Abstract
The human riboflavin (RF) transporter-3 (product of the SLC52A3 gene) plays an important role in intestinal RF absorption. Our aims in this study were to identify the minimal 5′-regulatory region of the SLC52A3 gene and the regulatory element(s) involved in its activity in intestinal epithelial cells, as well as to confirm promoter activity and establish physiological relevance in vivo in transgenic mice. With the use of transiently transfected human intestinal epithelial HuTu 80 cells and 5′-deletion analysis, the minimal SLC52A3 promoter was found to be encoded between −199 and +8 bp (using the start of the transcription start site as position 1). Although several putative cis-regulatory elements were predicted in this region, only the stimulating protein-1 (Sp1) binding site (at position −74/−71 bp) was found to play a role in promoter activity, as indicated by mutational analysis. Binding of Sp1 to the minimal SLC52A3 promoter was demonstrated by means of EMSA and supershift assays and by chromatin immunoprecipitation analysis. Studies with Drosophila SL2 cells (which lack Sp activity) confirmed the importance of Sp1 in driving the activity of the SLC52A3 minimal promoter; they further showed that Sp3 can also do the activation. Finally, with the use of luciferase gene fusions, the activity of the cloned SLC52A3 promoter was confirmed in vivo in transgenic mice. These studies report, for the first time, on the identification and characterization of the SLC52A3 promoter and also demonstrate the importance of Sp1 in regulating its activity in intestinal epithelial cells.
Keywords: riboflavin, riboflavin transporter-3, intestinal cells, promoter activity, minimal promoter region, stimulating protein-1
the micronutrient riboflavin (RF), a member of the B-family of vitamins, is important for many cellular functions. In the form of riboflavin-5-phosphate and flavin adenosine dinucleotide, it plays key metabolic roles in biological reduction-oxidation reactions involving fatty acid and amino acid metabolism. It participates in the conversion of the water-soluble vitamins folate and pyridoxine (B6) into their metabolically active forms (5), and it is required for proper protein folding inside the endoplasmic reticulum (34) and in reducing cellular oxidative stress (12, 19, 29). Recent studies have also reported a role for RF in immune function (18, 21, 28, 33) and that it has powerful anti-inflammatory properties (4, 12). Systemic RF deficiency leads to a range of clinical abnormalities that include anemia, degenerative changes in the nervous system, and growth retardation (5, 9), and it is also a risk factor for esophageal squamous cell carcinoma and gastric cardia adenocarcinoma (35). Deficiency and suboptimal levels of RF occur in chronic alcoholics, in patients with inflammatory bowel disease, and those with diabetes mellitus (2, 7, 8, 14, 15, 20, 27). Optimizing RF body levels, on the other hand, is effective in the treatment of patients suffering from riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency (16, 17, 39) and those with Brown-Vialetto-Van Laere and Fazio Londe syndrome (3); it is also protective against oxidative (ischemia-induced) injury (1, 19).
Humans and other mammals cannot synthesize RF endogenously; rather, they obtain the vitamin from external sources through intestinal absorption. Absorbed RF is distributed to different tissue compartments via circulation, and the vitamin is transported into cells across the plasma membrane by a specialized carrier-mediated process. Three specific RF transport (RFVT) systems have been shown to mediate RF uptake by mammalian cells; they are RFVT-1, -2, and -3, products of the SLC52A1, SLC52A2, and SLC52A3 genes, respectively (36–38). Of these transporters, the RFVT-3, a 468-amino-acid protein, shows a higher expression in the intestine compared with the other RFVTs under normal physiological condition and appears to play an important role in intestinal RF absorption as indicated by recent studies from our laboratory involving a gene-specific (siRNA) silencing approach (32). This transporter also appears to be involved in our recently observed differentiation-dependent regulation of the RF uptake process, an event that appears to be transcriptionally mediated (30). Other studies from our laboratory have utilized a live cell confocal imaging approach to show that the hRFVT-3 protein is exclusively expressed at the apical membrane domain of polarized absorptive epithelia (31, 32). However, there is little currently known about how the hRFVT-3 system is regulated at either the transcriptional or posttranscriptional levels in intestinal epithelial (and other) cells. Such knowledge is important for a better understanding of how internal and external factors and conditions (e.g., differentiation of intestinal epithelial cells, substrate deficiency, and oversupplementation) affect the function of this important transporter in health and disease. In this study, we characterized the basal transcriptional activity of the SLC52A3 gene in a model of intestinal epithelial cells (the human-derived intestinal epithelial Hutu 80 cells). This was achieved by determining the minimal promoter region that confers basal promoter activity, identifying regulatory elements involved in driving this activity, and validating promoter functionality in vivo in transgenic mice.
MATERIALS AND METHODS
Materials.
Biochemicals were of molecular biology grade and purchased from commercial vendors. Primers and oligonucleotides were acquired from Sigma Genosys (Woodlands, TX).
Cell culture, transfection, and luciferase assay.
The human-derived duodenal epithelial HuTu 80 cells and the Schneider Drosophila SL2 cells were obtained from American Type Culture Collection (ATCC, Rockville, MD) and maintained in MEM and Schneider's Drosophila medium, in the presence of 10% FBS as directed. Cultured cells were transfected with individual promoter constructs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA); 48 h after transfection, luciferase activity was measured. Luciferase data were normalized with pRL-TK expression (Promega, Madison, WI) for respective samples and expressed in arbitrary units of fold over basic. For Drosophila SL2 transfection, stimulating protein (Sp) constructs (Sp0, Sp1, and Sp3, respectively) cloned in pPac expression vector (Clontech, Palo Alto, CA) were used as described previously (23).
Generation of truncated and mutated constructs.
The 5′- and 3′-deletion clones were amplified by PCR from the full-length promoter construct using gene-specific primers (Table 1), followed by subcloning of the PCR fragments into pGL3-basic vector. Site-specific mutagenesis was done using wild-type minimal promoter (−199 to +8 bp) as a template by QuickChangeII XL (Agilent Technologies, Santa Clara, CA) kit. The mutation was verified through sequencing (Laragen, Los Angeles, CA), and the mutated constructs were used for transient transfection into HuTu 80 cells as mentioned above.
Table 1.
