Summary
Fragile X syndrome (FXS) is caused by the absence of the fragile X mental retardation protein (FMRP). We have previously generated FXS-induced pluripotent stem cells (iPSCs) from patients’ fibroblasts. In this study, we aimed at unraveling the molecular phenotype of the disease. Our data revealed aberrant regulation of neural differentiation and axon guidance genes in FXS-derived neurons, which are regulated by the RE-1 silencing transcription factor (REST). Moreover, we found REST to be elevated in FXS-derived neurons. As FMRP is involved in the microRNA (miRNA) pathway, we employed miRNA-array analyses and uncovered several miRNAs dysregulated in FXS-derived neurons. We found hsa-mir-382 to be downregulated in FXS-derived neurons, and introduction of mimic-mir-382 into these neurons was sufficient to repress REST and upregulate its axon guidance target genes. Our data link FMRP and REST through the miRNA pathway and show a new aspect in the development of FXS.
Highlights
-
•
FXS-derived neurons display downregulation of neural and axon guidance genes
-
•
These neural genes are regulated by the transcription repressor REST
-
•
REST is not silenced in FXS-derived neurons
-
•
REST is regulated by hsa-mir-382, which has lower levels in FXS-derived neurons
In this article, Benvenisty and colleagues show that fragile X-derived neurons are aberrantly regulating neural differentiation and axon guidance genes due to elevated expression of the REST transcription repressor. They further suggest that the high levels of REST result from low levels of hsa-mir-382. Introduction of mimic-mir-382 into fragile X-derived neurons repressed REST and upregulated its axon guidance target genes.
Introduction
Fragile X syndrome (FXS) affects approximately 1 in every 4,000 boys and 1 in 8,000 girls worldwide (Callan and Zarnescu, 2011; Penagarikano et al., 2007). It is now believed that FXS is the leading cause of inherited intellectual disability in males and one of the major monogenic causes for autism (Boyle and Kaufmann, 2010; Callan and Zarnescu, 2011; Penagarikano et al., 2007; Wang et al., 2012). The syndrome is caused primarily by an expansion of a CGG repeat at the 5′ untranslated region (UTR) of the fragile X mental retardation gene 1 (FMR1). This repeat expansion leads to CpG methylation, which spreads to the FMR1 promoter, modifications in chromatin conformation of the FMR1 gene, and silencing of the gene expression. Subsequently, the fragile X mental retardation protein (FMRP) is no longer produced (Coffee et al., 1999, 2002; Sutcliffe et al., 1992).
FMRP is a highly conserved protein, expressed in mammals mainly in the brain and testes (Devys et al., 1993; Santoro et al., 2012; Verkerk et al., 1991). In the brain, FMRP is found primarily in neurons, where it plays an important role in synaptic plasticity (Devys et al., 1993). FMRP is an RNA-binding protein that acts as a translation regulator by either stalling polyribosomes or inhibiting translation initiation (Ashley et al., 1993; Feng et al., 1997; Khandjian et al., 2004; Napoli et al., 2008; Stefani et al., 2004). It may also regulate mRNA levels through the microRNA (miRNA) pathway, as work on both Drosophila and mammalian cells revealed association of FMRP with components of the RNA-induced silencing complex and several miRNAs (Caudy et al., 2002; Ishizuka et al., 2002; Jin et al., 2004; Plante et al., 2006). FMRP was also shown to associate with specific miRNAs, which together select and repress target mRNAs to regulate neuronal morphology (Edbauer et al., 2010).
Several works have implicated a role for FMRP in neurogenesis, and although some of the results were contradicting, all of these studies have shown impairment in dendritic spine morphology, maturation or pruning, or abnormal gene expression during neural development that may persist to adulthood (Bhattacharyya et al., 2008; Castrén et al., 2005; Comery et al., 1997; Galvez et al., 2005; Irwin et al., 2001; Tessier and Broadie, 2008). Other studies have shown FMRP to be crucial for the regulation of timing and proliferation capacities of neural progenitor cells (NPCs), thus regulating the proper number of neurons (Callan et al., 2010; Egger et al., 2008; Luo et al., 2010). All of these data place FMRP as an important regulator of proper development and maturation of the neural network.
Another key factor important for proper brain development is the repressor element 1 silencing transcription factor (REST) (Chen et al., 1998). REST is considered a master negative regulator of neurogenesis, regulating the pool size and timing of differentiation of different neural lineages (Chen et al., 1998; Covey et al., 2012; Satoh et al., 2013; Schoenherr and Anderson, 1995). REST is expressed in embryonic stem cells (ESCs), NPCs, and nonneuronal cells, where it suppresses neuron-specific genes, in contrast to differentiated neurons where it is silenced (Chen et al., 1998; Schoenherr and Anderson, 1995). REST both regulates and is regulated by brain specific miRNAs and has been implicated to be involved in pluripotency and neurodegenerative pathologies (González-Castañeda et al., 2013; Gopalakrishnan, 2009; Hermanson, 2008; Marullo et al., 2010; Ooi and Wood, 2007; Zuccato et al., 2003).
We have previously generated both ESCs and induced pluripotent stem cells (iPSCs) derived from FXS patients (Bar-Nur et al., 2012; Eiges et al., 2007; Urbach et al., 2010). Although the functions of FMRP have been studied extensively, the underlying molecular mechanisms causing the severe neuronal phenotypes are still largely unknown. In this study, we aim to understand the molecular pathology underling FXS using FXS-derived iPSCs, NPCs, and neurons. Our study suggests a major role for REST in the molecular pathology of FXS neurons. A better understanding of the developmental processes dysregulated in FXS will help in the search for a treatment to alleviate or even correct some of the abnormal molecular phenotypes.
