M, del Solar G. A functional lagging strand origin does not stabilize plasmid pMV158 inheritance in Escherichia coli. Plasmid. 2000;43:49–58. doi: 10.1006/plas.1999.1430. [DOI] [PubMed] [Google Scholar]
- 101.del Solar G, Kramer G, Ballester S, Espinosa M. Replication of the promiscuous plasmid pLS1: a region encompassing the minus origin of replication is associated with stable plasmid inheritance. Mol. Gen. Genet. 1993;241:97–105. doi: 10.1007/BF00280206. [DOI] [PubMed] [Google Scholar]
- 102.Gruss AD, Ross HF, Novick RP. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci USA. 1987;84:2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kramer MG, Espinosa M, Misra TK, Khan SA. Characterization of a single-strand origin, ssoU, required for broad host range replication of rolling-circle plasmids. Mol. Microbiol. 1999;33:466–475. doi: 10.1046/j.1365-2958.1999.01471.x. [DOI] [PubMed] [Google Scholar]
- 104.Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, Johnson-Rollings AS, Jones DL, Lee NM, Otten W, Thomas CM, Williams AP. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 105.Fernandez-Lopez R, Machon C, Longshaw CM, Martin S, Molin S, Zechner EL, Espinosa M, Lanka E, de la Cruz F. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology. 2005;151:3517–3526. doi: 10.1099/mic.0.28216-0. [DOI] [PubMed] [Google Scholar]
- 106.Lin A, Jimenez J, Derr J, Vera P, Manapat ML, Esvelt KM, Villanueva L, Liu DR, Chen IA. Inhibition of bacterial conjugation by phage M13 and its protein g3p: quantitative analysis and model. PLOS ONE. 2011;6:e19991. doi: 10.1371/journal.pone.0019991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proc. Natl. Acad. Sci. USA. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Garcillán-Barcia MP, Jurado P, González-Pérez B, Moncalián G, Fernández LA, de la Cruz F. Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol. Microbiol. 2007;63:404–416. doi: 10.1111/j.1365-2958.2006.05523.x. [DOI] [PubMed] [Google Scholar]
- 109.Bron S, Bosma P, van Belkum MJ, Luxen E. Stability function in the Bacillus subtilis plasmid pTA1060. Plasmid. 1987;18:8–15. doi: 10.1016/0147-619x(87)90073-4. [DOI] [PubMed] [Google Scholar]
- 110.Janniere L, Bruand C, Ehrlich SD. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990;87:53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- 111.Jannière L, Gruss A, Ehrlich SD. Plasmids. In: Sonenshein AL, Hoch JA, Losick R, Washington. American Society for, Microbiology, editors. Bacillus subtilis and other Gram-positive bacteria: Biochemistry, physiology and molecular genetics. 1993. pp. 625–644. [Google Scholar]
- 112.López P, Espinosa M, Greenberg B, Lacks SA. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1984;81:5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leenhouts KJ, Tolner B, Bron S, Kok J, Venema G, Seegers JFML. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWV01. Plasmid. 1991;26:55–66. doi: 10.1016/0147-619x(91)90036-v. [DOI] [PubMed] [Google Scholar]
- 114.Miller WG, Lindow SE. An improved GFP cloning cassette designed for prokaryotic transcriptional fusions. Gene. 1997;191:149–153. doi: 10.1016/s0378-1119(97)00051-6. [DOI] [PubMed] [Google Scholar]
- 115.Puyet A, Ibañez AM, Espinosa M. Characterization of the Streptococcus pneumoniae maltosaccharide regulator MalR, a member of the LacI-GalR family of repressors displaying distinctive genetic features. J. Biol. Chem. 1993;268:25402–25408. [PubMed] [Google Scholar]
- 116.Nieto C, Fernández de Palencia P, López P, Espinosa M. Construction of a tightly regulated plasmid vector for Streptococcus pneumoniae: controlled expression of the Green Fluorescent Protein. Plasmid. 2000;43:205–213. doi: 10.1006/plas.2000.1465. [DOI] [PubMed] [Google Scholar]
