Abstract
Mice deficient in cholesterol 7α-hydroxylase (Cyp7a1) have a diminished bile acid pool (BAP) and therefore represent a useful model for investigating the metabolic effects of restoring the pool with a specific BA. Previously we carried out such studies in Cyp7a1−/−mice fed physiological levels of cholic acid (CA) and achieved BAP restoration, along with an increased CA enrichment, at a dietary level of just 0.03% (w/w). Here we demonstrate that in Cyp7a1−/− mice fed chenodeoxycholic acid (CDCA) at a level of 0.06 % (w/w), the BAP was restored to normal size and became substantially enriched with muricholic acid (MCA)(>70%), leaving the combined contribution of CA and CDCA to be <15%. This resulted in a partial to complete reversal of the main changes in cholesterol and BA metabolism associated with Cyp7a1 deficiency such as an elevated rate of intestinal sterol synthesis, an enhanced level of mRNA for Cyp8b1 in the liver, and depressed mRNA levels for Ibabp, Shp and Fgf15 in the distal small intestine. When Cyp7a1−/− and matching Cyp7a1+/+ mice were fed a diet with added cholesterol (0.2%) (w/w), either alone, or also containing CDCA (0.06%) (w/w) or CA (0.03%) (w/w) for 18 days, the hepatic total cholesterol concentrations (mg/g) in the Cyp7a1−/− mice were 26.9±3.7, 16.4±0.9 and 47.6±1.9, respectively, vs 4.9±0.4, 5.0±0.7 and 6.4±1.9, respectively in the corresponding Cyp7a1+/+ controls. These data affirm the importance of using moderate levels of dietary BA supplementation to elicit changes in hepatic cholesterol metabolism through shifts in BAP size and composition.
Keywords: bile acid pool composition, cholesterol 7α-hydroxylase, cholesterol absorption, cholesterol synthesis, hepatic cholesterol concentration, muricholic acid
1. Introduction
The liver plays a central role in maintaining net sterol balance across the whole animal because it receives essentially all of the cholesterol that is absorbed from the small intestine and is also the site for the degradation and secretion of cholesterol through the bile [1–3]. The major steps involved in the cholesterol absorption pathway and their regulation have been elucidated in great detail [4]. Research in recent years has focused on the role of the transporter NPC1L1 in facilitating the uptake of cholesterol and non-cholesterol sterols into the enterocyte [5, 6]. However, it has long been known that physico-chemical events within the lumen that lead to the solubilization of sterols in mixed micelles together constitute a major regulatory step in determining how much of the cholesterol entering the lumen ultimately reaches the liver [7, 8]. A key component of this regulatory effect is the size and composition of the BA pool [9, 10]. It is well documented that shifts in either of these parameters profoundly affect the amount of cholesterol that is absorbed. The family of proteins that regulates the synthesis, transport, and reabsorption of BA, and the size and composition of the BA pool, has been the subject of multiple comprehensive reviews [11–17].
Newer studies, and several related reviews, suggest that, quite apart from their regulatory roles in lipid absorption and cholesterol and triacylglycerol metabolism, bile acids and their receptors might exert more global effects on metabolic regulation including the control of hepatic insulin resistance and glucose homeostasis [18–28]. The exploration of such putative new roles is often conducted using animal models in which BA metabolism is profoundly altered through the use of levels of dietary BA supplementation that cause pharmacological, rather than physiological shifts in BA synthesis and transport, as well as in the size and composition of the intestinal BA pool. This concern prompted our earlier studies where we used the cholesterol 7α-hydroxylase-deficient (Cyp7a1−/−) mouse, which has an inherently small BA pool [29, 30], to investigate how pool replacement through the feeding of very low dietary levels of cholic acid (CA) impacted cholesterol and BA synthesis, storage and transport by the small intestine and liver at a biochemical and molecular level [31]. One reason why cholic acid was used first for pool restoration in this model is that CA mediates a clear regulatory effect on multiple aspects of cholesterol metabolism [32, 33]. In the present studies we have again used mice deficient in Cyp7a1 to determine what happens when pool restoration is achieved through low levels of chenodeoxycholic acid (CDCA) supplementation. This results in a BA pool in which muricholic acid (MCA) becomes the dominant species through the action of a 6α/β-hydroxylase [34]. Although this has been achieved in the past by feeding the three different isomeric forms of muricholic acid to normal mice, the dietary levels used were very high (0.5% w/w), and the main endpoint studied was the level of intestinal cholesterol absorption [10]. Data on pool sizes were not presented. However, it is likely that they were substantially greater than those in normal chow-fed mice because of the large amounts of MCA that were in the diet.
Another set of studies used mice in which the gene for sterol 12α-hydroxylase (Cyp8b1) had been deleted [33]. This deficiency prevented the conversion of CDCA to CA thus leading ultimately to a major enrichment of the pool with MCA. This compositional change was accompanied by a modest expansion of BA pool size. Here we describe a different approach to generating mice with BA pools consisting predominantly of MCA, but without a concomitant expansion of pool size beyond that seen in adult wildtype mice given a rodent chow diet. This was accomplished by giving Cyp7a1−/− mice graded amounts of CDCA in their diet. Bile acid pool size and composition, as well as several key parameters of intestinal and hepatic sterol and BA metabolism, were determined. In addition, we also compared, in both biochemical and molecular terms, the effects of low levels of CDCA and CA supplementation in the presence of an elevated dietary cholesterol content on intestinal and hepatic BA and cholesterol metabolism in Cyp7a1−/− mice and their matching Cyp7a1+/+ controls.
2. Materials and methods
2.1. Animals and diets
Cholesterol 7α-hydroxylase-deficient mice (Cyp7a1−/−) were generated and maintained as previously described [9]. The initial experiments involving the feeding of diets containing different levels of CDCA used both male and female mice in the age range of 6 to 9 months. For the studies involving the measurement of cholesterol absorption, neutral sterol excretion, rates of cholesterol synthesis, and mRNA expression levels at single level of CDCA supplementation, male mice at 3 to 5 months of age were used. The final experiment with cholesterol-fed mice given a single level of either CDCA or CA supplementation was carried out in females in the age range of 4 to 7 months. In all studies, a cereal-based rodent diet (Wayne Lab Blox, No. 8604; Harland Teklad, Madison, WI), was used. It had an inherent cholesterol content of 0.02% (w/w) and an approximate total lipid content of ~ 5% (w/w). For the dose response study, this regimen, referred as the basal diet, was made to contain varying levels of CDCA (0.015, 0.03 and 0.06% w/w). Thereafter, only one level of CDCA supplementation (0.06% w/w) was used. This dietary level provided an intake of ~9.6 mg (24.5 µmol) /day/100 g body weight (bw). The final study used the basal diet containing added cholesterol (0.2% w/w) either alone, or also containing CDCA (0.06% w/w) or CA (0.03% w/w). Both CDCA (C-9377) and CA (C-1129) were obtained from Sigma-Aldrich Corp (St Louis, MO). All mice were fed their diets ad libitum for either 15–18 days, or 21 days, as specified. Depending on the metabolic parameter being measured, the mice were housed either individually or in groups of three or four in plastic colony cages with wood shavings in a light-cycled room. All animals were studied in the fed state toward the end of the dark-phase of their light cycle. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center.
2.2. Bile acid pool size and composition, fractional cholesterol absorption, and fecal neutral sterol excretion
Pool size was determined as the total BA content of the small intestine, gallbladder, and liver combined. The bile acids were extracted in ethanol in the presence of an internal standard ([24-14C] taurocholic acid, PerkinElmer, Inc,Waltham, MA) and analyzed by HPLC [9]. Bile acids were detected by measurement of the refractive index and identified by comparison with authentic standards. For muricholic acids, no attempt was made to determine whether the major peak identified as β-muricholic acid might have also represented unknown amounts of α and ω-muricholic acid. However, other laboratories have shown that these latter isomers of MCA are either undetectable or present in only trace amounts in the liver and bile of the mouse, [10, 35] and that essentially all conjugation is with taurine [10, 35]. Pool size was expressed as µmol per 100 g body weight. Fractional cholesterol absorption was measured using a dual-isotope method as described [9]. For the fecal neutral sterol measurements, stools collected over 3 days from individually housed mice were dried, weighed, and ground to fine powder. A 1 -g aliquot of this material was used to determine total neutral sterol content [9]. The excretion rate of neutral sterols was expressed as µmol per day per 100 g body weight.