Results
Downregulation of Neuronal Differentiation and Axon Guidance Genes in FXS-Derived Neurons
In order to understand the molecular pathology in FXS, we differentiated FXS-derived iPSCs into either NPCs (FXS-derived NPCs) or neurons (FXS-derived neurons) using two different protocols (Bar-Nur et al., 2012; Kim et al., 2010). We next compared global gene expression profiles of two normal control cell lines with five different FXS clones generated from three different patients, in three different categories of cell types: fibroblasts, iPSCs, and neurons (derived from the aforementioned fibroblasts or iPSCs, respectively) using DNA microarray data. Principal component analysis (PCA) shows that FXS and control cells cluster according to cell type and not by genetic origin (Figure 1A). This observation was further supported by hierarchical clustering showing that FXS and control cells from each cell type category cluster together (Figure 1B). Next, ANOVA analysis was performed for each cell type category to detect differentially expressed genes (Figure S1A available online). Only genes that passed a threshold of p < 0.05 with fold change > 2 and are expressed in either FXS or control cells were further analyzed. FXS fibroblasts taken from patients exhibited a very mild difference, with 106 genes that were significantly downregulated in FXS cells compared with control fibroblasts (Figure 1C). Functional annotation analysis of the 106 downregulated genes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation clustering tool showed enrichment for metal binding proteins (Figure 1C). We next compared the global gene expression of iPSCs derived from the above fibroblasts. Surprisingly, control and FXS-derived iPSCs were almost identical in their total gene expression profiles, as only 21 genes showed a significant difference in expression levels (Figure 1C). In this case, we found no significant gene ontology (GO) term enrichment for any cellular process common to these genes (Figure 1C). Finally, we repeated the gene expression analysis with neurons derived from either the control iPSCs or FXS-derived iPSCs. Neurons showed the largest difference in global gene expression than all other cell types, with 198 genes being significantly downregulated (Figures 1C and S1A). Functional annotation analysis revealed that the FXS-derived neurons were downregulated in processes such as neuron differentiation, neuron projection development, and axonogenesis, all of which found to be statistically significant with a Benjamini correction for multiple tests of p < 5 × 10−4 (Figure 1C). We next wanted to analyze whether the downregulated genes in FXS-derived neurons are part of a specific cellular pathway. For this aim, we used the Kyoto Encyclopedia for Genes and Genomes (KEGG). The highest ranking pathway generated by KEGG revealed a significant enrichment of the downregulated genes for the axon guidance pathway, which is a key step in neural network formation and determines which way a newly formed growth cone will turn. Expression levels of representative downregulated axon guidance genes are shown in Figure 1D. To verify that the downregulation observed in neural genes in FXS-derived neurons is not a result of incomplete differentiation, we looked at various key neural genes. Expression levels of TUBB3, NESTIN, and HOMER3 were examined and showed similar or even higher expression levels in FXS-derived neurons compared with control cells (Figure S1B).
Figure 1.
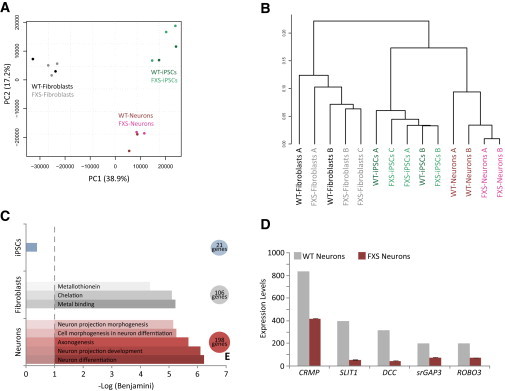
Differentially Expressed Genes in FXS versus Control Cells
(A) PCA on total gene expression profiles of control (WT) and FXS-derived cells shows clustering by cell type.
(B) The same profiles were used for hierarchical clustering using Pearson correlation exhibiting fibroblasts, iPSCs, and neurons to group apart.
(C) Downregulated genes in FXS iPSCs, fibroblasts, or neurons, which passed a statistically significant threshold of differential expression, were analyzed for different cellular processes or pathways using Functional annotation clustering with the DAVID functional annotation clustering tool (http://david.abcc.ncifcrf.gov/). FXS iPSCs showed no significant GO term enrichment for downregulated genes. Downregulated genes in FXS fibroblasts exhibit a significant enrichment for metal binding proteins while FXS-derived neurons display a significant enrichment for axonogenesis and neuron differentiation.
(D) DNA-microarray analysis revealed several axonal guidance genes with markedly lower expression levels in FXS-derived neurons compared with control (WT) (columns represent average values of two control microarrays for WT and three FXS microarrays for FXS; scale bars represent SE).
Most Downregulated Genes in FXS-Derived Neurons Are Regulated by REST
During neural differentiation, specific transcription factors act at different stages to orchestrate the process of turning on tissue specific genes and shutting off pluripotency genes. We searched for common promoter motifs within the downregulated genes of FXS-derived neurons. We used two different platforms for this analysis, the integrated Allegro/Amadeus motif discovery platform and the DAVID functional annotation clustering tool together with the UCSC transcription factor binding site data. Both platforms identified REST as a candidate transcription factor, regulating over half of the downregulated genes with a statistical significance by Benjamini correction of p value = 1.9 × 10−7 in DAVID and p value = 2.8 × 10−17 in Allegro/Amadeus. The Allegro/Amadeus software indicated most target genes to have the REST recognition sequence at a very close proximity to their transcription start site (Figure 2A).
