Abstract
Since the development of desorption electrospray ionization (DESI), many other ionization methods for ambient and atmospheric pressure mass spectrometry have been developed. Ambient ionization mass spectrometry has now been used for a wide variety of biological applications, including plant science, microbiology, neuroscience, and cancer pathology. Multimodal integration of atmospheric ionization sources with the other biotechnologies, as well as high performance computational methods for mass spectrometry data processing is one of the major emerging area's for ambient mass spectrometry. In this opinion article, we will highlight some of the most influential technological advances of ambient mass spectrometry in recent years and their applications to the life sciences.
Ambient Ionization Mass Spectrometry
In the late 20th century, acquiring label-free chemical information from the biological systems has became a reality due to the development of biomolecule-compatible ionization methods for mass spectrometry (MS), e.g. electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) [1,2]. The mass spectral data, when acquired in a spatial manner, can be intuitively reconstructed as and image and this has resulted in the booming filed of imaging mass spectrometry (IMS). A large number of ionization methods has been developed over the past decade that combine IMS with biological systems, demonstrating its profound impact on life sciences [3].
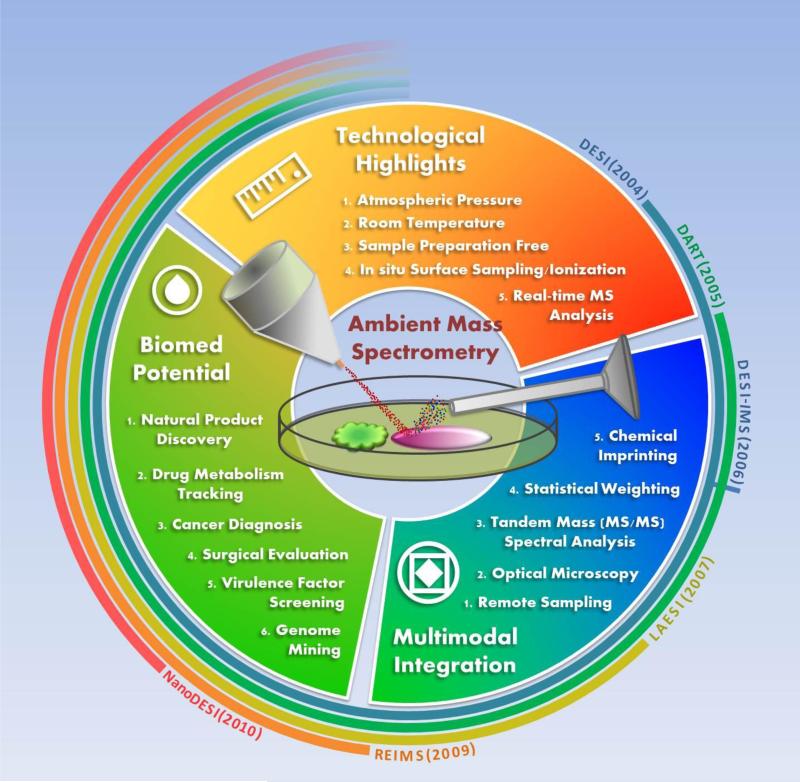
One can say that the technological development of ambient ionization methods has entered the golden age. The term “ambient mass spectrometry” was first introduced in 2006 [4]. In the past decade, the analytical science as well as biological and medical science communities have, extensively utilized ambient mass spectrometry, or ambient ionization mass spectrometry. Each ambient ionization MS technique follows three main principles: (1) ions are generation and maintenance under atmospheric pressure and room temperature (for most of the techniques) before introducing the ions into a mass spectrometer for analysis; (2) samples do not require any pretreatment or only require minimal preparation; (3) analytes are directly desorbed/ionized from the sample surface such that instantaneous or real-time MS measurements becomes possible (Figure 1).
Figure 1.
The cycle of major ambient mass spectrometry methods discussed in this opinion article. Center: Schematic diagram of desorption electrospray ionization (DESI) source obtaining chemical information from a biological sample surface. A stream of charged micro-droplets (in red) is sprayed onto the sample surface to desorb and ionize compounds (in multiple colors), during which molecules retain in their structural intact states. Outer strips: Time frame of the major development in ambient ionization mass spectrometry methods since DESI MS was reported in 2004. The year of the first publication using the indicated ion sources are listed in the parentheses.
Due to the simplicity and insightful molecular information it can provide, there has been a growing interest in ambient mass spectrometry to be used as new tools for exploration of the chemical dimensions such as searching for new drug leads and signatures of disease. This opinion article is not aimed to be a comprehensive review of ambient ionization methods rather it is aimed to highlight some of the recent tools and how we see they fit into major areas of the life science communities. In the first part of the article we highlight extractive-based ionization methods, including desorption electrospray ionization (DESI) and nanospray desorption electrospray ionization (nanoDESI). Further we highlight representative examples of applications that indicate they have high biomedical potential. We also describe recent technological advancements that strongly indicate one of the future directions of ambient mass spectrometry is towards integration with non-MS based multi-modal interfaces to improve the understanding of the true complexity of biology.
DESI
The Basics
The introduction of desorption electrospray ionization (DESI) in 2004 is a landmark development in the history of ambient ionization mass spectrometry [5]. DESI utilizes a nebulized electrospray of highly charged micro-droplets to pick up the analytes directly from the sample surface. The subsequent secondary droplets carry the dissolved analytes to the mass spectrometry without separation procedures (Figure 1). Similar to conventional ESI of bulk solution, gaseous ions are believed to be created by electrostatic explosion and evaporation [5].