- 117.Nieto C, Espinosa M. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the Green Fluorescent Protein. Plasmid. 2003;49:281–285. doi: 10.1016/s0147-619x(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 118.Li J, Busscher HJ, van der Mei HC, Sjollema J. Surface enhanced bacterial fluorescence and enumeration of bacterial adhesion. Biofouling. 2012;29:11–19. doi: 10.1080/08927014.2012.742074. [DOI] [PubMed] [Google Scholar]
- 119.Letiembre M, Echchannaoui H, Bachmann P, Ferracin F, Nieto C, Espinosa M, Landmann R. Toll-Like Receptor 2 deficiency delays pneumococcal phagocytosis and impairs oxidative killing by granulocytes. Infect. Immun. 2005;73:8397–8401. doi: 10.1128/IAI.73.12.8397-8401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ribes S, Ebert S, Regen T, Agarwal A, Tauber SC, Czesnik D, Spreer A, Bunkowski S, Eiffert H, Hanisch U-K, Hammerschmidt S, Nau R. Toll-like receptor stimulation enhances phagocytosis and intracellular killing of non encapsulated and encapsulated Streptococcus pneumoniae by murine microglia. Infection and Immunity. 2010;78:865–871. doi: 10.1128/IAI.01110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fernández de Palencia P, de la Plaza M, Mohedano ML, Martínez-Cuesta MC, Requena T, López P, Peláez C. Enhancement of 2-methylbutanal formation in cheese by using a fluorescently tagged Lacticin 3147 producing Lactococcus lactis strain. Int J. Food Microbiol. 2004;93:335–347. doi: 10.1016/j.ijfoodmicro.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 122.Fernández de Palencia P, Fernández M, Mohedano ML, Ladero V, Quevedo C, Alvarez MA, López P. Role of tyramine synthesis by food-borne Enterococcus durans in adaptation to the gastrointestinal tract environment. Appl. Environ. Microbiol. 2011;77:699–702. doi: 10.1128/AEM.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lorenzo-Díaz F, Espinosa M. Large-scale filter mating assay for intra-and inter-specific conjugal transfer of the promiscuous plasmid pMV158 in Gram-positive bacteria. Plasmid. 2009;61:65–70. doi: 10.1016/j.plasmid.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 124.DebRoy S, van der Hoeven R, Singh KV, Gao P, Harvey BR, Murray BE, Garsin DA. Development of a genomic site for gene integration and expression in Enterococcus faecalis. J. Microbiol. Meth. 2012;90:1–8. doi: 10.1016/j.mimet.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Askary A, Masoudi-Nejad A, Sharafi R, Mizbani A, Parizi SN, Purmasjedi M. N4: a precise and highly sensitive promoter predictor using neural network fed by nearest neighbors. Genes Genet. Syst. 2009;84:425–430. doi: 10.1266/ggs.84.425. [DOI] [PubMed] [Google Scholar]
- 126.Jacques PE, Rodrigue S, Gaudreau L, Goulet J, Brzezinski R. Detection of prokaryotic promoters from the genomic distribution of hexanucleotide pairs. BMC Bioinformatics. 2006;7:423–436. doi: 10.1186/1471-2105-7-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ross W, Gourse RL. Analysis of RNA polymerase-promoter complex formation. Methods: Methods Related to Bacterial Transcriptional Control. 2009;47:13–24. doi: 10.1016/j.ymeth.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ruiz-Cruz S, Solano-Collado V, Espinosa M, Bravo A. Novel plasmid-based genetic tools for the study of promoters and terminators in Streptococcus pneumoniae and Enterococcus faecalis. J. Microb. Meth. 2010;83:156–163. doi: 10.1016/j.mimet.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 129.Ruiz-Masó JA, López-Aguilar C, Nieto C, Sanz M, Burón P, Espinosa M, del Solar G. Construction of a plasmid vector based on the pMV158 replicon for cloning and inducible gene expression in Streptococcus pneumoniae. Plasmid. 