Figure 2.
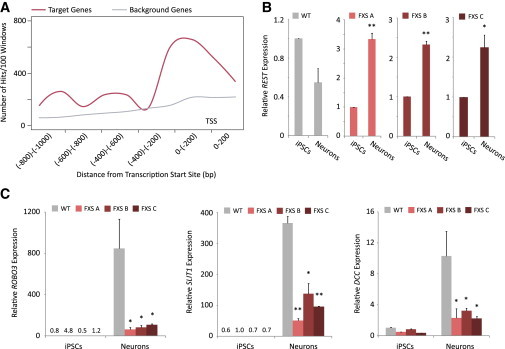
REST Regulates Neuron Differentiation and Axon Guidance Genes and Is Aberrantly Expressed in FXS-Derived Neurons
(A) Binding sites locations of REST on its target genes within the downregulated genes in FXS neurons as predicted by the Amadeus (Allegro/Amadeus) BPM v.1.0 software (http://acgt.cs.tau.ac.il/allegro/).
(B) REST mRNA levels were analyzed in iPSCs and neurons by qRT-PCR. Results show that in FXS-derived neurons from three independent patients (FXS-A, FXS-B, FXS-C), REST mRNA levels are upregulated in contrast to the downregulation seen in control (WT) neurons.
(C) REST target genes are upregulated during neural differentiation; however, their upregulated is limited in FX-derived neurons, as seen by qRT-PCR analysis.
qRT-PCR analyses were performed on three to four biological repeats; scale bars represent SE, ∗p < 0.05, ∗∗p < 0.005 using Student’s t test.
REST Fails to Undergo Downregulation as Differentiation Progresses in FXS-Derived Neurons
REST acts to suppress neural genes in nonneuronal tissues, and its expression levels must be downregulated as neural differentiation progresses (Ballas et al., 2005; Paquette et al., 2000; Schoenherr and Anderson, 1995). To investigate whether aberrant gene expression in FXS-derived neurons results from abnormal expression of REST during neural differentiation, we performed a qRT-PCR analysis on FXS-derived iPSCs, NPCs, and neurons. Our results show that REST mRNA levels increase during early differentiation of iPSCs into NPCs, both in control and FXS cells (Figures S2A and S2B). When differentiating the cells into neurons, REST mRNA levels decrease in control cells, as expected. In contrast, FXS-derived neurons fail to downregulate REST as differentiation progresses, resulting in a marked difference in REST mRNA levels between control and all FXS-derived neurons (Figure 2B). In order to verify a progressive differentiation process, we analyzed the FXS-derived NPCs and neurons for key neural markers by quantitative RT-PCR (qRT-PCR). Results show that FXS-derived NPCs express higher levels of NESTIN than neurons while neurons express higher levels of TUBB3 than NPCs (Figures S2C and S2D).
Our analysis of the downregulated genes in FXS-derived neurons using the KEGG pathway database revealed that some of the genes are key players in the axon guidance pathway. Genes in this pathway such as ROBO3, DCC, and SLIT1 were suggested to be direct targets of REST by large-scale chromatin immunoprecipitation assay or bioinformatically by the ENCODE project (Johnson et al., 2007; Myers et al., 2011). We have thus verified the differences in expression levels of the axon guidance genes in control and FXS-derived neurons using a qRT-PCR analysis. The analysis confirmed that the axon guidance genes are indeed significantly downregulated in all FXS lines-derived neurons compared with control neurons (Figure 2C). This sharp downregulation is not a result of failure to turn on the neural genes as differentiation progresses, but rather a result of suppression that occurs from high levels of the regulating repressor of these genes. This was verified by analyzing differences in expression of ROBO3, SLIT1, and DCC between iPSCs and neurons of FXS and control cells. Results show that expression of these axon guidance genes is upregulated in both control and FXS neurons compared with iPSCs. However, in control cells, the increase in expression following differentiation is very dramatic in contrast to FXS cells where the increase in expression after differentiation is subdued (Figure 2C). Furthermore, other neural markers such as NESTIN and TUBB3 show similar expression levels between control and FXS-derived neurons (Figures S2E and S2F).
FXS-Derived Neurons Exhibit Downregulation of Specific miRNAs
We next sought to decipher the association between FMRP and REST. As FMRP is associated with the miRNA machinery and REST is regulated by several brain specific miRNAs, we compared miRNA arrays from both control and FXS-derived neurons. When looking at brain-associated miRNAs, we could observe variations in the levels of some neural miRNAs. When comparing control and FXS-derived neurons, some miRNAs are present at the same levels, while others show either much lower or higher levels between the two cell types (Figure 3A). It was visible from the results, however, that overall, many miRNAs are downregulated in FXS-derived compared with control neurons (Figure 3B). Surprisingly, when we set a cutoff of 4-fold difference in mature miRNA levels, we could not detect any upregulated miRNAs associated with the FXS-derived neurons, and we were left only with miRNAs that are downregulated (Figure 3B).
Figure 3.