2.3. Rates of cholesterol synthesis in small intestine and liver, and tissue total cholesterol concentrations
Rates of cholesterol synthesis were measured in vivo using [3H]water, as described elsewhere [9]. At 1 hour after intraperitoneal administration of ~40 mCi of [3H]water to the mice, the liver and whole small intestine were removed, rinsed, blotted, and weighed. The organs were then saponified, and the labeled sterols were extracted and quantitated as described [9]. The rate of cholesterol synthesis in each organ was calculated as nanomoles of [3H]water incorporated into sterols per hour per g of tissue. Hepatic total cholesterol concentrations were determined in all mice by gas chromatography and expressed as mg/g of tissue [9]. For the small intestine, total cholesterol concentrations were similarly measured but only in those mice used for the sterol synthesis experiment. Plasma total cholesterol concentrations were measured by the same method used for tissue cholesterol levels. For the mice in the study using the high cholesterol diet, plasma alanine transaminase (ALT) activity was measured by a commercial laboratory.
2.4. Relative mRNA expression analysis
Small intestines were removed, flushed with ice-cold phosphate-buffered saline and then cut into three sections of similar length. The proximal and distal sections were opened longitudinally and the mucosae were removed by gentle scraping. These scrapings, along with aliquots of liver, were quickly frozen in liquid nitrogen. mRNA levels were measured using a quantitative real-time PCR assay [36]. All analyses were determined by the comparative cycle number at threshold method (User Bulletin No. 2, Perkin-Elmer Life Sciences) with cyclophilin as the invariant housekeeping gene [37]. Relative mRNA levels in individual animals were determined by expressing the amount of mRNA found relative to that obtained for Cyp7a1+/+ mice fed the basal diet alone (all studies with males), or the basal diet enriched with cholesterol (studies with females), which in each case was arbitrarily set at 1.0. The primer sequences used to measure RNA levels for all genes have been reported in two earlier publications [31, 38]. The names of all genes studied are as follows; Cyp7a1, Cholesterol 7α-hydroxylase; Cyp27a1, Sterol 27-hydroxylase; Cyp39a1, Oxysterol 7α-hydroxylase; Cyp7b1, Oxysterol 7α-hydroxylase; Cyp8b1, Sterol 12α-hydroxylase; Abca1, ATP binding cassette member A1; Abcb11, Bile salt export pump (Bsep); Abcg5, ATP-binding cassette G5; Fabp6, Ileal bile acid binding protein (Ibabp); Fgf15, Fibroblast growth factor 15; Hmgcs, Hydroxymethylglutaryl coenzyme A synthase; Npc1l1, Niemann-Pick Type C1 -like 1; and Nr0b2, Short heterodimer partner (Shp).
2.5. Analysis of data
All data are reported as means ± SEM for the specified number of animals. GraphPad Prism 6 software (GraphPad, San Diego, CA) was used to perform all statistical analyses. Differences between means were tested for statistical significance (p < 0.05) by one-way analysis of variance. Bars denoted by different letters are statistically different.
3. Results
3.1. Establishment of a level of dietary CDCA supplementation that normalized bile acid pool size in Cyp7a1−/− mice without expanding it in their Cyp7a1+/+ controls
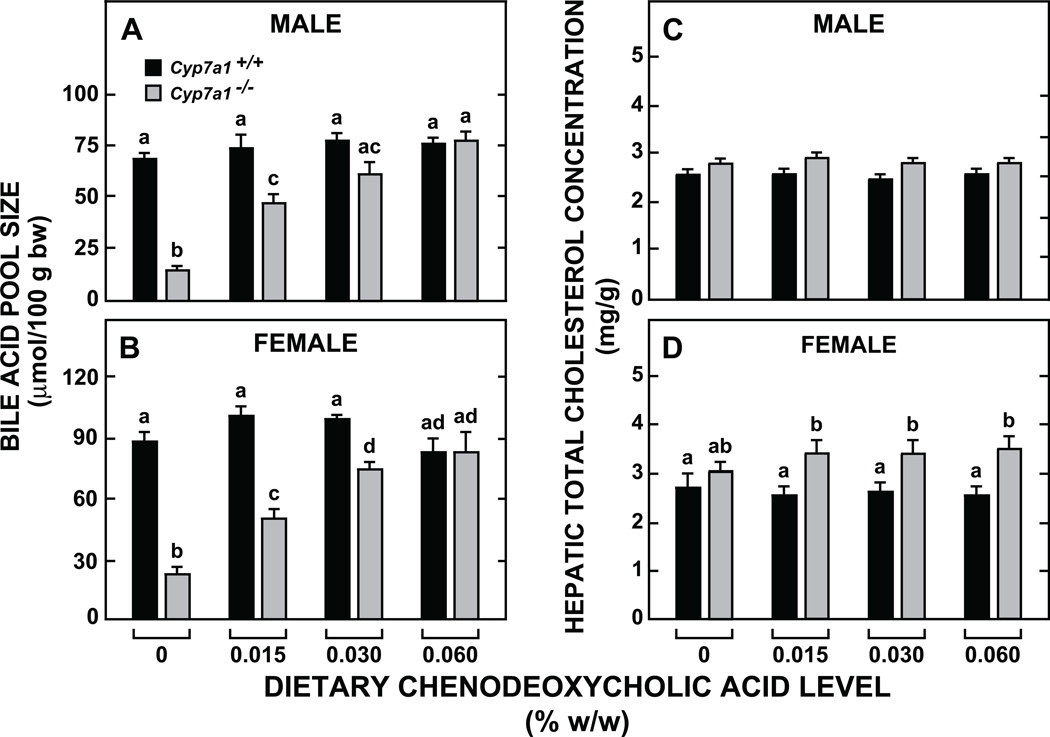
The dietary level of CDCA that restored BA pool size in both male and female Cyp7a1−/− mice to values comparable to those found in their Cyp7a1+/+ counterparts consuming the basal diet alone was 0.06% w/w (Figs 1A and 1B). This provided a daily intake of ~24 to 25 µmol/day/100g bw which equated to just 36% and 28% of the BA pool size in unsupplemented Cyp7a1+/+ male and female mice, respectively. For their male and female Cyp7a1−/− counterparts, this level of CDCA supplementation corresponded to about 160% and 110%, respectively, of their inherently small pools. The BA pool size in both the male and female Cyp7a1+/+ mice given the diet with 0.06% (w/w) CDCA was not significantly different than that in matching Cyp7a1+/+ mice given the basal diet with no added BA.
Fig. 1.
Bile acid pool size (A, B) and hepatic total cholesterol concentrations (C, D) in Cyp7a1+/+ and Cyp7a1−/− mice given varying levels of dietary chenodeoxycholic acid supplementation. The mice were fed their respective diets for 21 days. Values are the mean ± SEM, n=8 animals per group. Different letters above bars denote statistically significant differences between values (p < 0.05) as determined by one-way A NOVA.