Sequence of primers used in PCR to generate 5′-deletion constructs of the SLC52A3 promoter
| Position | Sequence (5′-3′) |
|---|---|
| −3,125 bp | CGACGCGTTTGATCAAATCCTGGTTG |
| −1,509 bp | CGACGCGTTGCAGCTACAAAGAAGG |
| −9,66 bp | CGACGCGTAGAAGACTCTGAAGCACAG |
| −199 bp | CGACGCGTAGTGGCTCCTCCCAG |
| −101 bp | CGACGCGTGTGAGGGAATTCCGTG |
| Reverse | CCGCTCGAGCTTCTTCCTTCTAGTACAAAGC |
MluI and XhoI restriction sites are used in the forward and reverse primers, respectively, as shown in underlined and bold underlined, respectively. The relative positions of the primers are assigned by setting start of transcription initiation of SLC52A3 as position +1.
EMSA.
Nuclear extract from HuTu 80 cells was isolated with the commercially available NE-PER Nuclear and Cytoplasmic extraction reagents (Thermo Scientific Pierce, Rockford, IL) according to manufacturer's instructions. A 35-bp region of the minimal promoter between −89 and −55 bp (5′-GTGTGGGAACCGTGGGGAGGAGCTGCCAGGATTCA-3′) was used. The promoter fragment was biotinylated using a 3′-end DNA labeling kit (Thermo Scientific Pierce). EMSA was performed using LightShift Chemiluminescent EMSA kit (Thermo Scientific). Binding reactions were performed at room temperature in 20 μl of reaction volume by incubating 5 μg of nuclear extract, 20 fmol of biotin-labeled DNA fragment, and 50 ng/μl poly(dIdC) for 20–30 min, as indicated in the manufacturer's protocol. Oligonucleotide competition was conducted using a 50-fold molar excess of the unlabeled oligonucleotides. For the supershift assays, we pretreated the nuclear extract with 2 μg anti-Sp1 antibodies (Millipore, Temecula, CA) (cat. no: 07-645). DNA-protein complexes were then separated on 6% DNA retardation gel (Invitrogen) in 0.5× Tris borate-EDTA, and the gel was transferred to nylon membrane followed by crosslinking of transferred DNA to membrane. Detection of biotin-labeled DNA was done using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) analysis was performed using the ChIP assay kit (Millipore) per the manufacturer's protocol. Briefly, HuTu 80 cells (1 × 106 cells) were treated with a final concentration of 1% formaldehyde at 37°C for 10 min, and the cells were lysed in SDS lysis buffer and then sonicated to shear DNA into fragments. The cell supernatant was centrifuged and further incubated with 10 μg of Sp1 or normal rabbit IgG overnight at 4°C. Protein A agarose was used to precipitate the DNA fragment bound to transcription factor and antibody complexes. To reverse protein/DNA crosslinks immunoprecipitates were incubated for 4 h at 65°C with NaCl as supplied by the manufacturer. Samples were treated with proteinase K, and the DNA was purified by phenol/chloroform extraction followed by ethanol precipitation. Purified DNA was used in PCR using specific primers [5′-GAAAGGCCACTGAGC-3′ (forward) and 5′-CCACGCCTCACCCAG-3′ (reverse)]. PCR conditions used were denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 45°C for 30 s, and extension at 72°C for 30 s.
Confirmation of promoter activity in vivo: Generation of transgenic mice.
We solicited the help of the transgenic mouse facility at the University of California Irvine (UCI-TMF) for creating transgenic founder mice carrying human SLC52A3 promoter fused to the luciferase reporter gene. This procedure uses pronuclear DNA injection as described below. A 974-bp SLC52A3 promoter construct fused to the luciferase reporter gene, as described previously (26), was PCR amplified, sequenced to verify the integrity of the sequence, and provided to the UCI-TMF. Eggs from super-ovulating females were injected and implanted into foster mothers. Pups were born from the implanted mothers, and tail tips were collected about 14 days after birth. Genomic DNA was extracted from the tail tip and used for genotyping using specific primers for the hSLC52A3 promoter (forward 5′-AGGTTAGGTCACTTGCTCAGC-3′) and luciferase gene (reverse 5′-CCCTTCTTGGCCTTTATGAG-3′) that would yield a 2,644-bp product. The result was further confirmed by performing another PCR with nested forward primer (forward 5′-ATGTGGACAGCAGAGG-3′) and the same primer from the luciferase gene sequence that would yield a product of 2,134 bp. The transgenic mice were bred to obtain homozygous transgenic mice for this study. The Institutional Animal Care and Use Committee (IACUC) of the Long Beach VA Medical Center and the IACUC at UCI approved the experimental procedures used for mice in this study.
Tissue isolation, determination of RNA levels, and analysis of luciferase activity in transgenic mice.
Specific tissue samples were collected from mice following euthanasia and were split into ice-cold Trizol (Invitrogen) for RNA isolation and passive lysis buffer (Promega) for further analysis. The tissue was lysed, and RNA was isolated, as directed by the manufacturer's protocol. cDNA was prepared, and real-time PCR was performed using specific primers to quantitate the relative expression of mRFVT-3 (5′- GGATCAGTGGAAGCCAGTG-3′ and 5′-GACCTGTTAGGCAGGAAGATG-3′) and luciferase mRNA (5′-ACCGCCTGAAGTCTCTGATT-3′ and 5′-CGACGTAATCCACGATCTCT-3′). Luciferase assays were performed using the manufacturer's protocol and a Promega 20/20 luminometer. We normalized the luciferase assays to total protein concentrations of each sample. Protein concentrations were measured by DC protein assay kit (Bio-Rad, Hercules, CA).
Isolation of intestinal epithelial crypt and villus cells.
Mouse intestinal epithelial villus and crypt cells were isolated as described previously using an established fractionation procedure (25, 30). Briefly, we collected 10 fractions with fractions 1 and 2 representing villus epithelial cells and fractions 9 and 10 representing crypt epithelial cells. Relative purity of these fractions has been established in our laboratory previously by measuring activity of marker enzymes (22).
Statistical analysis.
Data shown are means ± SE of at least three separate experiments and were analyzed for significance using the Student's t-test. P < 0.05 was assigned as the significance level.
RESULTS
Identification of the minimal SLC52A3 promoter region required for basal activity.