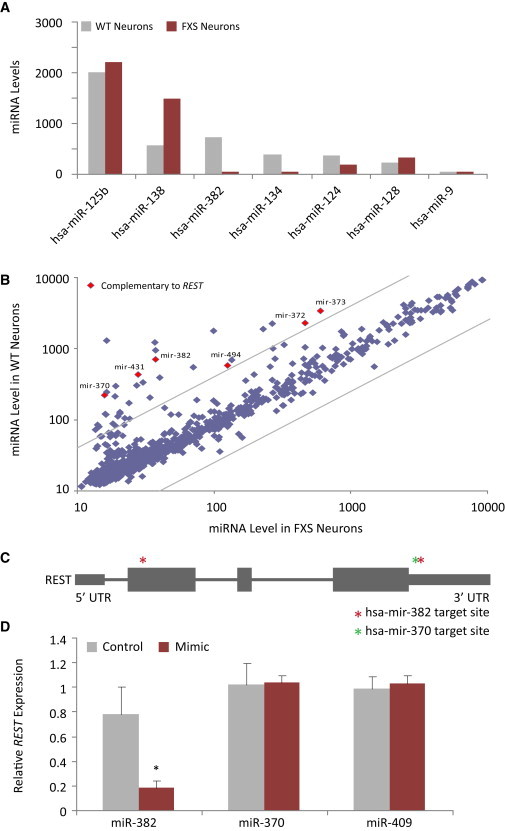
FXS Neurons Display Abnormal miRNAs Levels
(A) Analysis of expression of brain associated miRNAs in control (WT) and FXS neurons (columns represent average from two miRNA arrays of two biological repeats for each cell type [WT and FXS]).
(B) miRNAs arrays revealed significant differences in mature miRNAs levels between FXS-derived and control neurons (cutoff was set to 4-fold change). All miRNAs with target sites on the REST transcript are marked in red.
(C) Schematic representation of the REST gene and the predicted binding sites of hsa-mir-382 marked by red asterisks and hsa-mir-370 marked with a green asterisk.
(D) hsa-mir-382, hsa-mir-370, and hsa-mir-409 were transfected into iPSCs using mimic miRNAs. Cells were grown for 24 hr, harvested and RNA was purified. Only transfection of mimic-mir-382 was sufficient to significantly lower REST mRNA to 23% of its normal levels (three biological repeats; scale bars represent SE, ∗p < 0.05 using Student’s t test).
hsa-mir-382 Regulates REST mRNA Levels
From the group of miRNAs downregulated in FXS-derived neurons, we identified six miRNAs with target sites on the REST transcript (Figure 3B). We scanned the REST transcript for possible recognition sites for these miRNAs using the UCSC Genome Browser on Human February 2009 (GRCh37/hg19) Assembly using the miRcode miRNA sites track (UCSC miRcode) (Jeggari et al., 2012). Two of the six candidate miRNAs had two binding sites each, with at least one site at the 3′ UTR. One of those miRNAs was hsa-mir-382 that was previously associated with brain function and found to be expressed mainly in the brain (Mor et al., 2013). Hsa-mir-382 has two predicted binding sites on the REST transcript, one at the first exon and one at the 3′ UTR, as predicted by UCSC miRcode, and was found with high levels in control neurons and markedly low levels in FXS-derived neurons (Figures 3A–3C). To examine the association of hsa-mir-382 with REST, we transfected iPSCs with either mimic hsa-mir-382, mimic hsa-mir-370 that is also abundant in control compared with FXS-derived neurons and has one complementary site on the REST transcript (Figures 1B and 1C) or mimic hsa-mir-409 that behaves as the two former miRNAs but has no complementary sites on the REST transcript. qRT-PCR analysis of transfected iPSCs for REST mRNA levels shows that hsa-mir-382 was able to significantly decrease REST transcripts levels as opposed to the other two miRNAs, that did not exhibit any detectable differences (Figure 3D). After successful downregulation of REST in iPSCs by mimic hsa-mir-382, we aimed to see whether introduction of hsa-mir-382 could downregulate the high levels of REST in FXS-derived neurons and the effect it would have on the axon guidance genes. For this aim, we have again differentiated FXS-derived iPSCs into neurons. At day 25 of the differentiation process, the maturing neurons were transfected with the mimic hsa-mir-382. The transfected neurons were then analyzed for REST expression and REST protein content. It is apparent from the qRT-PCR results that introduction of mimic hsa-mir-382 for 30 hr into FXS-derived neurons caused a marked downregulation of REST (Figure 4A). Western blot analysis from protein extracts showed a downregulation of REST also at the protein level (Figure 4B). REST protein levels of control transfected neurons mimic transfected for 30 hr and mimic transfected for 45 hr show a progressive and marked decrease over time (Figure 4C). We next analyzed whether the downregulation of REST by the mimic hsa-mir-382 would affect its axon guidance target genes. We thus analyzed expression of neural genes by qRT-PCR in the 30 hr mimic-transfected neurons. The results clearly indicate significant upregulation of all three REST target axon guidance genes following overexpression of hsa-mir-382 (Figure 4D).
Figure 4.