3.2. Dietary CDCA supplementation resulted in muricholic acid being the dominant bile acid in the pool, especially in the Cyp7a1−/− mice
In addition to determining what level of CDCA supplementation would be needed to restore BA pool size to normal in the Cyp7a1−/− mice, there was also the parallel question of how BA pool composition might shift, not only in the Cyp7a1-deficient mice, but also in their Cyp7a1+/+ counterparts, given the remarkable capacity of the mouse to actively convert CDCA to muricholic acid through the action of a 6α/β-hydroxylase [34]. Consistent with our earlier findings [29], unsupplemented male Cyp7a1+/+ mice maintained a BA pool with a lower ratio of cholic acid (CA) to muricholic acid (MCA) (1:1.1) compared to female Cyp7a1+/+ controls (1:1.6) (Table 1). Even though CDCA supplementation did not change BA pool size in the Cyp7a1+/+ mice of either gender, there was a marked fall in the ratio of CA to MCA in both cases (to 1:0.4 for males and to 1:0.6 for females). More dramatic compositional changes were found in the BA pool of the Cyp7a1−/− mice given CDCA. In the Cyp7a1-deficient males, the ratio of CA to MCA in the pool was 1:0.6 on the basal diet but only 1:0.1 with the diet containing 0.06% CDCA. In the matching Cyp7a1−/− females, this ratio fell from 1:1.6 to 1:0.1. What this meant was that in Cyp7a1−/− mice given a modest level of CDCA supplementation, the bile acid pool consisted predominantly of muricholic acid with a greatly contracted proportion of cholic acid. In all mice, irrespective of genotype, gender, or dietary treatment, essentially all bile acids were taurine-conjugated.
Table 1.
Bile acid pool composition in Cyp7a1+/+ and Cyp7a1−/− mice fed a basal chow diet alone or containing a low level of chenodeoxycholic acid.
| MALE |
FEMALE |
|||||||
|---|---|---|---|---|---|---|---|---|
| Genotype |
Cyp7a1+/+ |
Cyp7a1−/− |
Cyp7a1+/+ |
Cyp7a1−/− |
||||
| Dietary CDCA level (% w/w) |
0 | 0.06 | 0 | 0.06 | 0 | 0.06 | 0 | 0.06 |
| Bile acid pool composition (% of total) CA |
50.9 ± 2.2 | 22.7 ± 1.4† | 36.8 ± 3.7* | 7.0 ± 0.8*† | 58.6 ± 2.3 | 31.3 ± 0.8† | 59.2 ± 1.7 | 9.5 ± 0.6*† |
| MCA# | 46.7 ± 2.0 | 63.4 ± 1.3† | 63.2 ± 1.3* | 74.0 ± 4.1*† | 37.0 ± 1.9 | 51.4 ± 2.5† | 37.0 ± 1.9 | 80.0 ± 1.1*† |
| CDCA | nd | 6.6 ± 0.7† | nd | 7.4 ± 1.6† | 1.2 ± 0.5 | 8.5 ± 0.8† | nd | 4.5 ± 0.9*† |
| UDCA | 1.1 ± 0.3 | 6.2 ± 0.2† | nd | 6.4 ± 0.5† | 1.8 ± 0.3 | 6.6 ± 0.6† | nd | 5.0 ± 0.5† |
| Others | 1.3 ± 0.6 | 1.3 ± 0.6 | nd | 5.2 ± 2.4 | 1.5 ± 0.5 | 2.4 ± 1.6 | 3.7 ± 2.1 | 1.1 ± 0.6 |
The bile acid composition is for the small intestine and gallbladder with their contents combined with most of the liver.
Values are the means ± SEM of data from 8 mice per group. CA, cholic acid; MCA, muricholic acid; CDCA, chenodeoxycholic acid; UDCA, ursodeoxycholic acid; nd, none detected.
The proportions of α-,β-, ω-muricholic acid were not determined.
Differences in mean values within gender were tested for statistical significance by one-way ANOVA.
P<0.05, Cyp7a1−/− vs. Cyp7a1+/+ mice of the same gender and on the same diet;
P<0.05, CDCA-fed mice vs. chow-fed mice of the same gender and genotype.
3.3. Dietary CDCA supplementation did not alter hepatic total cholesterol concentrations in either Cyp7a1−/− or Cyp7a1+/+ mice, but it did reverse, to a varying degree, some of the changes in hepatic and intestinal cholesterol metabolism associated with Cyp7a1 deficiency
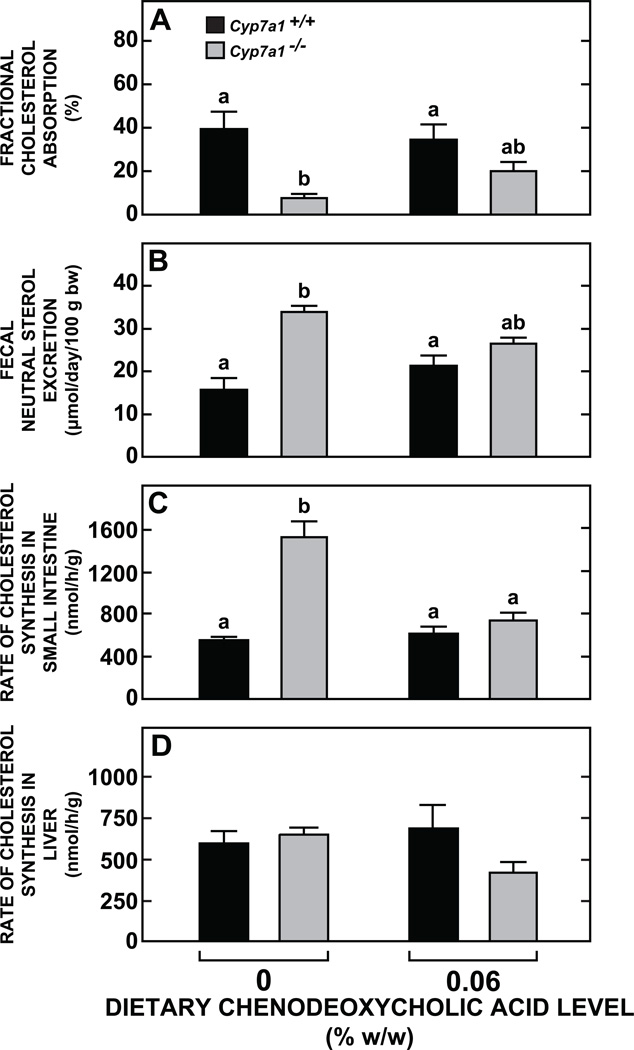
In the initial study using variable dietary levels of CDCA there was no change in hepatic total cholesterol concentrations in male or female mice of either Cyp7a1 genotype (Fig. 1C and 1D). The marginally higher levels evident in the female Cyp7a1−/−mice vs their matching Cyp7a1+/+ controls are consistent with previous findings for this model [29]. CDCA supplementation (0.06% w/w) to the Cyp7a1+/+ mice did not significantly alter their rates of cholesterol absorption, neutral sterol excretion, or cholesterol synthesis in their small intestine and liver (Fig. 2A-2D). In contrast, there were discernable changes in each of these parameters in the supplemented Cyp7a1−/−mice, although the only one that achieved statistical significance (p < 0.05) was the normalization of the rate of intestinal sterol synthesis (Fig. 2C). The concentration of total cholesterol in the small intestine was in the range of 2.8 to 3.0 mg/g in all four groups (data not shown).
Fig. 2.
Parameters of intestinal and hepatic cholesterol metabolism in Cyp7a1+/+ and Cyp7a1−/− mice given a fixed level of dietary chenodeoxycholic acid supplementation. Male mice were fed their respective diets for 15 to 18 days. The level of CDCA was 0.06% w/w. One set of animals was used to measure cholesterol absorption (A) and neutral sterol excretion (B), while a second set was used for determination of rates of intestinal (C) and hepatic (D) cholesterol synthesis. Values are the mean ± SEM, n=6 or 7 animals per group. Different letters above bars denote statistically significant differences between values (p < 0.05) as determined by one-way ANOVA.
