Abstract
Contemporary strategies that concentrate on only one or a handful of molecular targets limits the utility of the information gained for diagnostic and predictive purposes. Recent advances in the sensitivity, speed, and precision of measurements obtained from ion mobility coupled to mass spectrometry (IM-MS) have accelerated the utility of IM-MS in untargeted, discovery-driven studies in biology. Perhaps most evident is the impact that such wide-scale discovery capabilities have yielded in the areas of systems, synthetic, and chemical biology, where the need for comprehensive, hypothesis-driving studies from multidimensional and unbiased data is required.
Keywords: Ion mobility, Mass spectrometry, Ion mobility-mass spectrometry, Omics, Systems biology
Introduction
One of the clear paradigms from genomics and genome medicine is the potential of broad scale genome-wide association studies (GWAS) to correlate genetic alterations with phenotype. In tandem with advances in molecular characterization approaches with nuclear magnetic resonance (NMR) and mass spectrometry (MS), these broad scale concepts are more recently utilized in molecular or metabolome-wide association studies (MWAS) to correlate the dynamic metabolite complement in tissues or bodily fluids with phenotypic diversity [1,2]. The MWAS strategy is highly complementary with many systems biology strategies that entail characterizing, quantifying, and cataloging the biomolecular inventory of a sample at specific dimensions of space (e.g., cellular, tissue, or organism levels) and time (e.g., point in the life cycle, healthy vs. diseased state, longitudinal exposure). These largely hypothesis-independent data are then integrated with bioinformatics strategies to derive significant bimolecular signatures that describe the phenotype. Importantly, the generalized workflow of these strategies is well suited for many studies in systems, synthetic, and chemical biology in that although the specific query must be tailored to the question at hand, the prevailing analytics are one that conceptually requires the rapid generation of untargeted and rich datasets that are then interrogated to reveal those molecules most salient to the query based upon the systems-wide analysis.
Biological systems-wide analyses necessitate the acquisition of multi-dimensional datasets where individual dimensions represents molecular separations distinguishing different physical characteristics for orthogonal molecular selectivity. Oftentimes such datasets are challenging to obtain, in particular for limited or large numbers of samples, because of sacrifices in either molecular breadth, or sampling rate. While quantitation of gene transcription is dominated by array technology, many omics endeavors, such as metabolomics, proteomics, lipidomics, and glycomics are most commonly performed using MS or LC-MS [3]. In large part, this is attributed to the necessity of requiring massive numbers of experiments to understand metabolic and molecular networks under different conditions and the high throughput afforded by contemporary MS instrumentation that makes satisfying this requirement feasible [4,5]. Nevertheless, in many contemporary LC-MS or GC-MS omics studies, typically the class of molecule (e.g., proteomics, lipidomics, glycomics, etc.) is purified prior to analysis which simplifies the scope of the study, but restricts the molecular breadth in untargeted approaches. Clearly, large-scale systems-wide experiments motivate the development of measurement strategies that incorporate higher throughput, higher selectivity, are comprehensive, and require minimal sample manipulation.
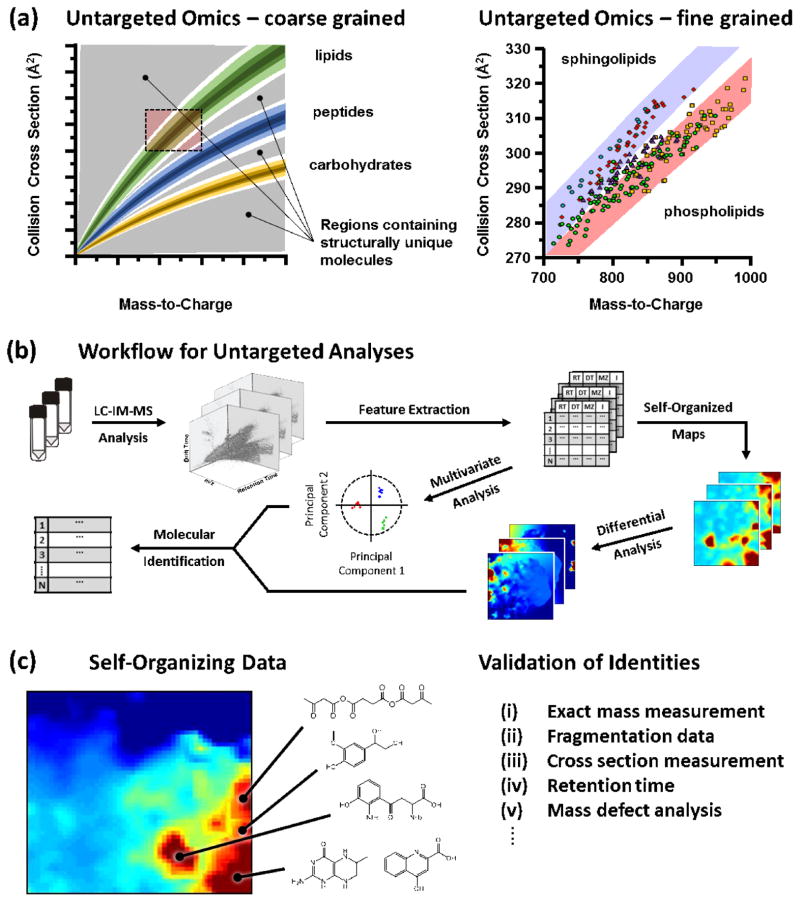
Recently, approaches using gas-phase electrophoresis, namely ion mobility spectrometry integrated with mass spectrometry (IM-MS), have been demonstrated to provide additional analyte selectivity without significantly compromising the speed of MS-based measurements. In these arrangements, the IM dimension provides molecular structural information, while the MS dimension affords accurate mass information. Importantly, the correlations of molecular density and mass obtained by the combination of IM-MS also permits the integration of omics measurements (Figure 1A), where very little sample pretreatment is necessary as the data is organized into well discerned patterns corresponding to the class of molecule to which a particular signal corresponds [6,7]. The structural and mass information afforded by IM-MS has found widespread utility in two primary areas: (i) elucidation of biomolecular tertiary and quaternary structure in structural biology [8–10], and (ii) rapid characterization of complex samples on the basis of structure and mass. Recent aspects of the latter, specifically for systems-wide analyses in systems, synthetic, and chemical biology is the focus of this report.
