Summary
When pluripotency factors are removed, embryonic stem cells (ESCs) undergo spontaneous differentiation, which, among other lineages, also gives rise to cardiac sublineages, including chamber cardiomyocytes and pacemaker cells. Such heterogeneity complicates the use of ESC-derived heart cells in therapeutic and diagnostic applications. We sought to direct ESCs to differentiate specifically into cardiac pacemaker cells by overexpressing a transcription factor critical for embryonic patterning of the native cardiac pacemaker (the sinoatrial node). Overexpression of SHOX2 during ESC differentiation upregulated the pacemaker gene program, resulting in enhanced automaticity in vitro and induced biological pacing upon transplantation in vivo. The accentuated automaticity is accompanied by temporally evolving changes in the effectors and regulators of Wnt signaling. Our findings provide a strategy for enriching the cardiac pacemaker cell population from ESCs.
Highlights
-
•
SHOX2 accentuates the molecular profile of pacemaker cells in differentiating ESCs
-
•
SHOX2 increases the frequency and rate of spontaneously active cardiac derivatives
-
•
SHOX2-overexpressing EBs function as biopacemakers when transplanted in vivo
-
•
Wnt signaling underlies SHOX2-mediated pacemaker cell specification
Cho and colleagues show that heterologous SHOX2 expression boosts the differentiation of embryonic stem cells to cardiac pacemaker cells from embryonic stem cells by enhancing pacemaker cell-specific electrophysiology. Implantation of SHOX2-overxpressing embryoid bodies to the rat heart in vivo creates biological pacing. The SHOX2-facilitated pacemaker cell phenotype correlates with specific Wnt modulation, suggesting that Wnt may be a negotiator for cardiac subtype specification.
Introduction
Since their first derivation (Thomson et al., 1998), embryonic stem cells (ESCs) have been validated to faithfully recapitulate early cardiogenesis (Boheler et al., 2002; Van Vliet et al., 2012) and touted for their potential as an unlimited source of de novo cardiomyocytes for replacement of diseased myocardium (Kehat et al., 2001). While the most commonly pursued therapeutic goal has been to boost contractile function, ESC-derived cardiac cells may also be useful as alternatives to electronic pacemakers (Cho and Marbán, 2010); we and others have exploited the automaticity of ESC-derived cardiomyocytes to create biological pacemakers (Kehat et al., 2004; Xue et al., 2005). The risk of teratoma may be diminished by technical refinements to increase general yield of ESC-derived cardiomyocytes (Dubois et al., 2011; Kattman et al., 2011; Nunes et al., 2013) and by attaining a “pure” cardiomyocyte population postdifferentiation (Dubois et al., 2011; Hattori et al., 2010). An outstanding issue, however, remains in the innate heterogeneity of ESC-derived (or any pluripotent stem cell) cardiac cells. The action potential (AP) profiles of de novo cardiomyocytes vary considerably from ventricular/atrial myocyte-like to nodal/Purkinje-like (He et al., 2003; Kolossov et al., 2005; Maltsev et al., 1993; Zhang et al., 2009). Such heterogeneity could result in unpredictable biological pacemakers, as reported in a subset of spontaneously contracting embryoid bodies (EBs) in which the beating rate either ceased or accelerated over time (Mandel et al., 2012).
We set out to develop a way to instruct the ESCs to differentiate into a cardiac pacemaker subtype with a factor relevant to embryonic pacemaker development. Native cardiac pacemaker cells are anatomically confined in the sinoatrial node (SAN), a diminutive structure comprising just a few thousand genuine pacemaker cells (Bleeker et al., 1980). During embryonic development, cardiac pacemaker cells originate from a subset of progenitors distinct from the first (marked by Nkx2.5) and second (marked by Isl1) heart fields not only in their genetic makeup (Christoffels et al., 2010; Wiese et al., 2009), but also in their anatomic location (Bressan et al., 2013). However, an area of Hcn4-positive primordial SAN is reported to express Isl1 (Mommersteeg et al., 2007), suggesting that second heart field progenitors may also contribute to the developing SAN. We have recently demonstrated that postnatal re-expression of an embryonic transcription factor, Tbx18, converts ventricular cardiomyocytes to pacemaker cells, recapitulating morphological as well as electrophysiological hallmarks of genuine SAN pacemaker cells (Kapoor et al., 2013). Elsewhere, transgenic overexpression of Tbx3 has been shown to elicit ectopic rhythm in mouse atrial myocardium (Bakker et al., 2012). Noting the powerful capacity of embryonic transcription factors in determining the fate of cardiac cell subtype, we hypothesized that overexpression of a SAN-specific transcription factor may steer ESC differentiation toward pacemaker cell subtype. Here, we report that heterologous expression of SHOX2 during early stages of mouse ESC (mESC) differentiation strongly favors a SAN-specific gene program, leading to enhanced pacemaker cell specification. The differentiated cells exhibit greater automaticity in vitro and perform biological pacemaker function when injected into the rat heart in vivo.