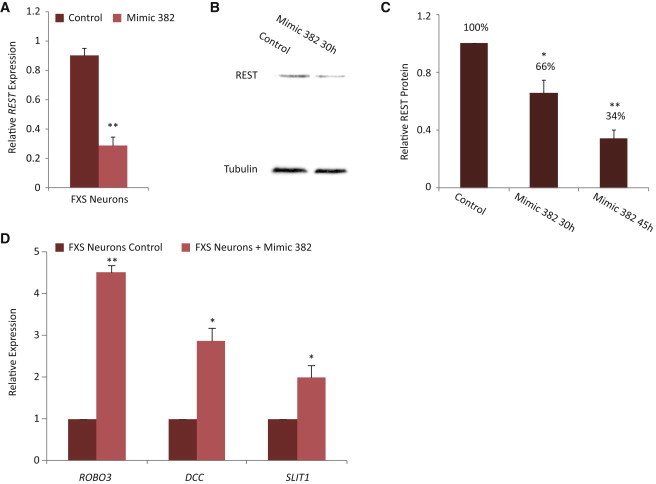
Correction of the Aberrant Molecular Phenotype by Overexpression of hsa-mir-382
Different FXS cell lines were transfected with mimic hsa-mir-382 or control transfection, cells were than harvested for either RNA or protein.
(A) qRT-PCR was performed to analyze REST expression and shows a marked downregulation of REST in mimic transfected cells after 30 hr.
(B) Western blot analysis of extract from the same time point shows lower levels of the REST protein.
(C) Quantification of western blots of extracts taken from control or mimic transfected cells after 30 and 45 hr show a progressive decrease of REST.
(D) REST axon guidance target genes show a significant upregulation after mimic treatment as observed by qRT-PCR.
All graphs represent three biological repeats; scale bars represent SE, ∗p < 0.05, ∗∗p < 0.005 using Student’s t test.
Discussion
FMRP plays a major role in the regulation of translation at the synapses. Several works have tried to scan for possible targets of FMRP that may explain the broad neurocognitive phenotype of FXS. Although many key neuronal proteins were found to be regulated by FMRP, there are still much data missing to explain the molecular pathology. In this work, we have chosen a different approach; rather than searching for FMRP targets, we have analyzed the global transcriptomic changes that occur in the absence of FMRP and linked these changes to the molecular phenotype of the disease. The strength of our model system enabled us to look at these changes at different developmental stages. FMRP may be expressed in human ESCs that carry a full mutation (Colak et al., 2014; Eiges et al., 2007). In this sense, the FXS-derived iPSCs are different, as the FMRP locus was found by us and others to be epigenetically resistant to the process of reprogramming (Alisch et al., 2013; Urbach et al., 2010). With this in mind, we have set to analyze the differences between FXS-derived and control iPSCs. We demonstrated that FXS and control derived iPSCs are highly similar, at the undifferentiated state (Figures 1A, 1B, and S1A). As FXS patients suffer from a neural pathology, we speculated that neurons derived from FXS-iPSCs would exhibit significant differences when compared with neurons derived from control iPSCs. We indeed found a large number of genes to be differentially expressed between the two groups (Figures 1C and S1A). The downregulated genes in FXS are mostly those related to neuron differentiation, axonogenesis, and the axon guidance pathway (Figures 1C and 1D). This result is in line with several other studies showing aberrations in neural development, abnormalities of dendritic spine morphologies, and deformities of growth cone development affecting axon guidance in the formation of FXS neurons (Antar et al., 2006; Bassell and Warren, 2008; Callan et al., 2010; Castrén et al., 2005; Comery et al., 1997; Egger et al., 2008; Irwin et al., 2001; Li et al., 2009; Luo et al., 2010; Tessier and Broadie, 2008).
Many of the genes that take part in these cellular and molecular processes were found to be regulated by REST. In fact, different studies aimed to identify REST target genes indicated that REST is involved in processes such as nervous system development, neuron projection, and axonal guidance signaling. Some of these studies have also suggested REST to play a part in glutamate receptor signaling (Bruce et al., 2004; Johnson et al., 2007; Satoh et al., 2013). This finding may in fact connect aberrant regulation of REST to the abnormal activity of the glutamate receptor signaling seen in FXS neurons (Dölen et al., 2007) or may cause an additive effect. It is becoming clear from recent studies that REST is a key regulator of proper neural differentiation and development, and any perturbation in the regulation of REST will eventually lead to abnormalities in creating the neural network (Ballas et al., 2005; Covey et al., 2012; Paquette et al., 2000). Constitutive expression of REST in differentiating neurons was found to disrupt neural gene expression and caused significantly higher frequencies of axon guidance errors but did not prevent neurogenesis (Paquette et al., 2000). As we could not detect differences in REST levels in NPCs, we believe that the abnormal regulation seen in FXS occurs at the stage of mature neuronal development and network formation (Figures S2A and S2B). In this sense, the molecular phenotypes seen in FXS-derived neurons mimic the molecular phenotypes seen in neurons expressing higher levels of REST and reinforce our finding of aberrant REST regulation in FXS cells.
As FMRP is involved in the miRNA machinery, we sought to explore the possibility that the regulation of REST by FMRP is mediated by miRNA levels in our iPSCs derived neurons. Global analysis of miRNAs expression in FXS cells and control cells revealed that at 4-fold cutoff we identify only miRNAs that are downregulated (and not upregulated) in FXS-derived neurons (Figure 3B). This finding may point to an important role for FMRP in the regulation of specific neural miRNAs. Of the candidate miRNAs identified, hsa-mir-382 was found to be enriched in the brain (Mor et al., 2013) and harbors two binding sites on the REST transcript (Figure 3C). Indeed, genetic manipulation of hsa-mir-382 affected REST expression in both iPSCs and FXS-derived neurons (Figures 3D and 4A–4C). Most importantly, overexpression of hsa-mir-382 was able to significantly upregulate the levels of the REST target axon guidance genes in FXS-derived neurons (Figure 4D). The specific role of FMRP in the maturation and function of neural miRNAs should be further studied in the future, as miRNAs are major posttranscriptional regulators affecting the levels of proteins, which are critical for proper neural differentiation and synaptic function.