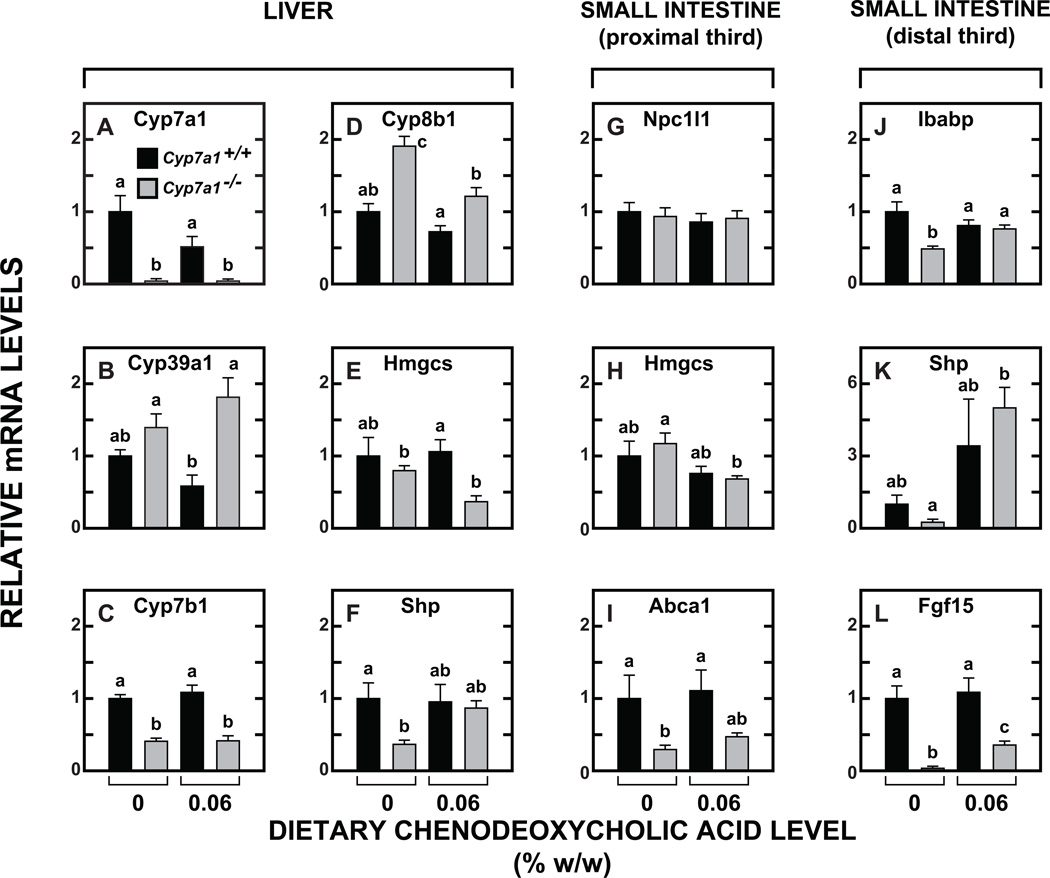
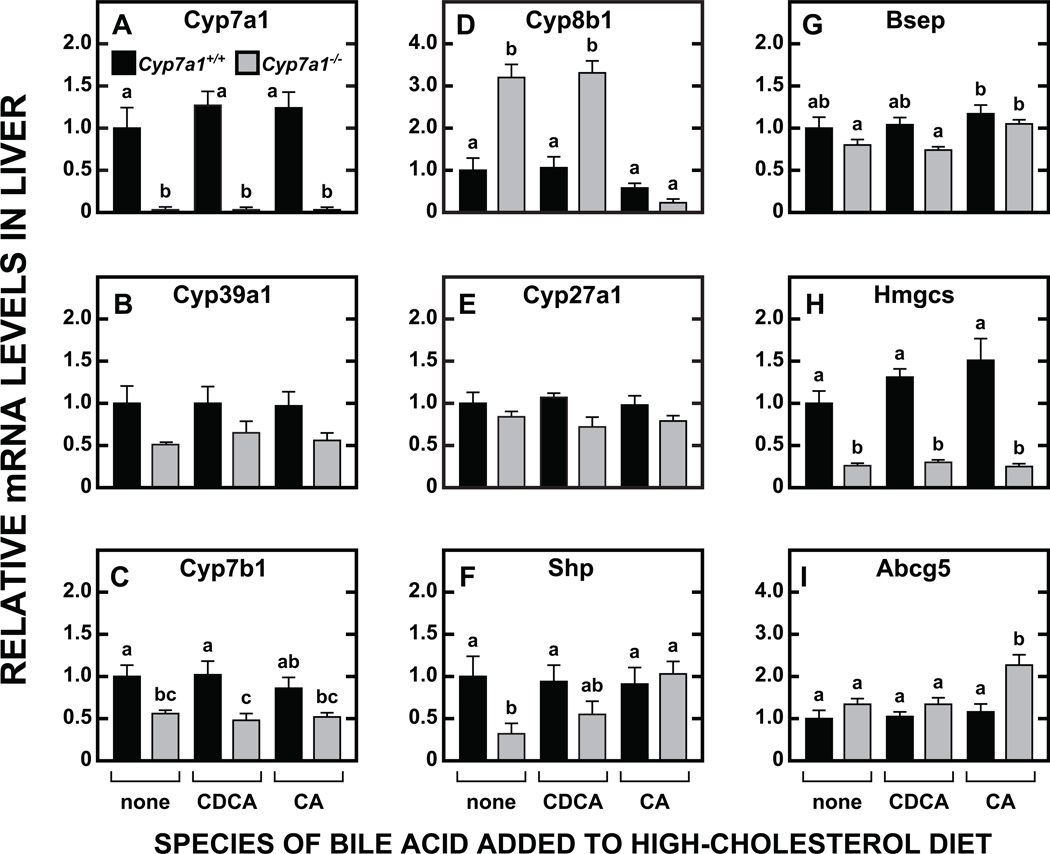
At a molecular level, CDCA supplementation effected modest changes in the relative mRNA expression for a number of genes in the liver (Fig. 3A-3F) and small intestine (Fig. 3G-3L). In the Cyp7a1+/+ mice given CDCA, the mRNA for Cyp7a1 (Fig. 3A), was clearly reduced (although the change was not statistically significant, p > 0.05), reflecting an anticipated lowering of the rate of bile acid synthesis in response to CDCA supplementation. There were also non-significant trends toward lower mRNA levels for both Cyp39a1 (Fig. 3B) and Cyp8b1 (Fig. 3D) in the Cyp7a1+/+ mice given CDCA. In the case of Cyp7b1 (Fig. 3C) there was no influence of CDCA feeding in mice of either genotype, but the level of mRNA in the Cyp7a1−/− mice was uniformly less than half of what it was in the Cyp7a1+/+ controls. In the small intestine, the fall in the mRNA for Hmgcs (Fig. 3H) in the Cyp7a1−/− mice given CDCA was consistent with the change in intestinal cholesterol synthesis for this group (Fig. 2C). The mRNA level for Abca1 (Fig. 3I) in the intestine of the Cyp7a1−/− mice was decisively lower than in their Cyp7a1+/+controls even with CDCA supplementation. In contrast, in the case of Ibabp, Shp, and Fgf15 (Fig 3J, 3K and 3L, respectively) the markedly reduced level of mRNA for these three genes in the Cyp7a1−/− mice given the basal diet was reversed to a varying degree with CDCA feeding.
Fig. 3.
Relative expression levels of mRNA for multiple genes in the liver and small intestine of Cyp7a1+/+ and Cyp7a1−/− mice given a fixed level of dietary chenodeoxycholic acid supplementation. The level of CDCA was 0.06% w/w. These tissues were from the male mice used for the cholesterol absorption and sterol excretion measurements. The gene names are given in Materials and Methods. Relative mRNA levels in individual animals were determined by expressing of the amount of mRNA found relative to that obtained for Cyp7a1+/+ mice not given bile acid supplementation which was arbitrarily set at 1.0. Values are the mean ± SEM, n=5 animals per group. Different letters above bars denote statistically significant differences between values (p < 0.05) as determined by one-way ANOVA.