Application on Tissues
DESI MS analysis of biological tissues has been used for direct chemical profiling since 2005, when the first ambient mass spectrometry analysis to animal tissue sections without any sample pretreatment was performed [6]. Strong ion signals from phospholipids were found at the mass-to-charge (m/z) range 700-900 from pancreas and brain tissues. The alteration of phospholipid profiles of tumor tissue was also first investigated and described in this paper [6]. The same group also applied DESI MS to the native and freshly cut plant tissue in the same year [7]. Dozens of alkaloids, coniine for example, were identified on different species. In 2006, the first whole-section surface IMS was demonstrated on rat brain tissue [8] showing spatial distributions of different lipid species that reflected the chemical-histological heterogeneity of a brain tissue. Automatic 2-dimensional (2-D) stages are employed with DESI ion sources to generate 2-D MS images [9]. Lateral resolution of a DESI IMS is usually around a few hundred microns, while ~40-μm spatial resolution has been reported [10]. Three-dimensional (3-D) DESI IMS could be constructed using 2-D overlays of serial sections [11].
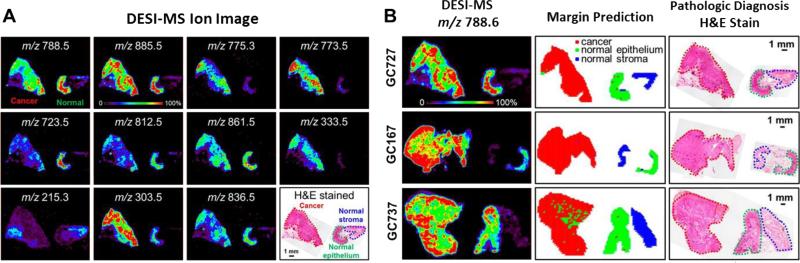
In general the most abundant and ionizable molecules, such as natural products from microbes or lipids present in most biological tissues are commonly detected when performing ambient mass spectrometry. Cell membranes are comprised of diverse lipids with different structures and functionalities that play important roles to cellular processes. DESI MS has been widely applied for the characterization and imaging of different classes of endogenous compounds especially lipids including fatty acid, phospholipids, sphingolipids, and sterol lipids in animal tissues [12-22]. Lipidomic profiling has become a mechanism to interrogate disease states in cancer cells [12-19]. Such molecular changes are readily detected by DESI IMS, featuring indicative disease states on morphological regions of tissues. Statistical and computational approaches for IMS-based cancer diagnosis have started to be implemented in medical investigations [13-20]. Moreover, the altered lipid metabolisms associated with cancer-causing genes can also be captured from DESI MS analysis [12.13]. The chemical disparity in lipid species among tumor and healthy tissues provides a fast and accurate marginal evaluation that could be used to improve the success rate of surgical resection (Figure 2) [16-20]. The majority of disease signature studies report on the shift in lipid metabolism or alteration of small metabolites. However, detection of large biomolecules on tissue sections using DESI-IMS, such as proteins and glycans, to date, have not yet been reported to be disease signatures. In addition to the endogenous compounds, DESI IMS can also be used for label-free tracing of drug and metabolites on histological sections [23-25].
Figure 2.
Representative DESI IMS of cancer tissue sections. (A) DESI IMS of gastric cancer tissue section showing relative ion abundances specifically observed in the regions of cancer or normal epithelial and stromal tissues. Molecular (ion) abundances are usually displayed in false colors. (B) Marginal evaluations (middle panel) of cancerous cells are made by statistical measurements using molecular data (identified as glycerophosphoserine) acquired by DESI-MS (left panel). Results of the computational assessment are compared with cancer diagnosis made by pathologists shown in optical images of H&E stained tissue sections (right panel). Figures are reproduced by permission [17].
Because the DESI IMS platform is operated in continuous scanning mode, smooth flat tissue sections are required for high-quality DESI IMS. Although DESI IMS has found uses in the molecular analysis of animal tissue sections, direct imaging of soft or irregular surfaces such as leaves, flowers, vegetable tissues, and microbial colonies remains challenging. To overcome this obstacle, indirect DESI IMS has to be made via imprinting techniques, where compounds of interests are transferred onto flat surfaces [26-28]. DESI IMS on microbial colonies from a Petri dish is not straightforward as the backpressure of the solvent stream in DESI damages the agar. Imprinting overcomes this challenge but there are alternative methods out there to get a molecular image of a microbial colony [26]. Imprinting of compounds from plant tissues and onto hard surface, e. g., porous Teflon, prior to DESI MS analysis is seeing wider utility for imaging with DESI [27-31].
Unlike what is encountered with animal tissues, microorganisms and plants produce a large variety of specialized metabolites. These are metabolites that are produced to carry out a specific functions such as growth simulation, biofilm formation, defense, cell differentiation and many other specific functions, as opposed to be involved in metabolism and storage of energy. Specialized metabolites also include molecules such as natural products, quorum sensors and virulence factors. Many microbe- and plant-derived natural products are of great medical value. Traditionally, detection and characterization of these specialized metabolites is very time-consuming as it involved laborious liquid extraction and isolation procedures. Ambient MS provides an alternative tool for a rapid exploration of these specialized metabolites [26-32]. The enormous structural diversity among the small molecules present in microorganisms and plants gives rise to the utilization of DESI MS analysis and imaging on a diverse array of plant metabolites. DESI MS of specialized metabolites in plant tissue is perhaps the second most common analysis performed with DESI, after animal or human tissues. For example, chemical cues, such as enzymatic conversion of glycosylated metabolites, were obtained at distinct phenotypes under environmental stresses [30-31], and DESI has been used to monitor environmental toxins that accumulate in plants, such as arsenic compounds in fern leaves [32].
In summary, DESI has an excellent ability to IMS of thin section of animal/human tissues, and we expect similar ease of use with many other thin sections, however further improvements are needed for non-even samples such as microbial colonies. Molecular information especially lipids and some abundant metabolites can be readily obtained by DESI MS, however analysis of large molecules have not been reported in diagnostic applications. Perhaps other methods that have improved analyte/ion transmission after desorption are needed to enable routine diagnostic analysis beyond lipids signatures.