In this work, we have shown the dramatic effect of the loss of FMRP on the gene expression profile of neurons. In the absence of FMRP, the neural hsa-mir-382 levels are decreased, preventing the differentiation-dependent downregulation of REST. The resulting higher levels of REST in FXS-derived neurons cause the suppression of neural genes important for proper axon development (Figure 5). We strengthen the importance of FMRP in the maturation and formation of the neural network through its interaction with the miRNA pathway and regulation of REST. Our work lays a foundation for identifying a biomarker for early detection of the syndrome in affected embryos and suggests several candidates for targeted therapy. Recent studies suggest that REST plays a key role in the pathological process of different human neurodegenerative diseases (González-Castañeda et al., 2013; Marullo et al., 2010; Zuccato et al., 2003). The role of REST in FXS should be furthered explored as well as the part that miRNAs play in the development of this pathology.
Figure 5.
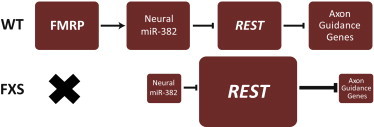
Schematic Representation of the Molecular Pathway Aberrant in FXS
In control (WT) neurons, FMRP is important for the regulation of the level of the mature hsa-mir-382. hsa-mir-382 in turn affects the regulation of REST during the maturation of the neurons, thus allowing the expression of the axon guidance genes. In FXS-derived neurons, FMRP is not produced; hsa-mir-382 levels are very low and insufficient for the downregulation of REST. REST levels do not decrease, resulting in the suppression of the axon guidance genes.
Experimental Procedures
Cell Culture
Cell culture and neuronal differentiation were previously described (Bar-Nur et al., 2012). We have differentiated iPSCs to NPCs according to a protocol published by Kim et al. (2010) with two inhibitors (Dorsomorphin and SB431542). At the end of the differentiation process, we stained the cells for NCAM1 and sorted only for positive cells using fluorescence-activated cell sorting. In this study, we have used two control cell lines, BJ-iPSCs 28 and BJ-iPSCs 94, and five different FXS-derived iPSC clones from three different patients: patient A with clones 47, 52, and 55; patient B with clone 40; and patient C with clone 2 (Bar-Nur et al., 2012; Urbach et al., 2010).
RNA Isolation and Reverse Transcription
RNA was isolated using PerfectPure RNA Cultured Cell Kit-50 (5 PRIME). One microgram of total RNA was used for reverse transcription reaction using ImProm-II reverse transcriptase (Promega). For sequencing and quantitative experiments, PCRs were performed with ReadyMix (Sigma); for overexpression experiments, PCR reactions used Herculase II Fusion DNA polymerase (Agilent Technologies). Quantitative real-time PCR was performed with 1 μg of RNA reverse transcribed to cDNA and TaqMan Universal Master Mix or SYBR Green qPCR Supermix (see the primer list in Table 1; Applied Biosystems) and analyzed with the 7300 real-time PCR system (Applied Biosystems).
Table 1.
Primers List
| Gene Name | Forward Primer | Reverse Primer |
|---|---|---|
| REST | ATTGGAATGGCCCTGCCTAA | CCAGTTAAGGCCACATTTGCC |
| ROBO3 | CGAGAGGAACCAAGATGACCCT | GCCAATTGAAATCGTGGAAACC |
| DCC | GGCAGACTTCCAGTTGCACTCT | CCCATGCCCCTGTGTTTATTAA |
| SLIT1 | GGAATCTGCCGCAAAAAGTCA | CACACTGAATCTCCTGGCCAA |
| GAPDH | AGCCACATCGCTCAGACACC | GTACTCAGCGGCCAGCATCG |
| NESTIN | ATCTGCAAACCCATCGGACTC | TGAGGCACCTTTTCTTCCTGG |
| TUBB3 | CCTCTTCTCACAAGTACGTGCC | AGGCCTGAAGAGATGTCCAAA |
| HOMER3 | AGATGCTGTTCAGAGGCAAAG | AGGCCATCATCAACAGCACTG |
DNA Microarray Analysis
Total RNA was extracted according to manufacturer’s protocol (Affymetrix). RNA was subjected to Human Gene 1.0 ST microarray platform (Affymetrix) analysis; washing and scanning were performed according to manufacturer’s protocol. Arrays were analyzed using Robust Multichip Analysis in the Affymetrix Expression Console.
miRNA Expression Analysis
Total RNA was extracted using MirVana miRNA isolation kit (Ambion) according to the manufacturer’s protocol. RNA was subjected to Human GeneChip miRNA array platform analysis (Affymetrix); washing and scanning were performed according to the manufacturer’s protocol. Arrays were analyzed using miRNA QC Tool.
Overexpression of miRNA
miRIDIAN miRNA Mimics by Thermo Scientific were transfected into control iPSCs or FXS-derived neurons using Lipofectamine 2000 by Invitrogen according to manufacturer protocol. At 30 or 45 hr after transfection, cells were harvested for RNA or lysed for protein (see the mimics sequences in Table 2).