Results
Shox2 Is Specific to Embryonic Development of the Cardiac SAN
mESCs were differentiated to form EBs by culturing them in suspension media for 6 days and then transferring them to adherent media (Wobus et al., 1991). The EBs were analyzed at three time points, based on the time course of electrophysiological maturation of mESC-derived cardiomyocytes (Maltsev et al., 1994): 4 days after transfer to adherent culture as an early time point of differentiation (D6+4), 7 days afterward (D6+7) as the mid phase of differentiation, and 14 days afterward (D6+14) as the terminal phase of differentiation (Figure 1A). A few transcription factors figure prominently in embryonic development of the SAN, notably the T box transcription factors Tbx18 and Tbx3 (Wiese et al., 2009), as well as the homeodomain transcription factor Shox2 (Espinoza-Lewis et al., 2009). We reasoned that overexpression of one of these transcription factors could steer ESCs to differentiate into cardiac pacemaker cells. To this end, we sought to identify a gene highly specific to the developing mouse SAN. Quantitative measurements of the mRNA levels of these transcription factors reveal that Shox2 expression is most specific to, and significant in, the SAN compared with the right atrium (RA), left atrium (LA), and left ventricle (LV) of the mouse heart at mouse embryonic day (ED) 18 (Figure 1B, top). The SAN-specific expression of Shox2 closely follows that of Hcn4, a marker of SAN pacemaker cells. Initially, we anticipated that Tbx18 expression might be the most specific to the SAN since mice deficient for Tbx18 fail to form sinus horns (Christoffels et al., 2006), bolstered by our recent demonstration that Tbx18 re-expression converts ordinary ventricular myocytes to native SAN-like induced pacemaker cells in vitro and in vivo (Kapoor et al., 2013). Yet, the present data indicate that Tbx18 is comparably expressed between the SAN and all major chambers, including the right atrium (RA), left atrium (LA), and left ventricle (LV) at ED 18 (Figure 1B, top). This may be due to the abundance of Tbx18+ proepicardial progenitor cells in the embryonic heart (Cai et al., 2008; Christoffels et al., 2009). Likewise, our data indicate that Tbx3 is comparably expressed in the SAN and LV (Figure 1B), which may be due to its expression in ventricular conduction system (Bakker et al., 2008). Analogous results were obtained at an earlier embryonic development time point (ED 15.5; Figure 1B, bottom). Guided by these insights, we selected Shox2 as the most SAN-specific transcription factor so as to boost SAN pacemaker cell-specific differentiation of the ESCs.
Figure 1.
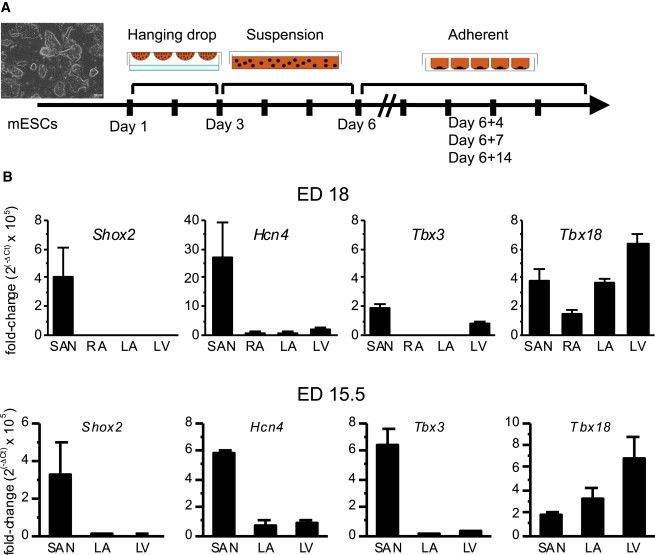
Shox2 Is Specific to the Developing Mouse SAN
(A) A schematic diagram of cardiac differentiation protocol. mESCs were dissociated into single cells on day 1(D1) and cultured in suspension for 2 days in hanging drops and then 3 days on a nonadhering culture plate. On D6, the cells were plated on an adherent plate. The differentiating cells were characterized 4, 7, and 14 days from D6.