NanoDESI
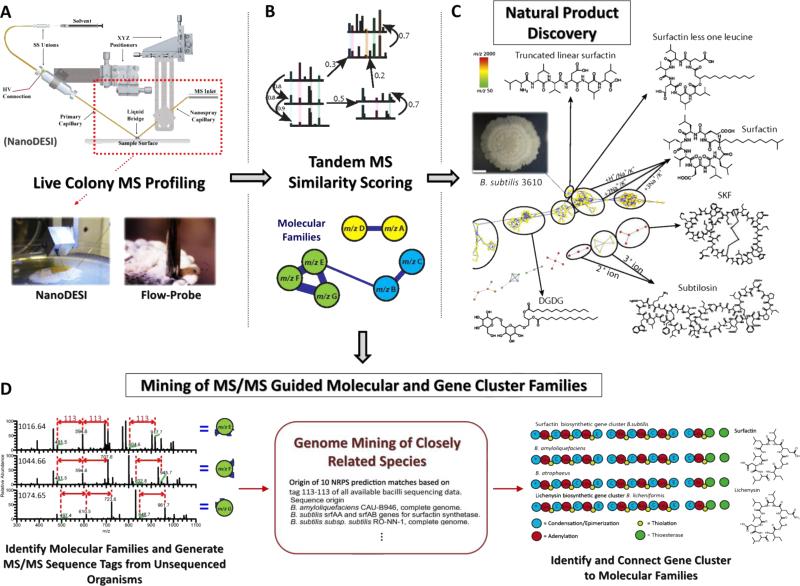
Another solution-phase-mobilized surface desorption/ionization approach that has made significant advances is nanospray desorption electrospray ionization (nanoDESI) [33]. The concept of nanoDESI is derived from DESI and can be considered a complementary of DESI. Any DESI MS platform can readily and cost-effectively be modified for use of a nanoDESI source. The nanoDESI source replaces the primary stream of micro-droplets of DESI with a micrometer-size fused silica capillary tube that is continuously infused by highly charged solvents. The second capillary tube is used to aspirate the analytes from the tissue surface via the micro-extraction from a tiny liquid junction sustained on the tissue surface (Figure 3A). The subsequent nanoelectrospray ionization occurs at the terminal end of the secondary capillary tube by which the concurrent ions of analytes as well as carrier solvent are pulled into the mass spectrometer.
Figure 3.
Overall real-time microbial metabolomics and peptidogenomics workflow basing on tandem mass spectral analysis. Molecular information is directly fetched from the surface of microbial colonies using nanoDESI [40] or continuous flow-probe [47] as depicted in step A. The massive MS/MS (molecular fragments) data are then grouped into multiple subsets (called “molecular families”) basing on the fragmentation patterns. Each node represents an individual ion and connects to each other showing the spectral similarity between each pair (B). The MS/MS-based computational pipelines have been extensively applied to high-throughput mining of microbe-derived natural products for an instantaneous insight into the molecular classes and structural elucidation (C). The pipelines can further be utilized to connect nonribosomal peptide synthetases (NRPSs) gene clusters to the molecular families (D).
The nanoDESI source was soon coupled with IMS scanning platform for a gentle surface lipidomic mapping on rat brain tissue [34,35]. Because the splash of secondary solvent micro-droplets in DESI is substituted with a self-aspirated capillary tube as an analyte carrier, the virtual sampling spot-size of nanoDESI is greatly reduced. A spatial resolution of ~10 μm was achieved using nanoDESI IMS [34], near the single human cell resolution. This resolution is compatible to the other laser-induced or -associated desorption ionization methods such as commercial MALDI sources. Similar to DESI MS, nanoDESI MS has the capability to desorb lipids including phospholipids and steroids [34-36]. In general, the MS sensitivity of nanoDESI is greater than DESI, as the splash of secondary microdroplets in DESI is substituted with a capillary tube analyte carrier in nanoDESI that results in better ion transfer efficiency. Although the efficiency of transfer is greater the probe and the liquid droplet at the interface always have to be in contact.
Moreover, distributions of small metabolites involved in energy metabolism and cell proliferation associated with pregnancy were recently revealed by nanoDESI IMS on uterine sections [37]. Bearing the ability to characterize compounds with smaller molecular weights, nanoDESI MS can act as the complementary tool with other mass spectrometry methodologies such as MALDI IMS for the characterization of key metabolites and metabolic exchanges in complex host-microbial systems [38]. Using germ-free mice guts as the control model, the interaction between gastrointestinal track and gut microbiota, as well as the chemical transformations yielded by the microbiota were revealed [38]. Similar to DESI IMS, it can also be applied to detect drugs and track drug metabolisms on a complex organ system, e.g. nicotine in rat brain tissue [39].
Among all its applications, as the technique does not have the destructive backpressure of DESI, nanoDESI MS has broad utility for the analysis and discovery of natural products from microbial colonies grown on Petri dishes [40,41]. The specialized metabolites from microbial communities have been a rich source of useful natural products including antibiotics, antifungals, anticancer agents, and immunosuppressants. However, more than 99% of the microorganisms on this planet have not yet been examined for their chemical outputs. In most cases, tedious procedures, including liquid-phase extraction and isolation of analytes using liquid chromatography, are required prior to MS analysis. These laborious steps become unrealistic costly when aiming for the analysis of a large set of microbial species. An ability that allows instantaneous MS analysis of microbial natural products bypassing the time-consuming preparations is therefore of significance in pharmaceutical science.