Figure 1.
Untargeted workflows for ion mobility-mass spectrometry analysis. (a) In 2-dimensional IM-MS analysis, biochemical classes partition in predicable regions, while outside of these regions contain structurally unique molecules such as conjugates of multiple classes. The right panel illustrates the capability for obtaining finer structural detail within the IM-MS measurement illustrated for lipids. (b) In one example of a discovery-driven IM-MS workflow, complimentary samples are subjected to LC-IM-MS analysis and molecular features representing scalar values of retention time, mass, collision cross section, and signal intensity are subsequently extracted from the 3-dimensional datasets. These tabulated features are subjected to one of several unsupervised statistical methods which seeks to reduce the dimensionality of the dataset in order to identify the most significant molecular features. Shown are two such methods: (1) clustering maps of self-organized data which groups related features based on correlations across individual scalar components, and (2) multivariate statistical analysis which reduces a highly-dimensional dataset into a binary comparison based on the two most descriptive components of the data. (c) From these statistical methods, the most descriptive molecular features are highlighted, and these features can be targeted for identification. Molecular identification proceeds by putative matching based on exact mass measurement which are then validated through other orthogonal pieces of information, such as retention time, cross section, and mass fragmentation data.
Data dimensionality and information content
There exist a multitude of arrangements for performing IM, many of which parallel strategies for MS mass-to-charge selectivity. In the context of structural biology and untargeted analyses using structure and mass correlations, IM-MS is commonly accepted to correspond to time-dispersive IM coupled with time-of-flight MS. Although a variety of implementations of this combination have been described since the 1960s, in recent years, the commercialization of IM-MS platforms based-on electrodynamic IM fields [11], and electrostatic IM fields [12] has fostered a considerable increase in IM-MS related publications and, congruently, advancement and expansion in IM-MS applications. This is especially true for applications centered on the analysis of complex biological samples.
One of the primary reasons time-dispersive IM-MS has been widely adopted is because the drift time across the ion mobility cell, analogous to LC retention time, can predictably be correlated to an observed collision cross section (Ω, Å2), which is a rotationally averaged apparent surface area of the ion. This is achieved through ion-neutral collisions with an inert background buffer gas as ions traverse a drift region under the influence of the defined electric field. Conformationally diffuse molecules experience a larger number of collisions relative to a conformationally dense molecule of the same mass, which results in a longer time spent in the mobility drift cell. Importantly for untargeted analyses, biomolecular classes distribute into unique regions of IM-MS separations space, or conformational space (Figure 1A). These mobility-mass correlations emerge as a result of the polymeric properties of biomolecules (e.g., amino acids comprising peptides, monosaccharides forming glycans) and the prevailing intramolecular and intermolecular forces for each biomolecular class [6,7]. These correlations have been expanded to finer-grain analysis within biomolecular classes, and for predictive purposes [12,13]. For example, Figure 1A (right) is an expanded region centered on lipid species by selection of the collision cross section and m/z region highlighted. The fine structure shows that for a large cohort of sphingolipids and glycerophospholipids that these two major classes of lipids separate into distinct regions within the coarse lipid region. Much recent attention has focused on the fine structure information that can be obtained for a wide variety of molecular classes, including those for peptides/proteins [14–16], lipids [17–19], and carbohydrates [20,21]. A sense of reproducibility for these collision cross section measurements for a recent interlaboratory study suggest that for 125 metabolite species, the precision of the collision cross sections measured to be better than 5% for the relative standard deviation [22].