Guided by our previous results of 5′-rapid amplification of cDNA ends (5′-RACE) that determined the transcription start site (TSS) of SLC52A3 (30), we aimed in this study at determining the minimal region required for basal activity of the SLC52A3 promoter. Our promoter analysis was performed using the human-derived intestinal epithelial HuTu 80 cells because they are easy to transfect (although similar promoter activity was noticed when the human intestinal epithelial Caco-2 cells were used; data not shown).
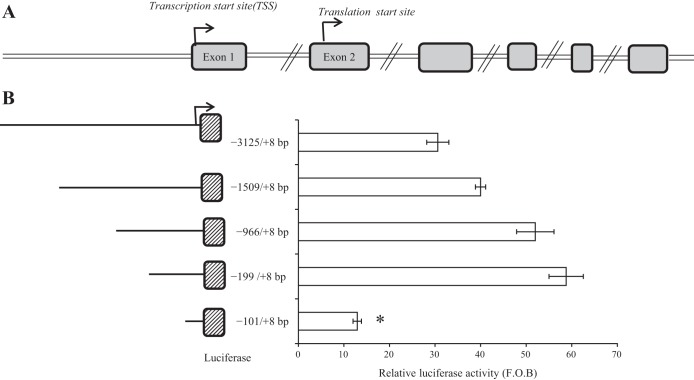
Genome analysis of the SLC52A3 gene showed that it has one 5′-untranslated exon and that the putative ATG TSS is located in exon 2 (Fig. 1A). We cloned the region −3,125 to +8 bp upstream of TSS, as this region was predicted by computational programs MatInspector (www.genomatix.de) and Alibaba2 (http://www.gene-regulation.com/pub/programs/alibaba2/index.html) to harbor the SLC52A3 promoter (i.e., it had many putative transcription factor binding sites). Indeed, the genomic fragment showed marked promoter activity (about 64-fold over basic) upon transfection into HuTu 80 cells (Fig. 2A). To determine the minimal region needed for basal activity of the SLC52A3 promoter, we generated a series of 5′-deletion constructs of the predicted full SLC52A3 promoter (−3,125 to +8 bp) and examined their promoter activity after transfection into HuTu 80 cells. The results showed that progressive 5′-deletion of the cloned SLC52A3 genomic fragment led to a gradual increase in promoter (luciferase) activity up to the regions of −966 and −199 bp (constructs of the last 2 regions showed similar activity). Promoter activity, however, significantly (P < 0.01) decreased upon further shortening of the cloned SLC52A3 genomic fragment to −101 bp (i.e., fragment −101/+8) (Fig. 1B). The latter suggests that the minimal SLC52A3 promoter region required for basal activity is encoded in the sequence between −199 and +8 bp.
Fig. 1.
Diagrammatic representation of exons (A) and functional analysis of 5′-regulatory region of the SLC52A3 gene (B). A: 5′-rapid amplification of cDNA ends (5′-RACE) and bioinformatics analysis revealed that the transcription start site (TSS) is positioned in untranslated exon 1, whereas the TSS is on exon 2. B: functional analysis of the putative promoters. Left: size and position of the different promoter constructs. Right: luciferase assay results of the different constructs following transient transfection into HuTu 80. Firefly luciferase activity was normalized relative to the activity of simultaneously expressed Renilla luciferase in the cellular extracts. Results are expressed as relative fold to the pGL3-basic (F.O.B.) vector and represent means ± SE of at least 3 independent experiments. *P < 0.01.
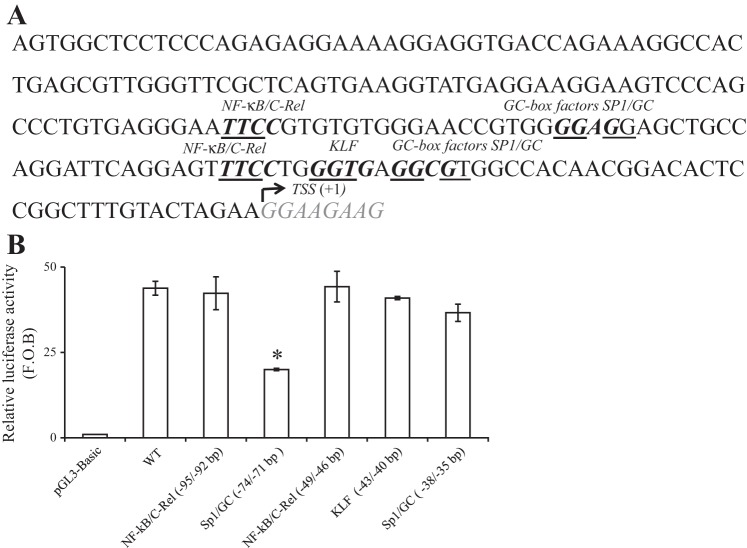
Fig. 2.
Schematic representation of the minimal promoter region of the SLC52A3 gene, and the effect of mutating putative cis-regulatory sites on promoter activity. A: nucleotide sequence of the minimal region required for basal activity of SLC52A3 is shown (38,119–37,913 on chromosome 20). Bold nucleotides indicate the positions of several identified putative cis-regulatory elements. TSS is underlined and numbered +1. B: effect of mutating specific putative sites in the SLC52A3 minimal promoter on promoter activity. Conserved nucleotides in the core consensus binding sites for stimulating protein-1 (Sp1), NF-κB/C-Rel, or Kruppel-like factor (KLF) were mutated. Mutated minimal regions were then tested for promoter activity using the luciferase assay system and HuTu 80 cells. The results are expressed as relative fold to the pGL3-basic vector and represent means ± SE of at least 3 independent experiments. *P < 0.01.
Identification of cis-regulatory elements and transcription factors involved in SLC52A3 promoter activity.
To identify putative cis-regulatory elements that may be involved in regulating the activity of the SLC52A3 minimal promoter, we subjected this genomic region to MatInspector and to Alibaba2 computational analyses. Results of these analyses revealed a lack of the typical TATA motif in the minimal region but the presence of a cluster of putative cis-regulatory elements that includes binding sites for NF-κB/cRel, Kruppel-like factor (KLF), and Sp1 (Fig. 2A). To investigate the possible role of these putative cis-regulatory elements in basal activity of the SLC52A3 promoter, we examined (by means of site-directed mutagenesis) the functional consequences of mutating these sites on the activity of the SLC52A3 minimal promoter in HuTu 80 cells. The results showed that mutating the putative cis-regulatory elements for NF-κB/C-Rel (−95/−92 bp and −49/−46 bp), Sp1 (−38/−35 bp), and KLF (−43/−40 bp) (considering TSS as +1) had no effect on the activity of the SLC52A3 promoter. In contrast, mutating the Sp1 site located at −74/−71 bp led to a significant (P < 0.01; 60%) reduction in promoter activity compared with the control (unmutated) minimal promoter (Fig. 2B). These findings demonstrate an important role for Sp1 in regulating the activity of the SLC52A3 promoter.