3.4. In cholesterol-fed Cyp7a1−/− mice hepatic cholesterol concentrations were either reduced or increased depending on whether CDCA or CA was added to the diet
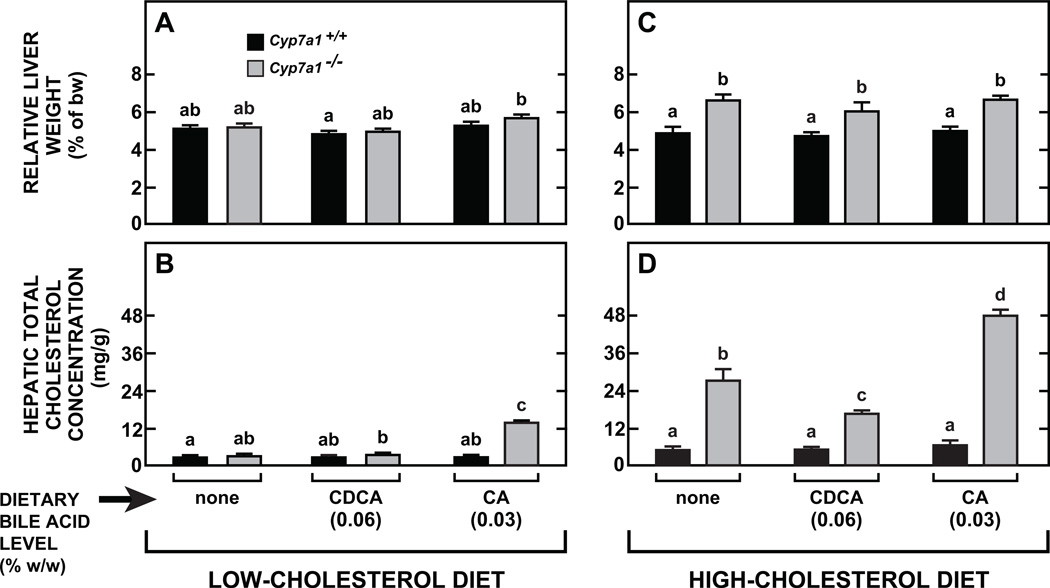
The objective of the study described in Fig. 4 was to investigate the extent to which we could modulate the response of both Cyp7a1−/− mice and their Cyp7a1+/+counterparts to a high-cholesterol diet by concurrent dietary supplementation with low levels of either CDCA (0.06% w/w) or CA (0.03% w/w). This study was preceded by a matching set of experiments in which Cyp7a1−/− and Cyp7a1+/+ mice received these same levels of BA supplementation but contained in the basal diet with no added cholesterol. Multiple parameters were measured in all mice, with hepatic total cholesterol concentrations serving as the main barometer of how the interplay between Cyp7a1 genotype and the species of bile acid added to the diet affected the enterohepatic flux and intrahepatic metabolism of cholesterol. Relative liver weights in the Cyp7a1+/+ mice showed little variation with either dietary cholesterol level or the species of bile acid in the diet (Fig. 4A and 4C). However, in all of the Cyp7a1−/− mice given the high cholesterol diet there was a significant and comparable increase in relative liver weight (Fig. 4C).
Fig. 4.
Comparison of effect of dietary supplementation with either chenodeoxycholic or cholic acid on hepatic total cholesterol concentrations in Cyp7a1+/+ and Cyp7a1−/− mice fed diets without and with added cholesterol. Female Cyp7a1+/+ and Cyp7a1−/− mice were fed for 18 days either the basal diet alone, or containing added cholest e rol (0.2% w/w), along with either CDCA or CA at a level known to be sufficient to fully restore BA pool size in Cyp7a 1−/− mice (Fig. 1B for CDCA, and Reference [31] for CA). Values are the mean ± SEM, n=5 animals per group. Within the groups given diets either without or with added cholesterol, different letters above bars denote statistically significant differences between values (p < 0.05), as determined by one-way ANOVA.
Of the four groups fed the low cholesterol diet with added BA, the only one to show significant change in hepatic cholesterol concentration were the Cyp7a1−/− mice given CA (Fig. 4B). More pronounced differences in liver cholesterol levels related to both genotype and species of bile acid in the diet were seen in the groups fed the high-cholesterol diet (Fig. 4D). This diet raised hepatic cholesterol levels to about 5 to 6 mg/g in all of the Cyp7a1+/+ mice irrespective of whether they were also given any BA. In marked contrast, for the Cyp7a1−/− mice given the cholesterol diet with no added bile acid, the hepatic cholesterol concentration was 26.9 ± 3.7 mg/g (Fig. 4D). This represented a 9.3-fold increase from the baseline level seen in Cyp7a1−/− mice fed the chow diet alone (Fig. 4B). The matching cholesterol-fed Cyp7a1−/− mice given CDCA had a liver cholesterol level of 16.4 ± 0.9 mg/g, which not only was well below that of the mutants fed cholesterol without added BA, but far less than the 47.6 ± 1.9 mg/g found in their Cyp7a1−/− counterparts given the diet containing CA (Fig. 4D). These differences in total cholesterol concentration in all cases reflected differing levels of cholesterol in the esterified fraction (data not shown). Despite the wide differences in hepatic cholesterol concentrations, the plasma total cholesterol levels across all groups in Fig. 4C and 4D averaged 134 to 145 mg/dl except the mutants fed the cholesterol diet with no added BA which had a mean concentration of 161 ± 11 mg/dl. In addition, plasma ALT activities were in the range of 25 to 35 units/L for all groups (data not shown).
3.5. Magnitude and direction of changes in the mRNA expression level for multiple genes in the liver of cholesterol-fed Cyp7a1−/− mice were broadly similar with CDCA and CA supplementation
The panel of genes for which relative mRNA expression levels are presented in Fig. 5 predominantly involve in BA synthesis. The other two, Hmgcs (Fig. 5H) and Abcg5 (Fig. 5I), are reflective of the pathways for cholesterol synthesis and efflux within and from the liver, respectively. In no case did the modest levels of CDCA or CA dietary supplementation elicit a significant change in the mRNA expression level for any of these genes in the Cyp7a1+/+ mice. However, in their Cyp7a1−/− counterparts there were several instances of clear differences in how mRNA expression levels shifted in response to cholesterol feeding together with either CDCA or CA supplementation. One of these pertains to the data for Cyp7a1. The data in Fig. 5A, like those in Fig. 3A, confirm the absence of this RNA species in the Cyp7a1−/− mice. However, in the case of Fig. 3A where all the mice were fed a low-cholesterol diet, there was a trend (p > 0.05) toward a lower mRNA level for Cyp7a1 in the Cyp7a1+/+ mice given CDCA. The finding of the opposite trend in the cholesterol-fed Cyp7a1+/+ mice receiving CDCA, or CA, signaled a modest overriding effect of high dietary cholesterol intake on that occurring in response to the entry of physiological amounts of exogenous BA into the pool. Another instance involves Cyp39a1, the gene for oxysterol 7α-hydroxylase which converts 24-hydroxycholesterol to 7α-hydroxylated oxysterols [29]. A comparison of the data in Fig. 5B with those in Fig. 3B shows a clear reduction in the mRNA for Cyp39a1 in the Cyp7a1−/− mice that paralleled their diet-driven increase in hepatic cholesterol content, but which was unrelated to dietary bile acid intake. For the other oxysterol 7α-hydroxylase, Cyp7b1, lower mRNA expression levels were evident in the cholesterol-fed Cyp7a1−/− mice, irrespective of whether they were without BA supplementation, or were given either CDCA or CA (Fig. 5C). This was also the case in Cyp7a1−/− mice maintained on a low-cholesterol diet containing either CDCA (Fig. 3C) or CA [31].
Fig. 5.