There are many recent directions being pursued to advance the structural measurements afforded by IM-MS. These include efforts to improve the instrumental figures of merit such as enhanced IM resolution [23], modular IM components for tailorable IM-MS platforms [24], and interfacing ion activation methods such as surface induced dissociation (SID) complementary to conventional CID [25]. Additional attractive strategies are actively pursued to modify the IM separation itself to provide additional structural characterization and quantification capabilities including the use of alternate IM drift gases and/or the effects of solvation on structure [26,27], energy resolved separations [28], and isotopic labeling strategies in the IM-MS [29], among others.
Unraveling untargeted data to targeted identifications in systems-wide analyses
One of the key advantages of IM-MS is the throughput of the analyses. The timescales of separation are uniquely suited for integrating LC (min separations) with IM (ms separations) with MS (us separations). However, in untargeted strategies, this results in a deluge of data. A single LC run of 10s of minutes easily results in the generation of >104 IM separations with >106 corresponding MS spectra. Thus, strategies such as that depicted in Figure 1B have been developed, whereby multidimensional feature extraction can be performed [30,31], followed by one of several strategies for self-organization of the data, with the goal of the latter to project high-dimensional data in a visually interpretive scheme in order to highlight the molecules of interest which are then targeted for identification and validation [32–35]. Such approaches have been demonstrated in a wide array of emerging applications ranging from systems diagnosis of drug addiction [32], to wound healing [36], to cancer [37,38], to drug discovery efforts [39–41]. Based on the success of these untargeted IM-MS approaches, new frontiers in synthetic biology using 3D organotypic cell culture to emulate human constructs on a chip based format have facilitated moving the boundries of molecular breadth and throughtput required for rapid analysis in these human-on-a-chip constructs [42–44].
Concluding remarks
The molecular breadth and throughtput of IM-MS separations provides a means for integrating untargeted omics measurements without sample pretreatment to isolate classes of molecules of interest. Importantly, these analytical advances permit rapid and unbiased characterization of extremely complex samples, which is opening new avenues of inquiry in biology using systems-wide analyses.
Highlights.
Ion mobility-mass spectrometry for untargeted omics studies.
Coarse and fine-grained IM-MS correlations reveal molecular class and sub-class.
Molecular breadth is enhanced through integrated omics strategies.
Bioinformatic strategies refine untargeted data for identification and validation.
Acknowledgments
This work is supported by the National Institutes of Health National Center for Advancing Translational Sciences (NIH-NCATS UH2TR000491); the National Science Foundation Major Research Instrumentation program (NSF/MRI CHE-1229341); the Vanderbilt Institute of Chemical Biology; the Vanderbilt Institute for Integrative Biosystems Research and Education; and the Vanderbilt College of Arts and Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature. 2012;491:384–392. doi: 10.1038/nature11708. [DOI] [PubMed] [Google Scholar]
- 2.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, Ebbels T, De Iorio M, Brown IJ, Veselkov KA, Daviglus ML, Kesteloot H, Ueshima H, Zhao L, Nicholson JK, Elliott P. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 4.Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr Opin Biotech. 2015;31:73–78. doi: 10.1016/j.copbio.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenn LS, McLean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 7.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder J, Heck AJ. Analytical approaches for size and mass analysis of large protein assemblies. Annu Rev Anal Chem. 2014;7:43–64. doi: 10.1146/annurev-anchem-071213-020015. [DOI] [PubMed] [Google Scholar]
- 9.Lanucara F, Holman SW, Gray CJ, Eyers CE. The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat Chem. 2014;6:281–294. doi: 10.1038/nchem.1889. [DOI] [PubMed] [Google Scholar]