To confirm that the nuclear factor Sp1 does indeed interact with the SLC52A3 in the minimal promoter region, we performed an EMSA using a 35-bp region of the minimal promoter (a sequence between −89 and −55 bp) and a nuclear extract from HuTu 80 cells. The EMSA revealed the formation of major DNA/protein complexes that led to a shift in band mobility (Fig. 3A, lane ii) compared with the control (lane i). We also found that formation of these complexes could be specifically inhibited by unlabeled probe (Fig. 3A, lane iii), which resulted in the disappearance of the band. Finally, direct evidence of Sp1 binding to the SLC52A3 minimal promoter was obtained from super-shift analysis with the use of specific Sp1 polyclonal antibodies (Fig. 3A; lane iv). For supershift assay, HuTu 80 nuclear extracts were preincubated with the individual antibodies, resulting in a dramatic decrease in mobility of the DNA/protein complexes (i.e., supershift in the gel).
Fig. 3.
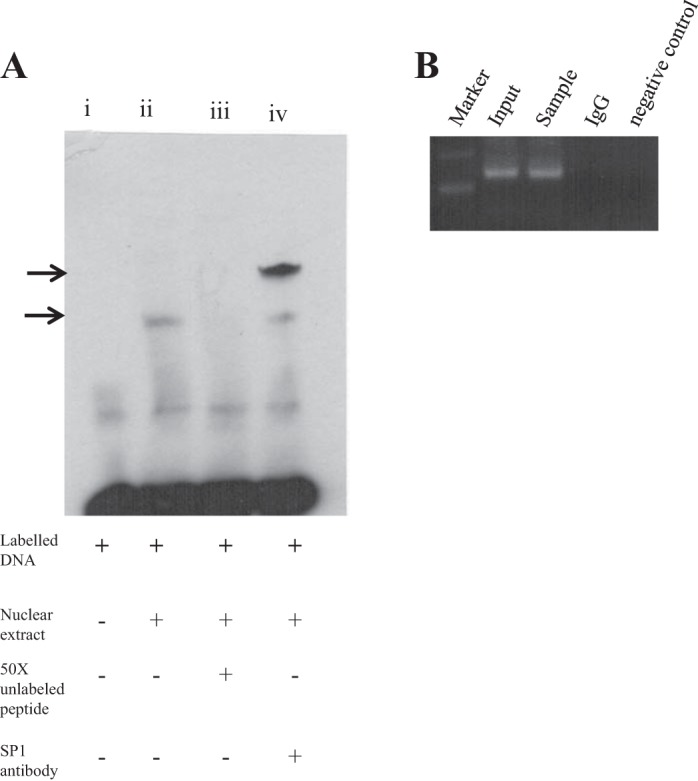
EMSA with oligonucleotide competition and chromatin immunoprecipitation (ChIP) analysis. A: gel shift assay was performed with a biotin-labeled 35-bp region of the SLC52A3 minimal promoter spanning a sequence between −89 and −55 and nuclear extract from HuTu 80 cells. A 50-fold molar excess of the same unlabeled fragment was used for oligonucleotide competition. Major DNA/protein complexes are indicated with arrows. Gel shift assay was performed by incubation with Sp1 antibody. B: binding of Sp1 transcription factor to the SLC52A3 promoter in vivo. ChIP assay was performed using the HuTu 80 cells and the antibodies against Sp1. The immunoprecipitated DNA was PCR amplified using primers specific for the region containing Sp1. A control reaction with normal IgG was used as a negative control.
In other studies, we examined the binding of Sp1 to the SLC52A3 promoter in vivo. This was done by ChIP analysis using the cross-linked chromatin prepared from HuTu 80 cells. Chromatin was immune precipitated with the antibody against Sp1 followed by cross-link reversal; the DNA was used for PCR amplification of the promoter region harboring both the upstream and downstream Sp1 binding site. Resulting PCR amplification in the sample that was pulled down by using Sp1 antibodies (and no amplification in negative control, where IgG was used) clearly suggested specific interaction of Sp1 protein in the minimal region of the promoter in vivo. In parallel, as a negative control, the same DNA sample was used for PCR amplification of the promoter region that does not contain any Sp1 cis-elements (region −3,410 to −3,182 bp) (data not shown).
Transactivation of the SLC52A3 minimal promoter by Sp1 and Sp3.
Previous studies have shown that the nuclear factor Sp3 can also bind to Sp1 cis-regulatory element sites (10, 24). The nuclear factor Sp1 is primarily a positive transactivating factor (24), whereas Sp3 can act as both an activator or a repressor depending on cell/promoter type (10, 11, 13, 23, 24). Thus, to further confirm the role of Sp1 in regulating the SLC52A3 minimal promoter and to determine whether Sp3 can also do the same, we performed cotransfection assays in Drosophila SL2 cells. The latter cells lack endogenous Sp activity, and thus they represent an in vitro model system to test the effects of exogenous Sp on the activity of Sp- dependent promoters (6, 23). In these studies, we transfected a promoter-luciferase construct containing the minimal SLC52A3 promoter region with vectors pPac Sp1 and pPac Sp3 (which express the human Sp1 and Sp3, respectively) into SL2 cells. The level of expression of the reporter gene detected in the absence of coexpression of exogenous Sp proteins was considered as control. The results showed that transfection with the pPac Sp1 and pPac Sp3 vectors led to a significant (P < 0.01 for both) increase in SLC52A3 promoter activity compared with controls (Fig. 4); the increase in the promoter activity with pPac Sp1, however, was greater than that caused by pPac Sp3.
Fig. 4.