One of the advantages the nanoDESI source, is that there is only slight damage to the sample that is caused during its sampling process. The bombardment on the sample surface initiated by charged micro-droplet in DESI is detrimental to microbial colonies and inevitably gives rise to breakdown of colony morphology and delocalization of analytes. The imprinting of compounds to a rigid substrate is essential for a high quality DESI IMS for microorganisms [26]. However, this indirect approach is challenging for the real-time MS analysis on agar media. Using nanoDESI MS, we successfully obtained spatiotemporal MS profiles of living microorganisms directly from Petri dishes (Figure 3A), which provided chemical information of microbe-derived natural products [40].
Another challenge for natural products is the expensive costs and personnel time that are required to identify and elucidate microbial metabolites. The work also demonstrated that by comparing the tandem mass (MS/MS) spectral similarity (Figure 3B), which is herein referred to as “molecular networking”, massive amount of secondary metabolites secreted from microbes as well as the metabolic exchanges between microbial communities are revealed and rapidly identified (Figure 3C) [40]. This nanoDESI-MS/MS networking platform has been applied to the metabolomics analysis of a large variety of microbial species [40-44]. Furthermore, the ease of this nanoDESI MS/MS-guided approach enables a new bioinformatics paradigm of genome mining called peptidogenomics [45], in which MS/MS profiles of peptide natural products from species of closely related microorganisms (identified as “molecular families”) are rapidly screened on living colonies, and relate to their gene cluster families (Figure 3D). This genome-mining method, that connects MS/MS spectral patterns of microbial peptide families to their biosynthetic gene cluster families, has been conceptually carried out directly on the top of living microbial colonies of 60 related strains [42]. Comprehensive studies for a larger scale of species using similar strategy are expected and may once and for all provide an answer of how many natural product classes are there for specific classes of microbes.
IMS using nanoDESI for microbial colonies or biofilms is complicated as the surfaces of colonies are usually rugged and display contours that can span millimeters. It is necessary for the use of z-axis adjustment to the probe position in order to follow the contour of the colony, so as to prevent clogging and loss of signal due to collision with or contact lose from the sample surface [44,46]. To compensate the vertical tolerance between the tip of the probe and sample surfaces, nanoDESI IMS requires larger droplets. This change results in poorer spatial resolution (usually ~ few-hundred μm) compared with the imaging on flat tissue sections [44]. At this point, full IMS of the microbial colonies by nanoDESI sources is still challenging, especially for species that can form biofilms with excessive extra-cellular matrix. Measuring natural products from microbial colonies at their intact states are challenging. Colonies can be imprinted onto a substrate for DESI MS, but the native morphological features are missing [26]. However clogging of the transfer capillary tubes due to surface roughness is common. In this regard, a coaxial geometry liquid micro-junction surface sampling probe (LMJSSP) was implemented for direct micro-extraction of microbe-derived natural products from colony surfaces for subsequent MS analysis. A real-time sampling was achieved pneumatically using a concentric nebulizer to transfer the neutral analytes to generate electrospray [47].
Multimodal Techniques Incorporating with Ambient Mass Spectrometry
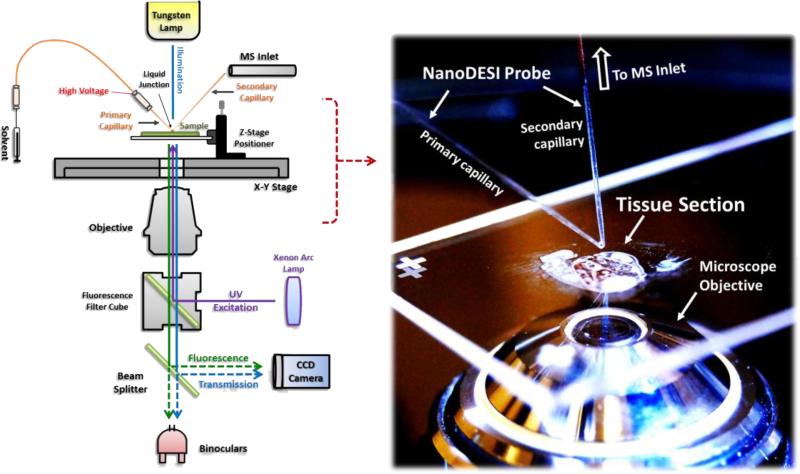
The unique nature of ambient mass spectrometry enables the interface with other imaging modalities. The use of the current MS-based multimodal tools allows for better visualization, portability, and flexibility. In the following section we will highlight some of newly developed techniques of the most recent years that are coupled with ambient ionization mass spectrometry. In 2013 our lab interfaced an inverted light microscope with nanoDESI MS into a single platform for a better visualization of samples and improved targeted MS analysis (Figure 4) [48]. Using this integrated platform, the area of interests on the biological sample surface can be targeted by the light microscope and the subsequent mass spectrometric analysis can be made in situ by nanoDESI. Amazingly, a broad range of endogenous molecules ranging from lipids to intact proteins associated with embryonic development of central nervous system could be ionized. Via the positioning over the microscope, a pixel-by-pixel data acquisition is achieved for the construction of a spatial map akin to IMS. It can be expected that over the next few years that many other atmospheric ionization strategies can be interfaced with other imaging modalities.
Figure 4.
Schematic diagram of microscopy ambient mass spectrometry combining a nanoDESI source with an inverted fluorescence microscope (left). The photo on the right shows a real-time snapshot of the microscope-nanoDESI platform about to make MS measurement from a mouse embryo section mounted on a regular microscope slide [48].