2012;67:53–59. doi: 10.1016/j.plasmid.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 130.Chan PF, O'Dwyer KM, Palmer LM, Ambrad JD, Ingraham KA, So C, Lonetto MA, Biswas S, Rosenberg M, Holmes DJ, Zalacain M. Characterization of a novel fucose-regulated promoter (PfcsK) suitable for gene essentiality and antibacterial mode-of-action studies in Streptococcus pneumoniae. J Bacteriol. 2003;185:2051–2058. doi: 10.1128/JB.185.6.2051-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weng L, Biswas I, Morrison DA. A self-deleting Cre-lox-ermAM cassette, Cheshire, for marker-less gene deletion in Streptococcus pneumoniae. J. Microbiol. Meth. 2009;79:353–357. doi: 10.1016/j.mimet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 133.Arends K, Celik E-K, Probst I, Goessweiner-Mohr N, Fercher C, Grumet L, Soellue C, Abajy MY, Sakinc T, Broszat M, Schiwon K, Koraimann G, Keller W, Grohmann E. TraG encoded by the pIP501 Type IV Secretion System is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J. Bacteriol. 2013;195:4436–4444. doi: 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnsen AR, Kroer N. Effects of stress and other environmental factors on horizontal plasmid transfer assessed by direct quantification of discrete transfer events. FEMS Microbiol. Ecol. 2007;59:718–728. doi: 10.1111/j.1574-6941.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 135.Zhang W, Rong C, Chen C, Gao GF, Schlievert PM. Type-IVC Secretion System: a novel subclass of Type IV Secretion System (T4SS) common existing in Gram-positive genus Streptococcus. PLoS ONE. 2012;7:e46390. doi: 10.1371/journal.pone.0046390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Melville S, Craig L. Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2013;77:323–341. doi: 10.1128/MMBR.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.DiPersio LP, DiPersio JR, Beach JA, Loudon AM, Fuchs AM. Identification and characterization of plasmid-borne erm(T) macrolide resistance in group B and group A Streptococcus. Diagn Microbiol Infect Dis. 2011;71:217–223. doi: 10.1016/j.diagmicrobio.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 138.Palmieri C, Magi G, Creti R, Baldassarri L, Imperi M, Gherardi G, Facinelli B. Interspecies mobilization of an erm(T)-carrying plasmid of Streptococcus dysgalactiae subsp. equisimilis by a coresident ICE of the ICESa2603 family. J Antimicrob Chemother. 2013;68:23–26. doi: 10.1093/jac/dks352. [DOI] [PubMed] [Google Scholar]
- 139.Nakamura M, Ogata K, Nagamine T, Tajima K, Matsui H, Benno Y. Characterization of the cryptic plasmid pSBO2 isolated from Streptococcus bovis JB1 and construction of a new shuttle vector. Curr. Microbiol. 2000;41:27–32. doi: 10.1007/s002840010086. [DOI] [PubMed] [Google Scholar]
- 140.Papadimitriou K, Ferreira S, Papandreou NC, Mavrogonatou E, Supply P, Pot B, Tsakalidou E. Complete genome sequence of the dairy isolate Streptococcus macedonicus ACA-DC 198. J Bacteriol. 2012;194:1838–1839. doi: 10.1128/JB.06804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takamatsu D, Osaki M, Sekizaki T. Sequence analysis of a small cryptic plasmid isolated from Streptococcus suis serotype 2. Curr. Microbiol. 2000;40:61–66. doi: 10.1007/s002849910012. [DOI] [PubMed] [Google Scholar]
- 142.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamine, and streptogramin type B antibiotics. J. Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kadlec K, Pomba CF, Couto N, Schwarz S. Small plasmids carrying vga(A) or vga(C) genes mediate resistance to lincosamides, pleuromutilins and streptogramin A antibiotics in methicillin-resistant Staphylococcus aureus ST398 from swine. J Antimicrob Chemother. 2010;65:2692–2693. doi: 10.1093/jac/dkq365. [DOI] [PubMed] [Google Scholar]