Relative expression levels of mRNA for multiple genes in the livers of cholesterol-fed Cyp7a1+/+ and Cyp7a1−/− mice given fixed levels of either chenodeoxycholic or cholic acid supplementation. The liver tissue for these analyses was derived from the same mice used in the study described in Fig. 4C and D. mRNA levels in individual animals were determined by expressing the amount of mRNA found relative to that obtained for Cyp7a1+/+ mice given the cholesterol-enriched diet but without any bile acid supplementation. Values are mean ± SEM, n=5 animals per group. Different letters above bars denote statistically significant differences between values (p < 0.05) as determined by one-way ANOVA.
The most striking changes in mRNA expression levels were those for Cyp8b1 (Fig. 5D). Cyp7a1-deficient mice showed a decisive increase in Cyp8b1 mRNA, irrespective of whether they are fed a low (Fig. 3D) or high (Fig. 5D) cholesterol diet. In the Cyp7a1−/− mice on the low cholesterol diet containing CDCA, the rise in the mRNA level for Cyp8b1 was blunted (Fig. 3D), but this was not the case when the mutants were maintained on the high cholesterol diet (Fig. 5D). A pronounced suppression of mRNA levels for Cyp8b1 in Cyp7a1-deficient mice was induced only with CA feeding, irrespective of whether the dietary cholesterol level was high (Fig. 5D) or low [31]. In marked contrast to the data for the other genes in the BA synthetic pathways that were studied, very little change in the mRNA for Cyp27a1, as a function of genotype or diet, was evident (Fig. 5E). For two other genes involved in BA metabolism, Shp (Fig. 5F) and Bsep (Fig. 5G), the mRNA expression levels in the Cyp7a1+/+ mice were unchanged with CDCA and CA supplementation. In the Cyp7a1−/− mice, however, the depressed mRNA level for Shp was reversed with CA feeding, as seen previously in Cyp7a1-deficient mice given CA in a low-cholesterol diet [31]. The mRNA level for Hmgcs (Fig. 5H), used as a barometer of the rate of hepatic cholesterol synthesis, showed marked suppression in all the Cyp7a1−/− mice, irrespective of the diet they were given. This was consistent with the much higher hepatic cholesterol levels evident in all the Cyp7a1−/− mice compared to their matching Cyp7a1+/+ controls (Fig. 4D). In the case of the mRNA level for Abcg5 (Fig. 5I), the significant elevation seen in the Cyp7a1−/−mice given CA was consistent with the hepatic cholesterol concentration being higher in these mice than in any other group (Fig. 4D).
4. Discussion
There is an extensive literature spanning more than five decades that describes the effects of dietary chenodeoxycholic acid (CDCA) supplementation on various aspects of cholesterol and bile acid metabolism in several animal models, including the mouse [10, 33, 39–45]. Within this time frame, CDCA was also evaluated for its potential as a therapy for gallstone dissolution [46] as well as for cerebrotendinous xanthomatosis in humans [47]. One distinguishing feature of the mouse is that, unlike most other species including humans, it has only minor amounts of CDCA in its pool [33, 44, 48, 49] because much of it is converted through the action of a 6α/β-hydroxylase to the isomeric forms of muricholic acid [34]. The impact of feeding CDCA has been explored in not only wildtype mice but also in several genetically manipulated models like those with a deficiency of sterol 27-hydroxylase (Cyp27a1), sterol 12α-hydroxylase (Cyp8b1), bile salt export pump (Bsep), and sodium-taurocholate cotransporter polypeptide (Ntcp) [33, 43, 49]. The level of CDCA added to the diet in these studies varied over the range of 0.1 % to 1.0% (w/w). A level of just 0.2% (w/w) in a plain rodent chow diet, when fed to a young adult wildtype mouse, provides a daily CDCA intake of around 80 µmol/100g bw which is equivalent to about one entire BA pool. In genetically manipulated mice that have inherently small BA pools, a dietary level of 0.2% (w/w) CDCA would result in a daily BA intake equating to multiple BA pools in such a model.
Our interest in giving CDCA to the cholesterol 7α-hydroxylase-deficient-mouse was driven not only by our previous studies on low levels of cholic acid supplementation in this model [31], but also by the possibility that we could potentially generate a mouse with a BA pool of normal size containing predominantly muricholic acid. In 2002, Li-Hawkins et al. showed that mice deficient in sterol 12α-hydroxylase (Cyp8b1) develop a moderately expanded BA pool with MCA as the dominant species [33]. Another laboratory subsequently achieved major enrichment of the bile acid pool in C57L mice with the three isomeric forms of muricholic acid by feeding each of these, as well as other species of bile acid, all at a level of 0.5% (w/w) [10]. BA pool enrichment with any of the three isoforms of muricholic acid, particularly β-MCA, resulted in very low levels of intestinal cholesterol absorption, especially compared to cholic acid. A preceding study found that gallstone-susceptible C57 mice given a lithogenic diet containing β-muricholic acid (0.5% w/w) manifested a marked reduction in gallstone prevalence [50].
Essentially, what the Wang laboratory demonstrated so clearly was that the hydrophobicity index of the bile acid pool is a very powerful determinant of how much luminal cholesterol is made available to the NPC1L1-transport system for internalization into the enterocyte and therefore potentially destined for processing by the liver. Unlike cholic acid, which is hydrophobic and promotes cholesterol absorption, muricholic acid, which is hydrophilic, has the opposite effect. In seeking to develop a mouse model that had a BA pool with a high degree of MCA enrichment, but which remained of normal size, we took advantage of the characteristically small pool that exists in Cyp7a1-deficient mice, and also of the efficient biotransformation of CDCA to MCA in this species [34]. In testing an optimum dietary CDCA level to use, we were guided by findings from earlier studies involving CA supplementation to Cyp7a1−/− mice [31].
Several aspects of the biochemical and molecular data from the model described here warrant discussion. These focus more on the Cyp7a1−/− mice because the shifts in BA pool composition produced by CDCA supplementation in their Cyp7a1+/+ controls were insufficient to significantly change any of the parameters that were measured. While CDCA feeding raised the proportion of MCA relative to CA in the pool of the Cyp7a1+/+ mice, the amount of CA in their pools remained much higher than was the case in the Cyp7a1−/− mice given CDCA (Table 1). Limited published data suggest dietary CDCA levels, several fold above 0.06% w/w, are needed to change mRNA expression levels for at least some genes in wildtype mice. Thus, Cheng et al. showed higher hepatic mRNA levels for Bsep in male mice given a dietary CDCA level of 0.3% [43], while another study found a significant rise in hepatic Shp mRNA levels in response to a diet containing 0.25% CDCA [33]. Despite the lack of statistically significant changes in the mRNA data for the CDCA-fed Cyp7a1+/+ mice, the findings specifically for Cyp7a1 and Cyp39a1 nevertheless are noteworthy. In male Cyp7a1+/+mice given low levels of supplementation with CA [31], or CDCA (Fig. 3A), the directional change in the mRNA level for Cyp7a1, while not statistically significant, was indicative of a compensatory reduction in BA synthesis. Not unexpectedly, this trend was reversed in the female Cyp7a1+/+ mice receiving the same levels of CDCA and CA supplementation, but maintained on a high cholesterol diet (Fig. 5A). The studies of Li-Hawkins et al. showed a decisive reduction in Cyp7a1 mRNA expression in wildtype mice given a low-cholesterol diet containing CA at a level of 0.1% w/w. An even higher level of CDCA was needed to markedly reduce Cyp7a1 mRNA levels [33].
The mRNA data for Cyp39a1 are of particular interest because of a marked sexual dimorphism in Cyp39a1 expression in mice [51]. Females exhibited higher levels of mRNA and protein for Cyp39a1 than males. In our CA feeding studies, the hepatic mRNA level for Cyp39a1 in female Cyp7a1−/− mice was significantly lower than in their Cyp7a1+/+ controls, irrespective of whether they were given CA supplementation or not [31]. This same pattern was evident in the current studies with the cholesterol-fed female mice receiving either the high cholesterol diet alone or together with CDCA or CA supplementation, although none of the genotypic differences were statistically significant (Fig. 5B). As shown by the data in Fig. 3B, the opposite genotypic difference in Cyp39a1 mRNA expression levels was apparent in males, irrespective of whether they were given the basal diet alone or with CDCA. Further explanation of this apparent gender-related difference in how Cyp7a1 deficiency affects Cyp39a1 expression will require male and female Cyp7a1−/− littermates and their matching Cyp7a1+/+ controls to be studied together.