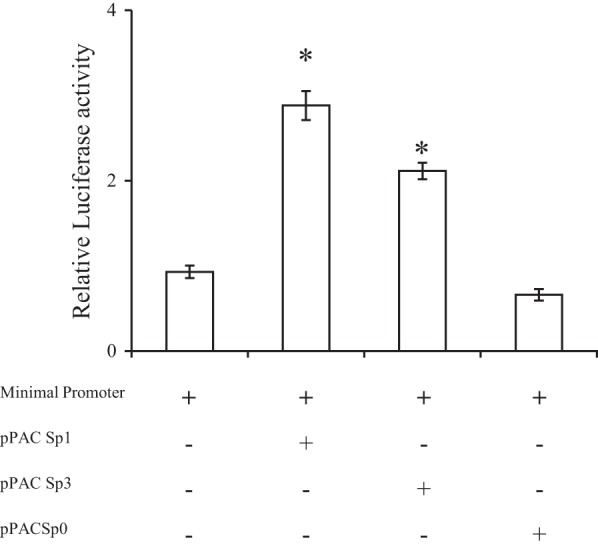
Cotransfection assays using either Sp1- or Sp3-containing vectors and SLC52A3 minimal promoter-luciferase construct in Drosophila SL2 cells. SL2 cells were cotransfected with 4 μg of the minimal promoter construct in pGL3-Basic and either control plasmid pPac Sp0 or Drosophila Sp expression plasmids (pPac Sp1, pPac Sp3). At 48 h posttransfection, cells were harvested, and firefly luciferase activity was assayed. Data are reported as relative firefly luciferase activity and represent means ± SE of at least 3 independent experiments. *P < 0.01.
Confirmation of activity of the cloned SLC52A3 promoter in vivo.
To confirm activity of the human SLC52A3 promoter in vivo, we generated transgenic mice expressing the −966/+8 nt region fused to the luciferase reporter gene. With the use of PCR on genomic DNA, mice carrying luciferase transgene were selected and used as founders (Fig. 5A); we also confirmed the presence of the transgene in mice found to be positive in the PCR assay using another set of nested PCR primers (Fig. 5B). The level of luciferase expression was then determined in the intestine (as well as in other tissues of the digestive system and the kidney), found to be considerable, and paralleled the level of expression in the endogenous mouse RFVT-3 (Fig. 6, A and B). These findings confirm the activity of the cloned SLC52A3 promoter in the in vivo setting; they also validate the suitability of this transgenic mouse model for further investigations on the effect of internal and external factors/conditions on activity of the SLC52A3 gene in vivo.
Fig. 5.
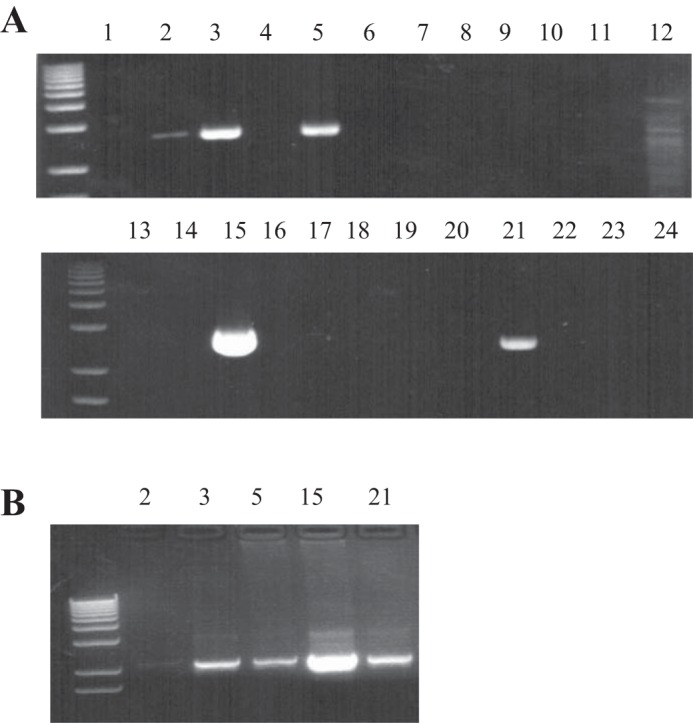
Identification of SLC52A3 promoter-luciferase transgenic mice. A: mouse genomic DNA was analyzed using PCR for the presence of the full-length promoter-luciferase construct. Lanes 2, 3, 5, 15, and 21 include PCR products that indicate mice considered positive for the transgene. B: positive samples were reconfirmed using a different set of nested primers.
Fig. 6.
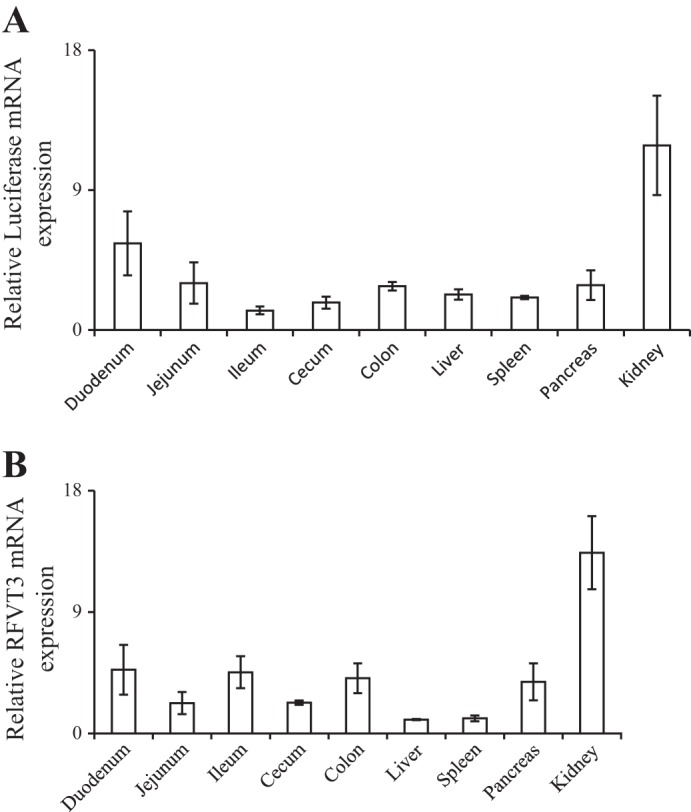
Levels of expression of luciferase and endogenous riboflavin transporter (RFVT)-3 mRNAs in the intestine (as well as other tissues of the digestive system and the kidney) of transgenic mice carrying SLC52A3 promoter-luciferase construct. Real-time PCR was performed (on the same tissue samples) using luciferase-specific primers (A) and SLC52A3-specific primers (B). Data are from at least 3 different mice from the same founders and are presented as means ± SE. β-Actin was used as a control for RNA normalization.