- 144.Sano K, Otani M, Okada Y, Kawamura R, Umesaki M, Ohi Y, Umezawa C, Kanatani K. Identification of the replication region of the Lactobacillus acidophilus plasmid pLA106. FEMS Microbiol. Lett. 1997;148:223–226. doi: 10.1111/j.1574-6968.1997.tb10292.x. [DOI] [PubMed] [Google Scholar]
- 145.Heinl S, Spath K, Egger E, Grabherr R. Sequence analysis and characterization of two cryptic plasmids derived from Lactobacillus buchneri CD034. Plasmid. 2011;66:159–168. doi: 10.1016/j.plasmid.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 146.Panya M, Lulitanond V, Tangphatsornruang S, Namwat W, Wannasutta R, Suebwongsa N, Mayo B. Sequencing and analysis of three plasmids from Lactobacillus casei TISTR1341 and development of plasmid-derived Escherichia coli-L. casei shuttle vectors. Appl Microbiol Biotechnol. 2012;93:261–272. doi: 10.1007/s00253-011-3503-0. [DOI] [PubMed] [Google Scholar]
- 147.Zhang H, Hao Y, Zhang D, Luo Y. Characterization of the cryptic plasmid pTXW from Lactobacillus paracasei TXW. Plasmid. 2011;65:1–7. doi: 10.1016/j.plasmid.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 148.Cho GS, Huch M, Mathara JM, van Belkum MJ, Franz CM. Characterization of pMRI 5.2, a rolling-circle-type plasmid from Lactobacillus plantarum BFE 5092 which harbours two different replication initiation genes. Plasmid. 2013;69:160–171. doi: 10.1016/j.plasmid.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 149.Feld L, Bielak E, Hammer K, Wilcks A. Characterization of a small erythromycin resistance plasmid pLFE1 from the food-isolate Lactobacillus plantarum M345. Plasmid. 2009;61:159–170. doi: 10.1016/j.plasmid.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 150.Bates EEM, Gilbert HJ. Characterization of a cryptic plasmid from Lactobacillus plantarum. Gene. 1989;85:253–258. doi: 10.1016/0378-1119(89)90491-5. [DOI] [PubMed] [Google Scholar]
- 151.Ammor MS, Gueimonde M, Danielsen M, Zagorec M, van Hoek AHAM, de los Reyes-Gavilan CG, Mayo B, Margolles A. Two Different Tetracycline Resistance Mechanisms, Plasmid-Carried tet(L) and Chromosomally Located Transposon-Associated tet(M), Coexist in Lactobacillus sakei Rits 9. Appl. Environ. Microbiol. 2008;74:1394–1401. doi: 10.1128/AEM.01463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhai Z, Hao Y, Yin S, Luan C, Zhang L, Zhao L, Chen D, Wang O, Luo Y. Characterization of a novel rolling-circle replication plasmid pYSI8 from Lactobacillus sakei YSI8. Plasmid. 2009;62:30–34. doi: 10.1016/j.plasmid.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 153.Aguado-Urda M, Gibello A, Blanco MM, Lopez-Campos GH, Cutuli MT, Fernandez-Garayzabal JF. Characterization of plasmids in a human clinical strain of Lactococcus garvieae. PLoS One. 2012;7:e40119. doi: 10.1371/journal.pone.0040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sanchez C, Mayo B. Sequence and analysis of pBM02, a novel RCR cryptic plasmid from Lactococcus lactis subsp cremoris P8-2-47. Plasmid. 2003;49:118–129. doi: 10.1016/s0147-619x(03)00013-1. [DOI] [PubMed] [Google Scholar]
- 155.Chae HS, Lee JM, Lee JH, Lee PC. Isolation and characterization of a cryptic plasmid, pMBLR00, from Leuconostoc mesenteroides subsp. mesenteroides KCTC 3733. J Microbiol Biotechnol. 2013;23:837–842. doi: 10.4014/jmb.1304.04009. [DOI] [PubMed] [Google Scholar]
- 156.Murray KD, Aronstein KA. Oxytetracycline-resistance in the honey bee pathogen Paenibacillus larvae is encoded on novel plasmid pMA67. J Apicultural Research. 2006;45:207–214. [Google Scholar]