One of the most creative interfaces of ambient ionization mass spectrometry is the interface of thermal desorption with atomic force microscopy (AFM) [49]. The platform is first operated at regular AFM mode to obtain topographical images with spatial resolution at sub-micrometer level. The same AFM proximal probe is then heated to ~350 °C to enable thermal desorption of the analytes on the sample surface, which is further ionized by a supplementary atmospheric pressure chemical ionization (APCI) source to give MS data. A MS image of the sample surface is made and co-registered with the topographical contour. However, its MS sensitivity and the subsequent ability to identify compounds are not comparable with other ambient ionization method. Although it might open up a new avenue by showing a high spatial-resolution MS profile, its capability for MS analysis on biological system remains unconvincing [47]. However we expect, as it has the highest spatial resolution among all atmospheric ionization technique to date, that as the technique improves that one can demonstrate, convincingly, that this method can indeed be employed to investigate biological samples.
Remote Sampling Using Long Distance Ion Transfer
Although the majority of mass spectrometry analysis is performed on thin sections or imprints, biology is not 2-D, rather it is 3-D. Ambient ionization methods that allow MS analysis from remote sampling can be used for studies on living entities, including human subjects. However, real-time MS measurement on large or uneven objects is difficult because a high-efficient long distance travelling of ions is difficult. The idea of distant MS sampling at ambient environment has been made possible using air flow carrier that enables a high efficient transfer of ions [50-54]. For example, a regular DESI source was adapted with a rigid 50-centimeter air flow-assisted ion transport tube for a whole-body section IMS of an adult rat [50]. Subsequently, an extended DESI source coupled with a 4-meter ion transmission Tygon tubing has been demonstrated for the distant ionization and MS analysis of polar lipids from a rat brain tissue [51]. The flexibility of the design and adequate ion transmission efficiency are promising for in vivo MS analysis compatible with surgical and endoscopic procedures.
This led to the development of rapid evaporative ionization mass spectrometry (REIMS) and its use in the surgical arena [52]. REIMS technology utilizes an electrosurgical dissector to generate aerosols containing gaseous ion from the surgical site. The thermoelectrically generated ions are transmitted to distant mass spectrometer via a flexible polymer transfer tubing, which is connecting to a Venturi gas jet pump to pull the ions pneumatically [53]. The coupling of REIMS with an electrosurgery for human patients using multivariate analysis is later known as intelligent knife (iKnife). Its capability of distant ionization/sampling is especially favored when cryosection is impossible. Using iKnife, real-time intraoperative MS characterization of cancerous tissues during surgical interventions in the operating theater is possible (Figure 5). It can determine the tumor margins for surgeons so as to improve the accuracy of surgical intervention in cancer treatments, and ideally could minimize the costly waiting time during intraoperative histological examination [53]. Excitingly, REIMS technology was also used to acquire MS profiles of microbial biomass. Microorganisms among different genus and species were categorized based on their mass spectral fingerprints using REIMS [54]. Commercial applications of these tools with associated signature databases are expected to emerge in the future and may become the standard clinical equipment of the next generation operating theaters.
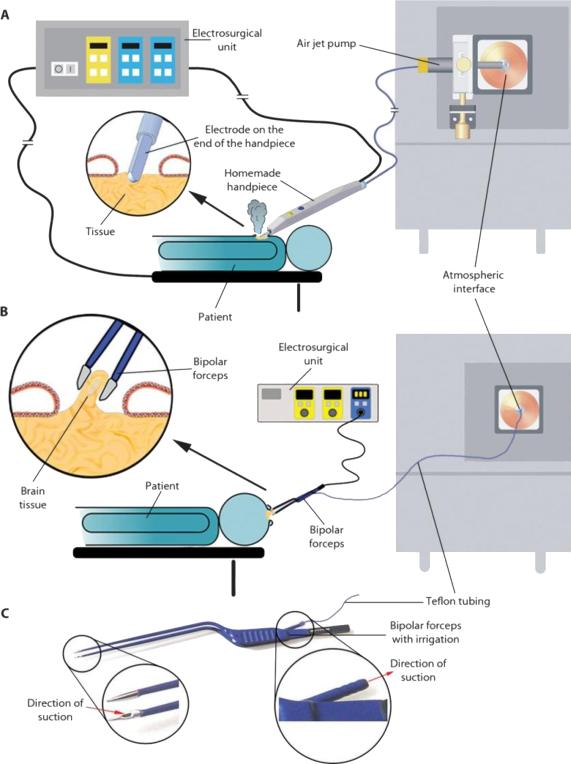
Figure 5.
Schematic diagram of REIMS-iKnife technology for real-time data collection in the operating theater using monopolar (A) or dipolar (B) electrosurgery. Evaporative products in REIMS experiment directly from patient tissues are remotely transferred via Teflon tubing into mass spectrometer. The real-time MS patterns provide surgeons an instantaneous indicator to determine tumor margins. Bipolar forceps (C) are commercially available for electrosurgical practices and are used as a REIMS ion source to generate evaporative aerosols. The figure is adapted by permission [53].
Perspective and Concluding Remarks
It is obvious ambient ionization has reached a Golden age. There is so much exploring to do in this area to really understand the utility of the tools that are being developed. The exploration is both in applications of existing tools as well as new ion source developments. Despite many ambient ionization methods in existence [55-57], we still anticipate three major directions of development in ambient mass spectrometry for life sciences applications in the next decade. First, invention of new ionization methods will continue, especially those aiming at sensitive ionization of large biomolecules and that push the boundaries of spatial resolution, eventually reaching the spatial resolution of light wavelengths. This is reasonable as the interface with AFM is only an order of magnitude away. Likely this development will also coincide with mass spectrometers that can routinely detect and quantify single ions. In addition many more ion sources that can be used for applications directly in the area needed will emerge as well. The iKnife is one such example. Second, integration of ambient ionization sources with other imaging modalities will evolve mass spectrometry to be more accessible for interpretation to biologists and clinicians. This is critical and will bring MS to be the routine tool to be used in the life sciences. Third, we are entering the Big Data era in mass spectrometry, ambient ionization is no exception. Over the next decade we can expect smart computational algorithms to manage and analyze large MS data sets to be developed. The data generated is increasing exponentially and we have to build the infrastructure to take on this emerging challenge. In the end, we hope that through the brief introduction we are able to highlight the ease of use and the exiting potential of ambient mass spectrometry to academic fields outside of mass spectrometry. More importantly, we hope that this perspective article will inspire the ambient mass spectrometry experts to develop the next-generation technology. Our lab is certainly looking forward to using and applying these emerging and exciting tools.