Table 2.
miRIDIAN miRNA Mimic List
| mimic-mir-382 | GAAGUUGUUCGUGGUGGAUUCG |
| mimic-mir-370 | GCCUGCUGGGGUGGAACCUGGU |
| mimic-mir-409 | AGGUUACCCGAGCAACUUUGCAU |
| mimic-negative-control | UCACAACCUCCUAGAAAGAGUAGA |
Western Blot Analysis
Ten percent polyacrylamide gel was used for protein separation. The gel was transferred to a nitrocellulose membrane, and antibody hybridization and chemiluminescence were performed according to the standard procedures. The primary antibodies used in this analysis were mouse anti-NRSF sc-374611 (SANTA CRUZ) and mouse antitubulin (Sigma). HRP-conjugated antirabbit and antimouse secondary antibodies were obtained from Jackson ImmunoResearch Laboratories. Western blot quantification was performed using the FUJIFILM Image Gauge software.
Functional Annotations and Motif Search
Functional annotations were achieved by subjecting differentially expressed genes to the DAVID functional annotation clustering tool (http://david.abcc.ncifcrf.gov/). Motifs were searched the same way by using both the DAVID functional annotation clustering tool (http://david.abcc.ncifcrf.gov/), the Amadeus BPM v.1.0 software (http://acgt.cs.tau.ac.il/allegro/), and PRIMA analysis (http://acgt.cs.tau.ac.il/prima/). Pathways search was performed using the KEGG (http://www.genome.jp/kegg/).
Acknowledgments
We thank members of the Stem Cell Unit at the Hebrew University and especially Uri Weissbein and Tamar Golan-Lev for their assistance with graphical design of the figures and Dr. Ofra Yanuka for her technical assistance and many advises. N.B. is the Herbert Cohn Chair in Cancer Research. This work was partially supported by a Hoffmann La Roche-Yissum Collaboration grant and the Israel Science Foundation-Morasha Foundation (grant number 1252/12).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
The Gene Expression Omnibus (GEO) accession number for the data reported in this paper is GSE62721.
Supplemental Information
References
- Alisch R.S., Wang T., Chopra P., Visootsak J., Conneely K.N., Warren S.T. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med. Genet. 2013;14:18. doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar L.N., Li C., Zhang H., Carroll R.C., Bassell G.J. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol. Cell. Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Ashley C.T., Jr., Wilkinson K.D., Reines D., Warren S.T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bar-Nur O., Caspi I., Benvenisty N. Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J. Mol. Cell Biol. 2012;4:180–183. doi: 10.1093/jmcb/mjs007. [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Warren S.T. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A., McMillan E., Wallace K., Tubon T.C., Jr., Capowski E.E., Svendsen C.N. Normal Neurogenesis but Abnormal Gene Expression in Human Fragile X Cortical Progenitor Cells. Stem Cells Dev. 2008;17:107–117. doi: 10.1089/scd.2007.0073. [DOI] [PubMed] [Google Scholar]
- Boyle L., Kaufmann W.E. The behavioral phenotype of FMR1 mutations. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:469–476. doi: 10.1002/ajmg.c.30277. [DOI] [PubMed] [Google Scholar]
- Bruce A.W., Donaldson I.J., Wood I.C., Yerbury S.A., Sadowski M.I., Chapman M., Göttgens B., Buckley N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan M.A., Zarnescu D.C. Heads-up: new roles for the fragile X mental retardation protein in neural stem and progenitor cells. Genesis. 2011;49:424–440. doi: 10.1002/dvg.20745. [DOI] [PubMed] [Google Scholar]
- Callan M.A., Cabernard C., Heck J., Luois S., Doe C.Q., Zarnescu D.C. Fragile X protein controls neural stem cell proliferation in the Drosophila brain. Hum. Mol. Genet. 2010;19:3068–3079. doi: 10.1093/hmg/ddq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrén M., Tervonen T., Kärkkäinen V., Heinonen S., Castrén E., Larsson K., Bakker C.E., Oostra B.A., Akerman K. Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. USA. 2005;102:17834–17839. doi: 10.1073/pnas.0508995102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy A.A., Myers M., Hannon G.J., Hammond S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.F., Paquette A.J., Anderson D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Warren S.T., Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat. Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- Coffee B., Zhang F., Ceman S., Warren S.T., Reines D. Histone modifications depict an aberrantly heterochromatinized FMR1 gene in fragile x syndrome. Am. J. Hum. Genet. 2002;71:923–932. doi: 10.1086/342931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak D., Zaninovic N., Cohen M.S., Rosenwaks Z., Yang W.-Y., Gerhardt J., Disney M.D., Jaffrey S.R. Promoter-Bound Trinucleotide Repeat mRNA Drives Epigenetic Silencing in Fragile X Syndrome. Science. 2014;343:1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery T.A., Harris J.B., Willems P.J., Oostra B.A., Irwin S.A., Weiler I.J., Greenough W.T. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc. Natl. Acad. Sci. USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey M.V., Streb J.W., Spektor R., Ballas N. REST regulates the pool size of the different neural lineages by restricting the generation of neurons and oligodendrocytes from neural stem/progenitor cells. Development. 2012;139:2878–2890. doi: 10.1242/dev.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D., Lutz Y., Rouyer N., Bellocq J.P., Mandel J.L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dölen G., Osterweil E., Rao B.S.S., Smith G.B., Auerbach B.D., Chattarji S., Bear M.F. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Neilson J.R., Foster K.A., Wang C.F., Seeburg D.P., Batterton M.N., Tada T., Dolan B.M., Sharp P.A., Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Chell J.M., Brand A.H. Insights into neural stem cell biology from flies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:39–56. doi: 10.1098/rstb.2006.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiges R., Urbach A., Malcov M., Frumkin T., Schwartz T., Amit A., Yaron Y., Eden A., Yanuka O., Benvenisty N., Ben-Yosef D. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Galvez R., Smith R.L., Greenough W.T. Olfactory bulb mitral cell dendritic pruning abnormalities in a mouse model of the Fragile-X mental retardation syndrome: further support for FMRP’s involvement in dendritic development. Brain Res. Dev. Brain Res. 2005;157:214–216. doi: 10.1016/j.devbrainres.2005.03.010. [DOI] [PubMed] [Google Scholar]