(B) Transcript expression levels of the transcription factors that govern cardiac pacemaker cell specification were quantified by real-time RT-PCR from microsurgically isolated SAN tissues from ED 18 mouse hearts (top). The bottom illustrates the gene expression level at ED 15.5. Expression analysis of RA was not performed at ED 15.5 because the tissue was too small to be reliably separated from the SAN. Purity of the SAN preparation is validated by robust Hcn4 expression limited to the SAN (second from left). Data are represented as mean ± SEM. All experiments were performed in three independent biological replicates.
Exogenous SHOX2 Expression Activates Endogenous Shox2
We set out to facilitate the pacemaker-specific gene program at a time when endogenous Shox2 expression is low, from D3 to D7 (Figure 2A). To this end, we transduced the EBs with an adenoviral vector expressing human SHOX2 at three time points (D3, D6, and D6+1). The use of human SHOX2 allows specific transcript detection of endogenous mouse Shox2 from exogenous human SHOX2. Human SHOX2 protein shares 98% amino acid sequence homology with mouse SHOX2 (Figure S1 available online), signifying likely bioactivity of human SHOX2 in a mouse context. The somatic gene transfer resulted in rapid expression of SHOX2, which waned steeply with time (Figure 2B), as expected from the transience of adenoviral vector-mediated somatic gene transfer (Michou et al., 1997; Tripathy et al., 1996). Importantly, exogenous SHOX2 expression increased endogenous Shox2 expression throughout the differentiation phases (Figure 2C), suggesting stimulation of the pacemaker cell gene expression program.
Figure 2.
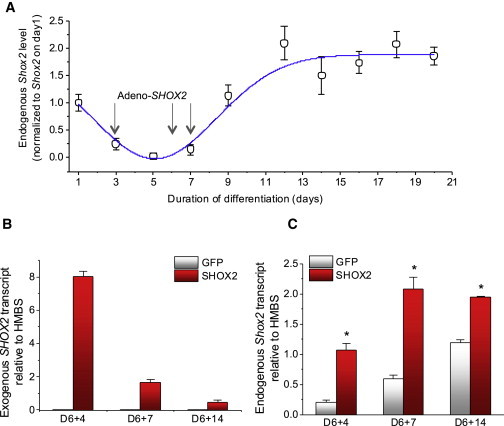
Exogenous Expression of Human SHOX2 Increases Endogenous Shox2 Expression
(A) Quantitative measurements of endogenous mouse Shox2 expression throughout the differentiation stages indicate that its expression is at the lowest during the first 6 days of differentiation. In SHOX2-treated group, the differentiating cells are transduced with an adenoviral vector expressing human SHOX2 on D3, D6, and D6+1 (D7) in order to maximize adenoviral somatic gene transfer. Experiments were performed on six independent mouse ESC cultures (i.e., six biological replicates).
(B) Human SHOX2-specific, quantitative PCR measurements of SHOX2 over time. No PCR product was detected in the GFP-EB group, indicating the specificity of human SHOX2 PCR primers.
(C) The level of endogenous Shox2 expression is higher in the SHOX2-EB group compared with control in all three time points. Data are represented as mean ± SEM. All experiments were performed on three independent biological replicates.
See also Figure S1.
SHOX2-EBs Exhibit Greater Automaticity with Concurrent Upregulation of Electrogenic Gene Program
We examined genes that play decisive roles for spontaneous diastolic depolarization as well as genes that underlie the propagation of electrical signals from cell to cell. Hyperpolarization-activated cyclic nucleotide-gated channel 4 (HCN4) is the molecular correlate of If in the SAN, which directly contributes to the linear rise of spontaneous depolarization during phase 4 of an AP (DiFrancesco, 2010; Yamamoto et al., 2006). Immunostaining of the whole EBs demonstrates that SHOX2 expression heightens HCN4 channel expression compared with control (Figure 3A). Total HCN4 protein expression was twice higher in SHOX2-EBs compared with control throughout the stages of differentiation (Figure 3B, left and middle). Na+-Ca2+exchanger (NCX) couples spontaneous intracellular Ca2+ release events to voltage oscillations in SA nodal pacemaker cells (Lakatta and DiFrancesco, 2009). The NCX1 protein level increased 4-fold in SHOX2-EBs at the later stages of differentiation (Figure 3B, right; Figure S2).
Figure 3.