Highlights.
Ambient ionization mass spectrometry provides chemical information from small metabolites to polypeptides directly on biological samples.
Ambient ionization coupled tandem mass analysis as a mean to natural product discovery and genome mining.
Ambient mass spectrometry imaging can be used to evaluate cancer.
Remote ionization methods provide new equipment to assist surgeons.
Acknowledgements
Dorrestein lab is funded by National Institution of Health (NIH) Grants GM094802, AI095125, GM097509 and S10RR029121.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 2.Hillenkamp F, Karas M, Beavis RC, Chait BT. Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 1991;63:A1193–A1202. doi: 10.1021/ac00024a002. [DOI] [PubMed] [Google Scholar]
- 3.Watrous JD, Alexandrov T, Dorrestein PC. The evolving field of imaging mass spectrometry and its impact on future biological research. J. Mass Spectrom. 2011;46:209–222. doi: 10.1002/jms.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooks RG, Ouyang Z, Takáts Z, Wiseman JM. Ambient mass spectrometry. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- 5.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 6.Wiseman JM, Puolitaival SM, Takáts Z, Cooks RG, Caprioli RM. Mass spectrometric profiling of intact biological tissue by using desorption electrospray ionization. Angew. Chem., Int. Ed. 2005;44:7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 7.Talaty N, Takáts Z, Cooks RG. Rapid in situ detection of alkaloids in plant tissue under ambient conditions using desorption electrospray ionization. Analyst. 2005;130:1624–1633. doi: 10.1039/b511161g. [DOI] [PubMed] [Google Scholar]
- 8.Wiseman JM, Ifa DR, Song Q, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew. Chem., Int. Ed. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 9.Ifa DR, Wiseman JM, Song Q, Cooks RG. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI). Int. J. Mass Spectrom. 2007;259:8–15. [Google Scholar]
- 10.Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Improved spatial resolution in the imaging of biological tissue using desorption electrospray ionization. Anal. Bioanal. Chem. 2012;404:389–398. doi: 10.1007/s00216-012-6173-6. [DOI] [PubMed] [Google Scholar]
- 11.Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew. Chem., Int. Ed. 2010;49:873–876. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry RH, Bellovin DI, Shroff EH, Ismail AI, Zabuawala T, Felsher DW, Zare RN. Characterization of MYC-induced tumorigenesis by in situ lipid profiling. Anal. Chem. 2013;85:4259–4262. doi: 10.1021/ac400479j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberlin LS, Gabay M, Fan AC, Gouw AM, Tibshirani RJ, Felsher DW, Zare RN. Alteration of the lipid profile in lymphomas induced by MYC overexpression. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1409778111. doi:10.1073/pnas.1409778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbassi-Ghadi N, Veselkov K, Kumar S, Huang J, Jones E, Strittmatter N, Kudo H, Goldin R, Takáts Z, Hanna GB. Discrimination of lymph node metastases using desorption electrospray ionisation-mass spectrometry imaging. Chem. Commun. 2014;50:3661–3664. doi: 10.1039/c3cc48927b. [DOI] [PubMed] [Google Scholar]
- 15.Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NYR. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72:645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Eberlin LS, Norton I, Orringer D, Dunn IF, Liu X, Ide JL, Jarmusch AK, Ligon KL, Jolesz FA, Golby AJ, et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1611–1616. doi: 10.1073/pnas.1215687110. [This work provides a new multimodal cancer diagnosis prototype that combines MRI (magnetic resonance imaging) with IMS using DESI. A brain tumor subtype classifier is developed and was used to discriminate normal and malignant tissues. This is a promising tool for future tumor surgery.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Eberlin LS, Tibshirani RJ, Zhang J, Longacre TA, Berry GJ, Bingham DB, Norton JA, Zare RN, Poultsides GA. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2436–2441. doi: 10.1073/pnas.1400274111. [In this work statistical weighting and classification of cancerous tissues are performed basing on the result of DESI IMS. The statistical assessments are also compared with diagnosis from pathologists showing high accuracy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calligaris D, Norton I, Feldman DR, Ide JL, Dunn IF, Eberlin LS, Cooks RG, Jolesz FA, Golby AJ, Santagata S, et al. Mass spectrometry imaging as a tool for surgical decision-making. J. Mass Spectrom. 2013;48:1178–1187. doi: 10.1002/jms.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Veselkov KA, Mirnezami R, Strittmatter N, Goldin RD, Kinross J, Speller AVM, Abramov T, Jones EA, Darzi A, Holmes E, et al. Chemo-informatic strategy for imaging mass spectrometry-based hyperspectral profiling of lipid signatures in colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1216–1221. doi: 10.1073/pnas.1310524111. [The authors propose a workflow for tissue-specific molecular exploration on cancer using an integrated bioinformatics DESI IMS platform which covers data-processing, co-registration of multimodal images, mining of tissue-specific MS patterns, and reconstruction of chemo-pathological annotation. The work covers state-of-the-art IMS strategy over the conventional paradigm of cancer pathology.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santagata S, Eberlin LS, Norton I, Calligaris D, Feldman DR, Ide JL, Liu X, Wiley JS, Vestal ML, Ramkissoon SH, et al. Intraoperative mass spectrometry mapping of an onco-metabolite to guide brain tumor surgery. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1404724111. doi: 10.1073/pnas.1404724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Serrano AF, Pirro V, Ferreira CR, Oliveri P, Eberlin LS, Heinzmann J, Lucas-Hahn A, Niemann H, Cooks RG. Desorption electrospray ionization mass spectrometry reveals lipid metabolism of individual oocytes and embryos. PloS One. 2013;8:e74981. doi: 10.1371/journal.pone.0074981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodzon-Kulakowska A, Cichon T, Golec A, Drabik A, Ner J, Suder P. DESI–MS as a tool for direct lipid analysis in cultured cells. Cytotechnology. 2014 doi: 10.1007/s10616-014-9734-z. doi: 10.1007/s10616-014-9734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiseman JM, Ifa DR, Zhu Y, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: imaging drugs and metabolites in tissues. Proc. Natl. Acad. Sci. U.S.A. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Gingras J, Ganley KP, Vismeh R, Teffera Y, Zhao Z. Whole-body tissue distribution study of drugs in neonate mice using desorption electrospray ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. 2014;28:185–190. doi: 10.1002/rcm.6775. [DOI] [PubMed] [Google Scholar]
- 25.Eberlin LS, Mulcahy JV, Tzabazis A, Zhang J, Liu H, Logan MM, Roberts HJ, Lee GK, Yeomans DC, Du Bois J, et al. Visualizing dermal permeation of sodium channel modulators by mass spectrometric imaging. J. Am. Chem. Soc. 2014;136:6401–6405. doi: 10.1021/ja501635u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watrous J, Hendricks N, Meehan M, Dorrestein PC. Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal. Chem. 2010;82:1598–1600. doi: 10.1021/ac9027388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thunig J, Hansen SH, Janfelt C. Analysis of secondary plant metabolites by indirect desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 2011;83:3256–3259. doi: 10.1021/ac2004967. [DOI] [PubMed] [Google Scholar]
- 28.Ifa DR, Srimany A, Eberlin LS, Naik HR, Bhat V, Cooks RG, Pradeep T. Tissue imprint imaging by desorption electrospray ionization mass spectrometry. Anal. Methods. 2011;3:1910–1912. [Google Scholar]
- 29.Mueller T, Oradu S, Ifa DR, Cooks RG, Kraeutler B. Direct plant tissue analysis and imprint imaging by desorption electrospray ionization mass spectrometry. Anal. Chem. 2011;83:5754–5761. doi: 10.1021/ac201123t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Li B, Knudsen C, Hansen NK, Jorgensen K, Kannangara R, Bak S, Takos A, Rook F, Hansen SH, Moller BL, et al. Visualizing metabolite distribution and enzymatic conversion in plant tissues by desorption electrospray ionization mass spectrometry imaging. Plant J. 2013;74:1059–1071. doi: 10.1111/tpj.12183. [We highlight this “non-technical” work because it shows a very successful case that ambient mass spectrometry can be applied to conventional life sciences and tackle fundamental biological question starting from hypotheses. The methodologies shown in this paper have a great potential on interdisciplinary biological studies involving molecular biology, biochemistry, and chemistry.] [DOI] [PubMed] [Google Scholar]
- 31.Hemalatha RG, Pradeep T. Understanding the molecular signatures in leaves and flowers by desorption electrospray ionization mass spectrometry (DESI MS) imaging. J. Agricult. Food Chem. 2013;61:7477–7487. doi: 10.1021/jf4011998. [DOI] [PubMed] [Google Scholar]
- 32.de Abreu LB, Augusti R, Schmidt L, Dressler VL, de Moraes Flores EM, Nascentes CC. Desorption electrospray ionization mass spectrometry (DESI-MS) applied to the speciation of arsenic compounds from fern leaves. Anal. Bioanal. Chem. 2013;405:7643–7651. doi: 10.1007/s00216-013-6986-y. [DOI] [PubMed] [Google Scholar]
- 33.Roach PJ, Laskin J, Laskin A. Nanospray desorption electrospray ionization: an ambient method for liquid-extraction surface sampling in mass spectrometry. Analyst. 2010;135:2233–2236. doi: 10.1039/c0an00312c. [DOI] [PubMed] [Google Scholar]
- 34.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanekoff I, Heath BS, Liyu A, Thomas M, Carson JP, Laskin J. Automated platform for high-resolution tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2012;84:8351–8356. doi: 10.1021/ac301909a. [DOI] [PubMed] [Google Scholar]
- 36.Laneko I, Thomas M, Laskin J. Shotgun approach for quantitative imaging of phospholipids using nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2014;86:1872–1880. doi: 10.1021/ac403931r. [DOI] [PubMed] [Google Scholar]
- 37.Lanekoff I, Burnum-Johnson K, Thomas M, Short J, Carson JP, Cha J, Dey SK, Yang P, Conaway MCP, Laskin J. High-speed tandem mass spectrometric in situ imaging by nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2013;85:9596–9603. doi: 10.1021/ac401760s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rath CM, Alexandrov T, Higginbottom SK, Song J, Milla ME, Fischbach MA, Sonnenburg JL, Dorrestein PC. Molecular analysis of model gut microbiotas by imaging mass spectrometry and nanodesorption electrospray ionization reveals dietary metabolite transformations. Anal. Chem. 2012;84:9259–9267. doi: 10.1021/ac302039u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanekoff I, Thomas M, Carson JP, Smith JN, Timchalk C, Laskin J. Imaging nicotine in rat brain tissue by use of nanospray desorption electrospray ionization mass spectrometry. Anal. Chem. 2013;85:882–889. doi: 10.1021/ac302308p. [DOI] [PubMed] [Google Scholar]