Confirmation of involvement of transcriptional mechanisms(s) in the differentiation-dependent regulation of intestinal SLC52A3.
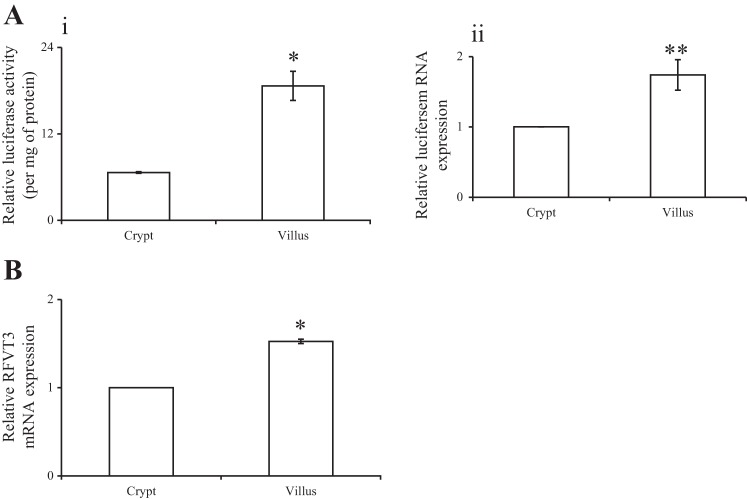
We have previously shown that expression of RFVT-3 in the intestine undergoes differentiation-dependent regulation, as it is higher in villus compared with crypt epithelial cells (30). Our results also suggested that this mode of regulation is mediated at the level of transcription of the SLC52A3 gene. To further confirm the latter, we used the newly generated transgenic mice described above to determine the level of activity of the SLC52A3 promoter (level of expression of the luciferase reporter gene) in epithelial cells of the intestinal villi and crypts. We also correlated the findings to the level of expression of RFVT-3 in these cells (Fig. 7B). The results showed a significantly higher level of expression of luciferase (P < 0.05) and of RFVT-3 (P < 0.01) in villus compared with crypt epithelial cells, confirming the involvement of transcriptional mechanisms in the differentiation-dependent regulation of the intestinal SLC52A3.
Fig. 7.
Luciferase activity and level of expression of luciferase and endogenous RFVT-3 mRNAs in jejunum crypt and villus epithelial cells of the transgenic mice carrying SLC52A3 promoter-luciferase construct. Luciferase activity was determined using luminometer, and data are expressed per milligram of protein (A, i). Real-time PCR analysis was performed using gene-specific primers against luciferase (A, ii) and RFVT-3 (B) from total RNA isolated from mice intestinal crypt and villus epithelial cells. Values are means ± SE from 3 separate animals. *P < 0.01 and **P < 0.05.
DISCUSSION
Our aims in this study were to clone the SLC52A3 promoter and to characterize its activity in intestinal epithelial cells, with special emphasis on identifying the minimal promoter region needed for basal activity and the cis-regulatory elements/transacting factors that are involved in this activation. The product of this gene, i.e., the RFVT-3 protein, is critical for intestinal absorption of RF and is abundantly expressed in the gut with expression being restricted to the apical membrane domain of the polarized epithelia (30, 32). Expression of RFVT-3 appears to be differentiation dependent with a higher level of expression in intestinal villus compared with crypt cells (30). There is little known at present about how this important transporter is regulated at the transcriptional and posttranscriptional levels. In this study, we focused on the basal transcriptional regulation of SLC52A3 in human intestinal epithelial (HuTu 80) cells. Guided by findings of 5′-RACE that identified the TSS of the SLC52A3 promoter (30) and by computer analysis, we cloned an ∼3.1-kb genomic fragment that represents the putative 5′-regulatory region of the SLC52A3 gene and then showed the cloned fragment to have a robust promoter activity in HuTu 80 cells. The minimal region required for basal activity of the SLC52A3 promoter was then determined by means of 5′-deletion approaches, and we found the region between −199 and +8 to fit that description. This region was TATA-less and contained a number of putative cis-regulatory elements including NF-κB/C-Rel, KLF, and Sp1. The role of these cis-regulatory elements in regulating the activity of the SLC52A3 minimal promoter was then studied by mutating these sites individually and examining the effect of such mutations on promoter activity. The results showed that, while mutating the NF-κB/C-Rel (−95/−92 bp and −49/−46 bp), Sp1 (−38/−35 bp), and KLF (−43/−40 bp) sites, there was no effect on promoter activity; mutating the Sp1 site located at (−74/−71 bp) led to a significant (P < 0.01) inhibition in promoter activity.
We further established the role for the Sp1 site (−74/−71 bp) and binding of the Sp1 transcription factor to it by performing EMSA and supershift analyses. A major specific DNA/protein complex that can be competed away with unlabeled promoter fragment was observed in the EMSA assay, whereas that of the supershift analysis (which used specific Sp1 antibodies) showed shifting to higher molecular weight. These findings indicate an interaction of the Sp1 transcription factor with its cis-regulatory element in the SLC52A3 promoter fragment. We further confirmed the latter by ChIP analysis, which showed amplification using a specific primer during PCR pulled down by Sp1 antibodies; no such amplification was found in the IgG negative control. Finally, the findings with the Drosophila SL2 cells (which lack Sp factors) showing that Sp1 and Sp3 can stimulate SLC52A3 promoter activity solidify our conclusion on the involvement of Sp1 in the activation of transcription of this important human gene.
To confirm activity of the cloned SLC52A3 promoter in an in vivo setting and to establish physiological relevance of our in vitro findings, we generated transgenic mice carrying the SLC52A3 promoter fused to the luciferase reporter gene and examined promoter (luciferase) activity in the intestine and other digestive tissues as well as the kidneys. A marked promoter activity was observed in all the tissues examined, which paralleled the expression pattern of the endogenous mouse RFVT-3. Using this transgenic mouse model, we then confirmed our recent observation on the involvement of transcriptional mechanism(s) in the differentiation-dependent regulation of RFVT-3 expression in the intestine (30). In the latter studies, a significantly higher luciferase activity was detected in the intestinal villus compared with the crypt epithelial cells, a pattern that paralleled the expression of the endogenous RFVT-3.