For the Cyp7a1−/− mice given CDCA, one particular set of interrelated biochemical and molecular changes across both the small intestine and liver require discussion. The elevated rate of intestinal cholesterol synthesis in the unsupplemented Cyp7a1−/−mice, a hallmark feature of this model (Fig. 2C), was abolished with CDCA feeding and this was reflected in a significant fall in the mRNA expression level for Hmgcs (Fig. 3H). Although not statistically significant, the changes seen in the rate of hepatic cholesterol synthesis, and the mRNA level for Hmgcs in the liver of the supplemented Cyp7a1−/−mice (Fig. 3E) were consistent with those in the small intestine. Coincident with these adaptive changes in intestinal and hepatic sterol synthesis was a discernable, although not statistically significant, rise in fractional cholesterol absorption (Fig. 2A) and fall in fecal neutral sterol excretion (Fig. 2B). While all of these changes are consistent with one another, they seem incongruous with the finding that the restored BA pool in the CDCA supplemented mutants consisted primarily of MCA. Although this hydrophilic BA dominated the pool composition, the size of the pool in the Cyp7a1−/− males given CDCA was about six times greater than in their unsupplemented mutant controls (Fig. 1B). Thus, here is what may be an unusual example of where size and compositional changes in the BA pool brought about by low levels of dietary CDCA supplementation effect changes in intestinal and hepatic cholesterol metabolism that cannot be predicted and so necessitate the direct measurement of such parameters in the intact animal. Ideally this should also include gene expression levels which in a number of cases were found to change dramatically in the Cyp7a1−/− mice given CDCA. Notable examples were the mRNA expression levels for Shp (Fig. 3K) and Fgf15 (Fig. 3L) in the distal small intestine of the Cyp7a1−/− mice.
There is also an additional critical point concerning the type of diet selected for enrichment with bile acid. Given that cereal-based rodent chow diets have a very low inherent cholesterol content, the impact that physiological levels of BA supplementation have on the enterohepatic flux of cholesterol and on intrahepatic cholesterol metabolism, should ideally be determined using a diet with a moderately elevated cholesterol content. This is well illustrated by the data from the study comparing hepatic cholesterol level in Cyp7a1−/− mice in which BA pool size had been restored using CDCA or CA (Fig. 4). In the group given CDCA, the total cholesterol concentration in the liver was only 34% of that in matching Cyp7a1−/− mice given CA. These findings are fully consistent with what would be predicted from the studies of Wang et al [10].
Finally, the CDCA-supplemented Cyp7a1−/− mouse model may potentially be of value for further investigations on the question of how muricholic acid affects FXR activity in vivo. Although CDCA is an FXR agonist [52], recent publications have identified tauro-βMCA and αMCA as FXR antagonists [53, 54]. Future studies aimed at preventing the conversion of CDCA to MCA (by inhibiting activity of the putative 6α/β hydroxylase), or manipulating the taurine conjugation of MCA, will be required to further delineate the role of the different isomeric forms of MCA in FXR-mediated control of bile acid and cholesterol metabolism.
Highlights.
Mice efficiently convert chenodeoxycholic acid (CDCA) to muricholic acid (MCA)
Bile acid (BA) pool size is diminished in cholesterol 7α-hydroxylase-deficient mice
A dietary CDCA level of just 0.06% (w/w) normalizes BA pool size in Cyp7a1−/− mice
MCA displaces cholic acid as the dominant BA in the pool of Cyp7a1−/− mice fed CDCA
Hepatic cholesterol levels in cholesterol-fed Cyp7a1−/− mice rise less with CDCA
Acknowledgements
This research was supported by National Institutes of Health Grants R01HL009610 (S. D. Turley) and R01DK078592 (J. J. Repa). We thank Mario Saucedo, Carolyn Crumpton, Taylor Wagner, Stephen Ostermann and Monti Schneiderman for excellent technical assistance.
Abbreviations
- BAP
bile acid pool
- Cyp7a1
cholesterol 7α-hydroxylase
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- MCA
muricholic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ryan D. Jones, Email: ryan.jones@UTSouthwestern.edu.
Adam M. Lopez, Email: adam.lopez@utsouthwestern.edu.
Ernest Y. Tong, Email: eytong@utmb.edu.
Kenneth S. Posey, Email: kenneth.posey@utsouthwestern.edu.
Jen-Chieh Chuang, Email: jen-chieh.chuang@utsouthwestern.edu.
Joyce J. Repa, Email: joyce.repa@utsouthwestern.edu.
References
- 1.Turley SD, Dietschy JM. The Metabolism and Excretion of Cholesterol by the Liver. In: Arias IM, Jakoby WB, Popper H, Schachter D, Shafritz DA, editors. The Liver: Biology and Pathobiology. New York: Raven Press; 1988. pp. 617–641. [Google Scholar]
- 2.Grundy SM. Absorption and metabolism of dietary cholesterol. Annu. Rev. Nutr. 1983;3:71–96. doi: 10.1146/annurev.nu.03.070183.000443. [DOI] [PubMed] [Google Scholar]
- 3.Cooper AD. Hepatic uptake of chylomicron remnants. J. Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 4.Wang DQ-H. Regulation of intestinal cholesterol absorption. Annu. Rev. Physiol. 2007;69:221–248. doi: 10.1146/annurev.physiol.69.031905.160725. [DOI] [PubMed] [Google Scholar]
- 5.Davis HR, Jr, Altmann SW. Niemann-Pick C1 Like 1 (NPC1L1) an intestinal sterol transporter. Biochim. Biophys. Acta. 2009;1791:679–683. doi: 10.1016/j.bbalip.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Jia L, Betters JL, Yu L. Niemann-pick C1-like 1 (NPC1L1) protein in intestinal and hepatic cholesterol transport. Annu. Rev. Physiol. 2011;73:239–259. doi: 10.1146/annurev-physiol-012110-142233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woollett LA, Wang Y, Buckley DD, Yao L, Chin S, Granholm N, et al. Micellar solubilisation of cholesterol is essential for absorption in humans. Gut. 2006;55:197–204. doi: 10.1136/gut.2005.069906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissinen M, Gylling H, Vuoristo M, Miettinen TA. Micellar distribution of cholesterol and phytosterols after duodenal plant stanol ester infusion. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;282:G1009–G1015. doi: 10.1152/ajpgi.00446.2001. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz M, Russell DW, Dietschy JM, Turley SD. Marked reduction in bile acid synthesis in cholesterol 7α-hydroxylase-deficient mice does not lead to diminished tissue cholesterol turnover or to hypercholesterolemia. J. Lipid Res. 1998;39:1833–1843. [PubMed] [Google Scholar]
- 10.Wang DQ, Tazuma S, Cohen DE, Carey MC. Feeding natural hydrophilic bile acids inhibits intestinal cholesterol absorption: studies in the gallstone-susceptible mouse. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285:G494–G502. doi: 10.1152/ajpgi.00156.2003. [DOI] [PubMed] [Google Scholar]
- 11.Zwicker BL, Agellon LB. Transport and biological activities of bile acids. Int. J. Biochem. Cell Biol. 2013;45:1389–1398. doi: 10.1016/j.biocel.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Lorbek G, Lewinska M, Rozman D. Cytochrome P450s in the synthesis of cholesterol and bile acids--from mouse models to human diseases. FEBS J. 2012;279:1516–1533. doi: 10.1111/j.1742-4658.2011.08432.x. [DOI] [PubMed] [Google Scholar]