- 40•.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [This work can be credited in many aspects in terms of natural product discover. The work shows for the first time that real-time metabolomics on living microorganisms are possible. The authors also present a MS/MS platform that provides a quick overview of the microbial metabolome and their structural similarity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traxler MF, Kolter R. A massively spectacular view of the chemical lives of microbes. Proc. Natl. Acad. Sci. U.S.A. 2012;109:10128–10129. doi: 10.1073/pnas.1207725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Nguyen DD, Wu C-H, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, et al. MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E2611–E2620. doi: 10.1073/pnas.1303471110. [A novel concept, molecular families (MFs) of microbe-derived natural product peptides, is introduced and determined by their MS/MS patterns. The authors further show that MFs can connect to gene cluster families. This paper shows that ambient mass spectrometry combining MS/MS networking is a powerful tool to genome mining and metabolite screening of closely related microorganisms.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the streptomyces coelicolor secreted metabolome. mBio. 2013;4:e00459–13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watrous J, Roach P, Heath B, Alexandrov T, Laskin J, Dorrestein PC. Metabolic profiling directly from the Petri dish using nanospray desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 2013;85:10385–10391. doi: 10.1021/ac4023154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kersten RD, Yang Y-L, Xu Y, Cimermancic P, Nam S-J, Fenical W, Fischbach MA, Moore BS, Dorrestein PC. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanekoff I, Geydebrekht O, Pinchuk GE, Konopka AE, Laskin J. Spatially resolved analysis of glycolipids and metabolites in living Synechococcus sp PCC 7002 using nanospray desorption electrospray ionization. Analyst. 2013;138:1971–1978. doi: 10.1039/c3an36716a. [DOI] [PubMed] [Google Scholar]
- 47.Hsu C-C, ElNaggar MS, Peng Y, Fang J, Sanchez LM, Mascuch SJ, Moller KA, Alazzeh EK, Pikula J, Quinn RA, et al. Real-time metabolomics on living microorganisms using ambient electrospray ionization flow-probe. Anal. Chem. 2013;85:7014–7018. doi: 10.1021/ac401613x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Hsu C-C, White NM, Hayashi M, Lin EC, Poon T, Banerjee I, Chen J, Pfaff SL, Macagno ER, Dorrestein PC. Microscopy ambient ionization top-down mass spectrometry reveals developmental patterning. Proc. Natl. Acad. Sci. U.S.A. 2013;110:14855–14860. doi: 10.1073/pnas.1310618110. [The authors present an integration of optical microscopy with ambient mass spectrometry. Area of interests can be targeted by optical microscopy for nanoDESI MS analysis directly above the lens of an inverted microscope. This paper also shows for the first time that top-down MS protein analysis on tissue section using ambient mass spectrometry.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oychinnikova OS, Kjoller K, Hurst GB, Pelletier DA, Van Berkel GJ. Atomic force microscope controlled topographical imaging and proximal probe thermal desorption/ionization mass spectrometry imaging. Anal. Chem. 2014;86:1083–1090. doi: 10.1021/ac4026576. [DOI] [PubMed] [Google Scholar]
- 50.Luo Z, He J, Chen Y, He J, Gong T, Tang F, Wang X, Zhang R, Huang L, Zhang L, et al. Air flow-assisted ionization imaging mass spectrometry method for easy whole-body molecular imaging under ambient conditions. Anal. Chem. 2013;85:2977–2982. doi: 10.1021/ac400009s. [DOI] [PubMed] [Google Scholar]
- 51•.Chen C-H, Lin Z, Garimella S, Zheng L, Shi R, Cooks RG, Ouyang Z. Development of a mass spectrometry sampling probe for chemical analysis in surgical and endoscopic procedures. Anal. Chem. 2013;85:11843–11850. doi: 10.1021/ac4025279. [Besides the surgical uses mentioned by the authors, this remote ion sampling technique for MS is possible to extend to many other studies involving chemical analysis on large bulk sample surfaces especially objects that must be preserved in their intact states.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schäfer K-C, Dénes J, Albrecht K, Szaniszló T, Balog J, Skoumal R, Katona M, Tóth M, Balogh L, Takáts Z. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew. Chem., Int. Ed. 2009;48:8240–8242. doi: 10.1002/anie.200902546. [DOI] [PubMed] [Google Scholar]
- 53••.Balog J, Sasi-Szabo L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezso B, Damjanovich L, Darzi A, et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Trans. Med. 2013;5:194ra93. doi: 10.1126/scitranslmed.3005623. [Takáts laboratory officially announced their “iKnife” in this paper. They are aiming at developing standard equipment of next generation in operating theaters.] [DOI] [PubMed] [Google Scholar]
- 54.Strittmatter N, Rebec M, Jones EA, Golf OMH, Abdolrasouli A, Balog J, Behrends VD, Veselkov KA, Takáts Z. Rapid characterization and identification of clinically relevant microorganisms using rapid evaporative ionization mass spectrometry. Anal. Chem. 2014;86:6555–6562. doi: 10.1021/ac501075f. [DOI] [PubMed] [Google Scholar]
- 55.Gross JH. Direct analysis in real time-a critical review on DART-MS. Anal. Bioanal. Chem. 2014;406:63–80. doi: 10.1007/s00216-013-7316-0. [DOI] [PubMed] [Google Scholar]
- 56.Venter AR, Douglass KA, Shelley JT, Hasman G, Jr., Honarvar E. Mechanisms of real-time, proximal sample processing during ambient ionization mass spectrometry. Anal. Chem. 2014;86:233–249. doi: 10.1021/ac4038569. [DOI] [PubMed] [Google Scholar]
- 57.Monge ME, Harris GA, Dwivedi P, Fernandez FM. Mass spectrometry: recent advances in direct open air surface sampling/ionization. Chem. Rev. 2013;113:2269–2308. doi: 10.1021/cr300309q. [DOI] [PubMed] [Google Scholar]