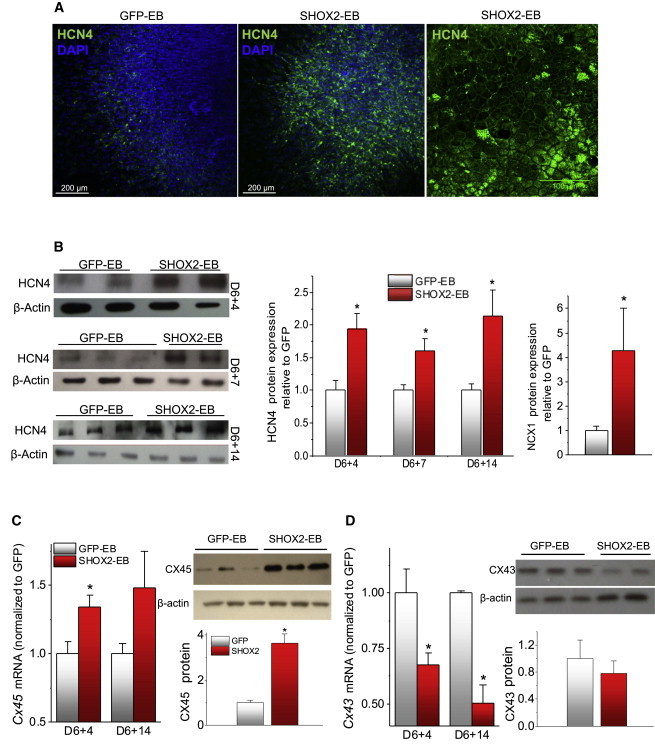
Changes to Ion Channel and Gap Junction Profiles in SHOX2-EBs Favor Automaticity
(A) Immunostaining images of GFP-EB (left) and SHOX2-EB (middle and right) with an antibody against HCN4 channel proteins. DAPI (blue), nuclear staining.
(B) Quantitative measurements of HCN4 (left and middle) and NCX1 (right) protein levels. D6+14 for NCX1.
(C) Transcript (left) and protein (right) level measurements of Cx45 in GFP-EBs and SHOX2-EBs. Protein level was determined with D6+14 samples.
(D) Transcript (left) and protein (right) level measurements of Cx43 in GFP-EBs and SHOX2-EBs. The protein level was determined with D6+14 samples. Data are represented as mean ± SEM. All experiments were performed on three independent biological replicates.
See also Figures S2 and S3.
The space constant at the core of the SAN is significantly lower than that of the neighboring atrial myocardium or the ventricles (<500 μm versus 1–2 mm, respectively) (Bonke, 1973). The high intercellular electrical resistance in the SAN allows its automaticity to propagate into the right atrium, which due to its highly hyperpolarized membrane potential would otherwise dissipate the electrogenic signals from the SAN (Joyner and van Capelle, 1986). The weak electrical coupling in the SAN pacemaker cells is mediated by a low-conductance gap junction protein, CX45, while the chamber myocardium predominantly expresses a large-conductance gap junction, CX43 (Jansen et al., 2010). SHOX2 overexpression increased the transcript level of Gja7, the gene that encodes CX45, throughout the differentiation phases. This is complemented by an ∼4-fold increase in CX45 protein expression (Figure 3C). Conversely, SHOX2 overexpression decreased CX43 expression at the transcript (33% ± 5%, p < 0.05) and protein (30% ± 10%, p < 0.05) levels compared with control (Figure 3D). In order to gauge the extent of total cardiac myocyte differentiation, we examined the expression levels of pan-cardiac myocyte markers, cardiac troponin T (Tnnt2), and cardiac actin (Actc1), as well as Nkx2-5, a negative marker of SAN but positive for ventricular and atrial cardiomyocytes. Quantitative RT-PCR data demonstrate that the expression level of Nkx2-5 is lower in SHOX2-EBs, while that of the pan-cardiac myocyte genes remains similar (Figure S3). The data indicate that the total cardiac differentiation efficiency remains similar in SHOX2-EBs and EBs transduced with a green fluorescent protein (GFP-EBs). Furthermore, the increased automaticity in SHOX2-EBs is likely to be a direct result of heightened pacemaker cell differentiation at the expense of diminished ventricular cardiomyocyte content. Thus, exogenous SHOX2 expression upregulates a number of electrophysiological hallmarks of SAN pacemaker cells during cardiac differentiation of ESCs.