- 10.Zhong Y, Hyung S-J, Ruotolo BT. Ion mobility-mass spectrometry for structural proteomics. Expert Rev Proteomics. 2012;9(1):47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 12.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA. Conformational ordering of biomolecules in the gas-phase: nitrogen collision cross-sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86:2107–2116. doi: 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLean JA. The mass-mobility correlation redux: the conformational landscape of anhydrous biomolecules. J Am Soc Mass Spectrom. 2009;20:1775–1781. doi: 10.1016/j.jasms.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Bush MF, Campuzano IDG, Robinson CV. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem. 2012;84:7124–7130. doi: 10.1021/ac3014498. [DOI] [PubMed] [Google Scholar]
- 15.Shliaha PV, Bond NJ, Gatto L, Lilley KS. Effects of traveling wave ion mobility separation on data Independent acquisition in proteomics studies. J Proteome Res. 2013;12:2323–2339. doi: 10.1021/pr300775k. [DOI] [PubMed] [Google Scholar]
- 16.Jia C, Lietz CB, Yu Q, Li L. Site-specific characterization of (D)-amino acid containing peptide epimers by ion mobility spectrometry. Anal Chem. 2014;86:2972–2981. doi: 10.1021/ac4033824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kliman M, May JC, McLean JA. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim Biophys Acta. 2011;1811:935–945. doi: 10.1016/j.bbalip.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castro-Perez J, Roddy TP, Nibbering NM, Shah V, McLaren DG, Previs S, Attygalle AB, Herath K, Chen Z, Wang SP, Mitnaul L, Hubbard BK, Vreeken RJ, Johns DG, Hankemeier T. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J Am Soc Mass Spectrom. 2011;22:1552–1567. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenk MR. Lipidomics: new tools and applications. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 20•.Both P, Green AP, Gray CJ, Sardzík R, Voglmeir J, Fontana C, Austeri M, Rejzek M, Richardson D, Field RA, Widmalm G, Flitsch SL, Eyers CE. Discrimination of epimeric glycans and glycopeptides using IM-MS and its potential for carbohydrate sequencing. Nat Chem. 2014;6:65–74. doi: 10.1038/nchem.1817. The utility of IM-MS to distinguish carbohydrate epimers in fine structural analyses. [DOI] [PubMed] [Google Scholar]
- 21•.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, Scanlan CN, Scarff CA, Thalassinos K, Scrivens JH. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. J Am Soc Mass Spectrom. 2010;22:568–581. doi: 10.1007/s13361-010-0053-0. Use of conformation space and selectivity from chemical noise to characterize glycans from microgram quantities of expressed glycoprotein. [DOI] [PubMed] [Google Scholar]
- 22.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldórsson S, Rolfsson O, Moseley A, Grant D, Langridge J, Palsson BO, Astarita G. Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem. 2014;86:3985–3993. doi: 10.1021/ac500405x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zucker SM, Ewing MA, Clemmer DE. Gridless overtone mobility spectrometry. Anal Chem. 2013;85:10174–10179. doi: 10.1021/ac401568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Webb IK, Garimella SVB, Tolmachev AV, Chen TC, Zhang X, Cox JT, Norheim RV, Prost SA, LaMarche B, Anderson GA, Ibrahim YM, Smith RD. Mobility-resolved ion selection in uniform drift field ion mobility spectrometry/mass spectrometry: dynamic switching in structures for lossless ion manipulations. Anal Chem. 2014;86:9632–9637. doi: 10.1021/ac502139e. This manuscript describes the development of modular units comprised of printed circuit board components for performing time selective ion mobility integrated with mass spectrometry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou M, Wysocki VH. Surface induced dissociation: dissecting noncovalent protein complexes in the gas phase. Acc Chem Res. 2014;47:1010–1018. doi: 10.1021/ar400223t. [DOI] [PubMed] [Google Scholar]
- 26.Jurneczko E, Kalapothakis J, Campuzano ID, Morris M, Barran PE. Effects of drift gas on collision cross sections of a protein standard in linear drift tube and traveling wave ion mobility mass spectrometry. Anal Chem. 2012;84:8524–8531. doi: 10.1021/ac301260d. [DOI] [PubMed] [Google Scholar]