In summary, this study represents the first characterization of the SLC52A3 promoter in vitro and in vivo and reports the identification of the minimal promoter and a critical role of Sp1 in driving its activity in intestinal epithelial cells. These findings should serve as the basis for future investigations into the molecular mechanisms involved in the regulation of RFVT-3 expression in health and disease.
GRANTS
This work was supported by grants from the Department of Veterans Affairs and the National Institutes of Health (National Institute on Alcohol Abuse and Alcohol Grant AA018071 and National Institute of Diabetes and Digestive and Kidney Diseases Grants DK56061 and DK58057).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: A.G. and H.M.S. conception and design of research; A.G. and S.S. performed experiments; A.G., S.S., and H.M.S. analyzed data; A.G., S.S., and H.M.S. interpreted results of experiments; A.G. and S.S. prepared figures; A.G. drafted manuscript; A.G., S.S., and H.M.S. edited and revised manuscript; A.G., S.S., and H.M.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We are grateful to Dr. Nabendu S. Chatterjee for valuable suggestions.
REFERENCES
- 1.Betz AL, Ren XD, Ennis SR, Hultquist DE. Riboflavin reduces edema in focal cerebral ischemia. Acta Neurochir Suppl Wien 60: 314–317, 1994. [DOI] [PubMed] [Google Scholar]
- 2.Bonjour JP. Vitamins and alcoholism. V. Riboflavin, VI. Niacin, VII. Pantothenic acid, and VIII. Biotin. Int J Vitam Nutr Res 50: 425–440, 1980. [PubMed] [Google Scholar]
- 3.Bosch AM, Abeling NG, Ijlst L, Knoester H, van der Pol WL, Stroomer AE, Wanders RJ, Visser G, Wijburg FA, Duran M, Waterham HR. Brown-Vialetto-Van Laere and Fazio Londe syndrome is associated with a riboflavin transporter defect mimicking mild MADD: a new inborn error of metabolism with potential treatment. J Inherit Metab Dis 34: 159–164, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cobianchi L, Fornoni A, Pileggi A, Molano RD, Sanabria NY, Gonzalez-Quintana J, Bocca N, Marzorati S, Zahr E, Hogan AR, Ricordi C, Inverardi L. Riboflavin inhibits IL-6 expression and p38 activation in islet cells. Cell Transplant 17: 559–566, 2008. [DOI] [PubMed] [Google Scholar]
- 5.Cooperman JM, Lopez R, editor Riboflavin R. In: Handbook of Vitamins: Nutritional, Biochemical and Clinical Aspects. New York, NY: Dekker, 1984, p. 299–327. [Google Scholar]
- 6.Courey AJ, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell 55: 887–898, 1988. [DOI] [PubMed] [Google Scholar]
- 7.Fennelly J, Frank O, Baker H, Leevy CM. Peripheral neuropathy of the alcoholic: I. Aetiological role of aneurin and other B-Complex vitamins. Br Med J 2: 1290–1292, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Banares F, Abad-Lacruz A, Xiol X, Gine JJ, Dolz C, Cabre E, Esteve M, Gonzalez-Huix F, Gassull MA. Vitamin status in patients with inflammatory bowel disease. Am J Gastroenterol 84: 744–748, 1989. [PubMed] [Google Scholar]
- 9.Goldsmith GA. Riboflavin deficiency. In: Riboflavin. New York, NY: Plenum, 1975. [Google Scholar]
- 10.Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res 20: 5519–5525, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J 13: 3843–3851, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwanaga K, Hasegawa T, Hultquist DE, Harada H, Yoshikawa Y, Yanamadala S, Liao H, Visovatti SH, Pinsky DJ. Riboflavin-mediated reduction of oxidant injury, rejection, and vasculopathy after cardiac allotransplantation. Transplantation 83: 747–753, 2007. [DOI] [PubMed] [Google Scholar]
- 13.Kiela PR, LeSueur J, Collins JF, Ghishan FK. Transcriptional regulation of the rat NHE3 gene. Functional interactions between GATA-5 and Sp family transcription factors. J Biol Chem 278: 5659–5668, 2003. [DOI] [PubMed] [Google Scholar]
- 14.Kodentsova VM, Vrzhesinskaia OA, Sokol'nikov AA, Alekseeva IA, Spirichev VB. [Metabolism of riboflavin and B group vitamins functionally bound to it in insulin-dependent diabetes mellitus]. Vopr Med Khim 39: 33–36, 1993. [PubMed] [Google Scholar]
- 15.Kodentsova VM, Vrzhesinskaia OA, Trofimenko EV, Sokol'nikov AA, Beketova NA, Blazheevich NV, Isaeva VA, Aleinik SI, Trofimenko LS, Dronova VI. [Vitamin status of children with diabetes mellitus]. Vopr Med Khim 40: 45–48, 1994. [PubMed] [Google Scholar]
- 16.Law LK, Tang NL, Hui J, Fung SL, Ruiter J, Wanders RJ, Fok TF, Lam CW. Novel mutations in ETFDH gene in Chinese patients with riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency. Clin Chim Acta 404: 95–99, 2009. [DOI] [PubMed] [Google Scholar]
- 17.Liang WC, Ohkuma A, Hayashi YK, Lopez LC, Hirano M, Nonaka I, Noguchi S, Chen LH, Jong YJ, Nishino I. ETFDH mutations, CoQ10 levels, and respiratory chain activities in patients with riboflavin-responsive multiple acyl-CoA dehydrogenase deficiency. Neuromuscul Disord 19: 212–216, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D, Zempleni J. Low activity of LSD1 elicits a pro-inflammatory gene expression profile in riboflavin-deficient human T Lymphoma Jurkat cells. Genes Nutr 9: 422, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mack CP, Hultquist DE, Shlafer M. Myocardial flavin reductase and riboflavin: A potential role in decreasing reoxygenation injury. Biochem Biophys Res Commun 212: 35–40, 1995. [DOI] [PubMed] [Google Scholar]