- González-Castañeda R.E., Sánchez-González V.J., Flores-Soto M., Vázquez-Camacho G., Macías-Islas M.A., Ortiz G.G. Neural restrictive silencer factor and choline acetyltransferase expression in cerebral tissue of Alzheimer’s Disease patients: A pilot study. Genet. Mol. Biol. 2013;36:28–36. doi: 10.1590/S1415-47572013000100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V. REST and the RESTless: in stem cells and beyond. Future Neurol. 2009;4:317–329. doi: 10.2217/fnl.09.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson O. Stem cells have different needs for REST. PLoS Biol. 2008;6:e271. doi: 10.1371/journal.pbio.0060271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin S.A., Patel B., Idupulapati M., Harris J.B., Crisostomo R.A., Larsen B.P., Kooy F., Willems P.J., Cras P., Kozlowski P.B. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M.C., Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggari A., Marks D.S., Larsson E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics. 2012;28:2062–2063. doi: 10.1093/bioinformatics/bts344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Zarnescu D.C., Ceman S., Nakamoto M., Mowrey J., Jongens T.A., Nelson D.L., Moses K., Warren S.T. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Khandjian E.W., Huot M.-E., Tremblay S., Davidovic L., Mazroui R., Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc. Natl. Acad. Sci. USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-S., Lee J.S., Leem J.W., Huh Y.J., Kim J.Y., Kim H.-S., Park I.-H., Daley G.Q., Hwang D.-Y., Kim D.-W. Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 2010;6:270–281. doi: 10.1007/s12015-010-9138-1. [DOI] [PubMed] [Google Scholar]
- Li C., Bassell G.J., Sasaki Y. Fragile X Mental Retardation Protein is Involved in Protein Synthesis-Dependent Collapse of Growth Cones Induced by Semaphorin-3A. Front Neural Circuits. 2009;3:11. doi: 10.3389/neuro.04.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Shan G., Guo W., Smrt R.D., Johnson E.B., Li X., Pfeiffer R.L., Szulwach K.E., Duan R., Barkho B.Z. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo M., Valenza M., Mariotti C., Di Donato S., Cattaneo E., Zuccato C. Analysis of the repressor element-1 silencing transcription factor/neuron-restrictive silencer factor occupancy of non-neuronal genes in peripheral lymphocytes from patients with Huntington’s disease. Brain Pathol. 2010;20:96–105. doi: 10.1111/j.1750-3639.2008.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor E., Kano S., Colantuoni C., Sawa A., Navon R., Shomron N. MicroRNA-382 expression is elevated in the olfactory neuroepithelium of schizophrenia patients. Neurobiol. Dis. 2013;55:1–10. doi: 10.1016/j.nbd.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R.M., Stamatoyannopoulos J., Snyder M., Dunham I., Hardison R.C., Bernstein B.E., Gingeras T.R., Kent W.J., Birney E., Wold B., ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I., Mercaldo V., Boyl P.P., Eleuteri B., Zalfa F., De Rubeis S., Di Marino D., Mohr E., Massimi M., Falconi M. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Ooi L., Wood I.C. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Paquette A.J., Perez S.E., Anderson D.J. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O., Mulle J.G., Warren S.T. The pathophysiology of fragile x syndrome. Annu. Rev. Genomics Hum. Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- Plante I., Davidovic L., Ouellet D.L., Gobeil L.-A., Tremblay S., Khandjian E.W., Provost P. Dicer-derived microRNAs are utilized by the fragile X mental retardation protein for assembly on target RNAs. J. Biomed. Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M.R., Bray S.M., Warren S.T. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Satoh J., Kawana N., Yamamoto Y. ChIP-Seq Data Mining: Remarkable Differences in NRSF/REST Target Genes between Human ESC and ESC-Derived Neurons. Bioinform. Biol. Insights. 2013;7:357–368. doi: 10.4137/BBI.S13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C.J., Anderson D.J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Stefani G., Fraser C.E., Darnell J.C., Darnell R.B. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Tessier C.R., Broadie K. Drosophila fragile X mental retardation protein developmentally regulates activity-dependent axon pruning. Development. 2008;135:1547–1557. doi: 10.1242/dev.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A., Bar-Nur O., Daley G.Q., Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk A.J.M.H., Pieretti M., Sutcliffe J.S., Fu Y.-H., Kuhl D.P.A., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Wang T., Bray S.M., Warren S.T. New perspectives on the biology of fragile X syndrome. Curr. Opin. Genet. Dev. 2012;22:256–263. doi: 10.1016/j.gde.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