- 27.Silveira JA, Fort KL, Kim D, Servage KA, Pierson NA, Clemmer DE, Russell DH. From solution to the gas phase: stepwise dehydration and kinetic trapping of substance P reveals the origin of peptide conformations. J Am Chem Soc. 2013;135:19147–19153. doi: 10.1021/ja4114193. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann W, Hofmann J, Pagel K. Energy-resolved ion mobility-mass spectrometry – a concept to improve the separation of isomeric carbohydrates. J Am Soc Mass Spectrom. 2014;25:471–479. doi: 10.1007/s13361-013-0780-0. [DOI] [PubMed] [Google Scholar]
- 29.Sturm RM, Lietz CB, Li L. Improved isobaric tandem mass tag quantification by ion mobility mass spectrometry. Rapid Commun Mass Spectrom. 2014;28:1051–1060. doi: 10.1002/rcm.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crowell KL, Slysz GW, Baker ES, LaMarche BL, Monroe ME, Ibrahim YM, Payne SH, Anderson GA, Smith RD. LC-IMS-MS feature finder: detecting multidimensional liquid chromatography, ion mobility and mass spectrometry features in complex datasets. Bioinformatics. 2013;29(1):2804–2805. doi: 10.1093/bioinformatics/btt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivalingam GN, Yan J, Sahota H, Thalassinos K. Amphitrite: A program for processing travelling wave ion mobility mass spectrometry data. Int J Mass Spectrom. 2013;345–347:54–62. doi: 10.1016/j.ijms.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Goodwin CR, Sherrod SD, Marasco CC, Bachmann BO, Schramm-Sapyta N, Wikswo JP, McLean JA. Phenotypic mapping of metabolic profiles using self-organizing maps of high-dimensional mass spectrometry data. Anal Chem. 2014;86:6563–6571. doi: 10.1021/ac5010794. A workflow for untargeted to targeted analysis for phenotypic mapping of metabolomics profiles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendall SC, Nolan GP. From single cells to deep phenotypes in cancer. Nat Biotech. 2012;30:639–647. doi: 10.1038/nbt.2283. [DOI] [PubMed] [Google Scholar]
- 34.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotech. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, Gonzalez FJ, Idle JR. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hines KM, Ashfaq S, Davidson JM, Opalenik SR, Wikswo JP, McLean JA. Biomolecular signatures of diabetic wound healing by structural mass spectrometry. Anal Chem. 2013;85:3651–3659. doi: 10.1021/ac303594m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, Orton DJ, Piehowski PD, Purdy DE, Moore RJ, Danielson WF, 3rd, Monroe ME, Crowell KL, Slysz GW, Gritsenko MA, Sandoval JD, Lamarche BL, Matzke MM, Webb-Robertson BJ, Simons BC, McMahon BJ, Bhattacharya R, Perkins JD, Carithers RL, Jr, Strom S, Self SG, Katze MG, Anderson GA, Smith RD. Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol Cell Proteomics. 2014;13:1119–1127. doi: 10.1074/mcp.M113.034595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hines KM, Ballard BR, Marshall DR, McLean JA. Structural mass spectrometry of tissue extracts to distinguish cancerous and non-cancerous breast diseases. Mol Biosyst. 2014;10:2827–2837. doi: 10.1039/c4mb00250d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodwin CR, Fenn LS, Derewacz DK, Bachmann BO, McLean JA. Structural mass spectrometry: rapid methods for separation and analysis of peptide natural products. J Nat Prod. 2012;75:48–53. doi: 10.1021/np200457r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esquenazi E, Daly M, Bahrainwala T, Gerwick WH, Dorrestein PC. Ion mobility mass spectrometry enables the efficient detection and identification of halogenated natural products from cyanobacteria with minimal sample preparation. Bioorg Med Chem. 2011;19:6639–6644. doi: 10.1016/j.bmc.2011.06.081. [DOI] [PubMed] [Google Scholar]
- 41.Derewacz DK, Goodwin CR, McNees CR, McLean JA, Bachmann BO. Antimicrobial drug resistance affects broad changes in metabolomic phenotype in addition to secondary metabolism. Proc Natl Acad Sci USA. 2013;110:2336–2341. doi: 10.1073/pnas.1218524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alcendor DJ, Block FE, Cliffel DE, Daniels JS, Ellacott KLJ, Goodwin CR, Hofmeister LH, Li D, Markov DA, May JC, McCawley LJ, McLaughlin BA, McLean JA, Niswender KD, Pensabene V, Seale KT, Sherrod SD, Sung HJ, Tabb DL, Webb DJ, Wikswo JP. Neurovascular unit on a chip: implications for translational applications. Stem Cell Res Ther. 2013;4(S18):1–5. doi: 10.1186/scrt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi M, Majumdar D, Gao Y, Brewer BM, Goodwin CR, McLean JA, Li D, Webb DJ. Glia co-culture with neurons in microfluidic platforms promotes the formation and stabilization of synaptic contacts. Lab on a Chip. 2013;13:3008–3021. doi: 10.1039/c3lc50249j. [DOI] [PMC free article] [PubMed] [Google Scholar]