- 20.Majumdar SK, Shaw GK, Thomson AD. Vitamin utilization status in chronic alcoholics. Int J Vitam Nutr Res 51: 54–58, 1981. [PubMed] [Google Scholar]
- 21.Mazur-Bialy AI, Buchala B, Plytycz B. Riboflavin deprivation inhibits macrophage viability and activity - a study on the RAW 264.7 cell line. Br J Nutr 110: 509–514, 2013. [DOI] [PubMed] [Google Scholar]
- 22.Nabokina SM, Reidling JC, Said HM. Differentiation-dependent up-regulation of intestinal thiamin uptake: cellular and molecular mechanisms. J Biol Chem 280: 32676–32682, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Nabokina SM, Said HM. Characterization of the 5'-regulatory region of the human thiamin transporter SLC19A3: In vitro and in vivo studies. Am J Physiol Gastrointest Liver Physiol 287: G822–G829, 2004. [DOI] [PubMed] [Google Scholar]
- 24.Philipsen S, Suske G. A tale of three fingers: The family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res 27: 2991–3000, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinkus L. Separation and use of enterocytes. Methods Enzymol 77: 154–162, 1981. [DOI] [PubMed] [Google Scholar]
- 26.Reidling JC, Said HM. In vitro and in vivo characterization of the minimal promoter region of the human thiamin transporter SLC19A2. Am J Physiol Cell Physiol 285: C633–C641, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal WS, Adham NF, Lopez R, Cooperman JM. Riboflavin deficiency in complicated chronic alcoholism. Am J Clin Nutr 26: 858–860, 1973. [DOI] [PubMed] [Google Scholar]
- 28.Schramm M, Wiegmann K, Schramm S, Gluschko A, Herb M, Utermohlen O, Kronke M. Riboflavin (vitamin B2) deficiency impairs NADPH oxidase 2 (Nox2) priming and defense against Listeria monocytogenes. Eur J Immunol 44: 728–741, 2014. [DOI] [PubMed] [Google Scholar]
- 29.Seekamp A, Hultquist DE, Till GO. Protection by vitamin B2 against oxidant-mediated acute lung injury. Inflammation 23: 449–460, 1999. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian VS, Ghosal A, Subramanya SB, Lytle C, Said HM. Differentiation-dependent regulation of intestinal vitamin B(2) uptake: Studies utilizing human-derived intestinal epithelial Caco-2 cells and native rat intestine. Am J Physiol Gastrointest Liver Physiol 304: G741–G748, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian VS, Rapp L, Marchant JS, Said HM. Role of cysteine residues in cell surface expression of the human riboflavin transporter-2 (hRFT2) in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol 301: G100–G109, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian VS, Subramanya SB, Rapp L, Marchant JS, Ma TY, Said HM. Differential expression of human riboflavin transporters -1, -2, and -3 in polarized epithelia: A key role for hRFT-2 in intestinal riboflavin uptake. Biochim Biophys Acta 1808: 3016–3021, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyosawa T, Suzuki M, Kodama K, Araki S. Highly purified vitamin B2 presents a promising therapeutic strategy for sepsis and septic shock. Infect Immun 72: 1820–1823, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 290: 1571–1574, 2000. [DOI] [PubMed] [Google Scholar]
- 35.Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, Li JM, Kong GQ, Qi H, Cui J, Zhang LQ, Yang JZ, Li JL, Li XC, Ren JL, Liu ZC, Gao WJ, Yuan L, Wei W, Zhang YR, Wang WP, Sheyhidin I, Li F, Chen BP, Ren SW, Liu B, Li D, Ku JW, Fan ZM, Zhou SL, Guo ZG, Zhao XK, Liu N, Ai YH, Shen FF, Cui WY, Song S, Guo T, Huang J, Yuan C, Huang J, Wu Y, Yue WB, Feng CW, Li HL, Wang Y, Tian JY, Lu Y, Yuan Y, Zhu WL, Liu M, Fu WJ, Yang X, Wang HJ, Han SL, Chen J, Han M, Wang HY, Zhang P, Li XM, Dong JC, Xing GL, Wang R, Guo M, Chang ZW, Liu HL, Guo L, Yuan ZQ, Liu H, Lu Q, Yang LQ, Zhu FG, Yang XF, Feng XS, Wang Z, Li Y, Gao SG, Qige Q, Bai LT, Yang WJ, Lei GY, Shen ZY, Chen LQ, Li EM, Xu LY, Wu ZY, Cao WK, Wang JP, Bao ZQ, Chen JL, Ding GC, Zhuang X, Zhou YF, Zheng HF, Zhang Z, Zuo XB, Dong ZM, Fan DM, He X, Wang J, Zhou Q, Zhang QX, Jiao XY, Lian SY, Ji AF, Lu XM, Wang JS, Chang FB, Lu CD, Chen ZG, Miao JJ, Fan ZL, Lin RB, Liu TJ, Wei JC, Kong QP, Lan Y, Fan YJ, Gao FS, Wang TY, Xie D, Chen SQ, Yang WC, Hong JY, Wang L, Qiu SL, Cai ZM, Zhang XJ. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet 42: 759–763, 2010. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto S, Inoue K, Ohta KY, Fukatsu R, Maeda JY, Yoshida Y, Yuasa H. Identification and functional characterization of rat riboflavin transporter 2. J Biochem 145: 437–443, 2009. [DOI] [PubMed] [Google Scholar]
- 37.Yao Y, Yonezawa A, Yoshimatsu H, Masuda S, Katsura T, Inui K. Identification and comparative functional characterization of a new human riboflavin transporter hRFT3 expressed in the brain. J Nutr 140: 1220–1226, 2010. [DOI] [PubMed] [Google Scholar]
- 38.Yonezawa A, Masuda S, Katsura T, Inui K. Identification and functional characterization of a novel human and rat riboflavin transporter, RFT1. Am J Physiol Cell Physiol 295: C632–C641, 2008. [DOI] [PubMed] [Google Scholar]
- 39.Zhu M, Zhu X, Qi X, Weijiang D, Yu Y, Wan H, Hong D. Riboflavin-responsive multiple Acyl-CoA dehydrogenation deficiency in 13 cases, and a literature review in mainland Chinese patients. J Hum Genet 59: 256–261, 2014. [DOI] [PubMed] [Google Scholar]