- 13.Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang JY. Bile acids: regulation of synthesis. J. Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J. Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J. Lipid Res. 2009;50:S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2014 doi: 10.1194/jlr.R054114. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 19.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 20.Staels B, Fonseca VA. Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care. 2009;32(Suppl. 2):S237–S245. doi: 10.2337/dc09-S355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe M, Horai Y, Houten SM, Morimoto K, Sugizaki T, Arita E, et al. Lowering bile acid pool size with a synthetic farnesoid X receptor (FXR) agonist induces obesity and diabetes through reduced energy expenditure. J. Biol. Chem. 2011;286:26913–26920. doi: 10.1074/jbc.M111.248203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Jadhav K, Zhang Y. Bile acid receptors in non-alcoholic fatty liver disease. Biochem. Pharmacol. 2013;86:1517–1524. doi: 10.1016/j.bcp.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Gerspach AC, Steinert RE, Keller S, Malarski A, Schulte FH, Beglinger C. Effects of chenodeoxycholic acid on the secretion of gut peptides and fibroblast growth factors in healthy humans. J. Clin. Endocrinol. Metab. 2013;98:3351–3358. doi: 10.1210/jc.2012-4109. [DOI] [PubMed] [Google Scholar]
- 24.Sonne DP, Hansen M, Knop FK. Bile acid sequestrants in type 2 diabetes: potential effects on GLP1 secretion. Eur. J. Endocrinol. 2014;171:R47–R65. doi: 10.1530/EJE-14-0154. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, Hylemon PB. Bile acids are nutrient signaling hormones. Steroids. 2014;86:62–68. doi: 10.1016/j.steroids.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang JLY. Bile Acid Metabolism. In: Monga SPS, Cagle PT, editors. Molecular Pathology of Liver Diseases, Springer. 2010. pp. 165–179. [Google Scholar]
- 28.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014;966:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz M, Russell DW, Dietschy JM, Turley SD. Alternate pathways of bile acid synthesis in the cholesterol 7α-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J. Lipid Res. 2001;1542:1594–1603. [PubMed] [Google Scholar]
- 30.Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, Nguyen L, et al. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J. Lipid Res. 2003;44:1001–1009. doi: 10.1194/jlr.M200489-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Jones RD, Repa JJ, Russell DW, Dietschy JM, Turley SD. Turley, Delineation of biochemical, molecular, and physiological changes accompanying bile acid pool size restoration in Cyp7a1−/− mice fed low levels of cholic acid. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G263–G274. doi: 10.1152/ajpgi.00111.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy C, Parini P, Wang J, Bjorkhem I, Eggertsen G, Gafvels M. Cholic acid as key regulator of cholesterol synthesis, intestinal absorption and hepatic storage in mice. Biochim. Biophys. Acta. 2005;1735:167–175. doi: 10.1016/j.bbalip.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, et al. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J. Clin. Invest. 2002;110:1191–1200. doi: 10.1172/JCI16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiang JY. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 35.Alnouti Y, Csanaky IL, Klaassen CD. Quantitative-profiling of bile acids their conjugates in liver, bile, plasma, and urine using LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;873:209–217. doi: 10.1016/j.jchromb.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurrasch DM, Huang J, Wilkie TM, Repa JJ. Quantitative real-time polymerase chain reaction measurement of regulators of G-protein signaling mRNA levels in mouse tissues. Meth. Enzymol. 2004;389:3–15. doi: 10.1016/S0076-6879(04)89001-3. [DOI] [PubMed] [Google Scholar]
- 37.Turley SD, Valasek MA, Repa JJ, Dietschy JM. Multiple mechanisms limit the accumulation of unesterified cholesterol in the small intestine of mice deficient in both ACAT2 and ABCA1. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;299:G1012–G1022. doi: 10.1152/ajpgi.00190.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang JC, Valasek MA, Lopez AM, Posey KS, Repa JJ, Turley SD. Sustained and selective suppression of intestinal cholesterol synthesis by Ro 48–8071, an inhibitor of 2,3-oxidosqualene:lanosterol cyclase, in the BALB/c mouse. Biochem. Pharmacol. 2014;88:351–363. doi: 10.1016/j.bcp.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spady DK, Stange EF, Bilhartz LE, Dietschy JM. Bile acids regulate hepatic low density lipoprotein receptor activity in the hamster by altering cholesterol flux across the liver. Proc. Natl. Acad. Sci. U. S. A. 1986;83:1916–1920. doi: 10.1073/pnas.83.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bergman F, van der Linden W. Liver morphology and gallstone formation in hamsters and mice treated with chenodeoxycholic acid. Acta Pathol. Microbiol. Scand. A. 1973;81:213–221. doi: 10.1111/j.1699-0463.1973.tb00014.x. [DOI] [PubMed] [Google Scholar]
- 41.Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, et al. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab. Pharmacokinet. 2006;21:315–323. doi: 10.2133/dmpk.21.315. [DOI] [PubMed] [Google Scholar]
- 42.Beher WT, Baker GD, Penney DG. A comparative study of the effects of bile acids cholesterol on cholesterol metabolism in the mouse, rat, hamster and guinea pig. J. Nutr. 1963;79:523–530. doi: 10.1093/jn/79.4.523. [DOI] [PubMed] [Google Scholar]
- 43.Cheng X, Buckley D, Klaassen CD. Regulation of hepatic bile acid transporters Ntcp and Bsep expression. Biochem. Pharmacol. 2007;74:1665–1676. doi: 10.1016/j.bcp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Klaassen CD. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J. Lipid Res. 2010;51:3230–3242. doi: 10.1194/jlr.M007641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sprinkle DJ, Hassan AS, Subbiah MT. Effect of chenodeoxycholic acid feeding during gestation in the rat on bile acid metabolism and liver morphology. Proc. Soc. Exp. Biol. Med. 1984;175:386–397. doi: 10.3181/00379727-175-41811. [DOI] [PubMed] [Google Scholar]
- 46.Thistle JL, Hofmann AF. Efficacy and specificity of chenodeoxycholic acid therapy for dissolving gallstones. N. Engl. J. Med. 1973;289:655–659. doi: 10.1056/NEJM197309272891303. [DOI] [PubMed] [Google Scholar]
- 47.Berginer VM, Salen G, Shefer S. Long-term treatment of cerebrotendinous xanthomatosis with chenodeoxycholic acid. N. Engl. J. Med. 1984;311:1649–1652. doi: 10.1056/NEJM198412273112601. [DOI] [PubMed] [Google Scholar]
- 48.Beher WT, Filus AM, Rao B, Beher ME. A comparative study of bile acid metabolism in the rat, mouse, hamster, and gerbil. Proc. Soc. Exp. Biol. Med. 1969;130:1067–1074. doi: 10.3181/00379727-130-33722. [DOI] [PubMed] [Google Scholar]
- 49.Uchida K, Makino S, Akiyoshi T. Altered bile acid metabolism in nonobese, spontaneously diabetic (NOD) mice. Diabetes. 1985;34:79–83. doi: 10.2337/diab.34.1.79. [DOI] [PubMed] [Google Scholar]
- 50.Wang DQ, Tazuma S. Effect of beta-muricholic acid on the prevention and dissolution of cholesterol gallstones in C57L/J mice. J. Lipid Res. 2002;43:1960–1968. doi: 10.1194/jlr.m200297-jlr200. [DOI] [PubMed] [Google Scholar]
- 51.Li-Hawkins J, Lund EG, Bronson AD, Russell DW. Expression cloning of an oxysterol 7α-hydroxylase selective for 24-hydroxycholesterol. J. Biol. Chem. 2000;275:16543–16549. doi: 10.1074/jbc.M001810200. [DOI] [PubMed] [Google Scholar]
- 52.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 53.Hu X, Bonde Y, Eggertsen G, Rudling M. Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J. Intern. Med. 2014;275:27–38. doi: 10.1111/joim.12140. [DOI] [PubMed] [Google Scholar]
- 54.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-betamuricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]