To quantify the automaticity of SHOX2-EBs, we scored the number of spontaneously beating EBs and the number of individual beating foci in each EB. At the early stage of differentiation, the majority of SHOX2-EBs beat spontaneously, while the majority of GFP-EBs were quiescent (83% ± 7% versus 15% ± 6%, respectively, n = 7 independent experiments; Figure 4A, left). Over time, most of the GFP-EBs eventually began to beat (Figure 4A, left). Still, SHOX2-EBs exhibited more than twice the number of spontaneously beating foci within each beating EB compared with control, a pattern that persisted throughout differentiation (Figure 4A, right). The beating rates of the EBs were analyzed by recording extracellular field potentials (FPs) from the EBs on multielectrode arrays (MEAs) (Figure 4B). The beating rates of SHOX2-EBs were similar to those of control at the beginning of differentiation and then became significantly faster compared with control beyond the mid phase of differentiation (Figures 4C and 4D). Taken together, the data demonstrate that SHOX2-EBs develop greater numbers of pacemaker foci with faster beating rates and that the increased automaticity persists throughout differentiation.
Figure 4.
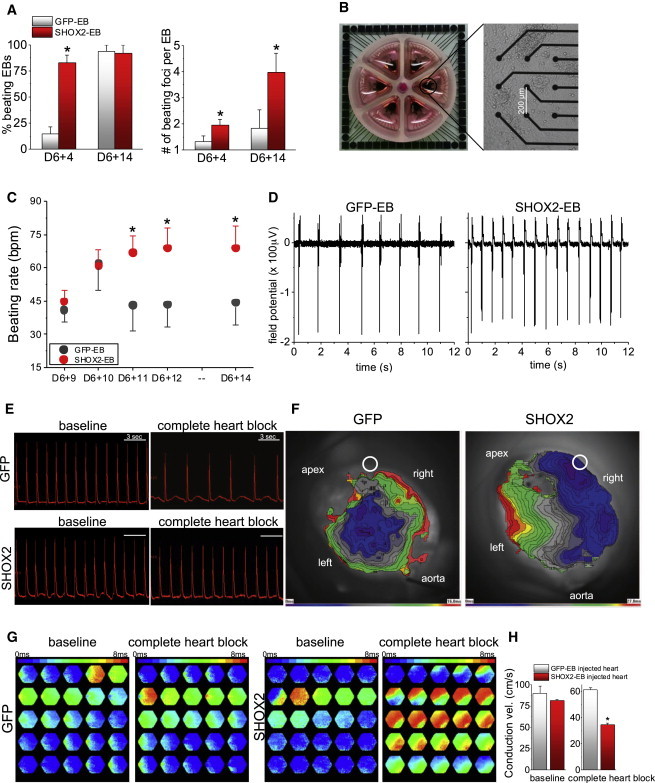
SHOX2-EBs Exhibit Intensified Automaticity In Vitro and Function as Biological Pacemakers when Implanted In Vivo
(A) The percentages of spontaneously contracting EBs were measured at D6+4 and D6+14 (left) by direct visualization. More than one beating focus could be detected in some EBs, and thus, the number of spontaneously beating foci per EB was measured at the same time points (right).
(B) The spontaneous beating rates were measured by culturing the EBs in MEAs and measuring the extracellular FPs from the GFP-EBs and SHOX2-EBs. Shown is a six-well MEA (left) in which two to three EBs were cultured per well (right).
(C) The average beating rates were measured at five time points by recording the spontaneous FPs for 20 min at 37°C in normal Tyrode’s (n > 10 for each data point).
(D) Representative raw FP traces are shown for a GFP-EB and a SHOX2-EB at D6+14.
(E) Whole-heart optical mapping recordings were performed on the rat hearts injected with GFP-EBs or SHOX2-EBs in vivo 2–3 days prior to the heart harvest. Raw AP traces reported as di-8-ANNEPS signal from one photodiode (123) are shown before (baseline) and after complete heart block. Upon complete heart block, the hearts injected with SHOX2-EBs demonstrated significantly higher heart rate compared to control (right).
(F) Optically recorded isochronal maps superimposed on the whole-heart injected with GFP-EBs (left) or SHOX2-EBs (right). A white circle indicates the EB implantation site, and the aorta is positioned at the bottom of the image. The anterior side of the heart was positioned to face the CMOS camera. Note the differences in the time scale of the isochronal map; 16 and 27 ms for end-to-end AP propagation for GFP-heart and SHOX2-heart, respectively.
(G) Frame-by-frame analysis of the optical mapping exhibiting AP propagation. The data illustrate slower AP propagation upon complete heart block in the hearts injected with SHOX2-EBs compared with control. Each hexagonal frame, recorded with a photodiode array, measures to a small area of about 8 mm2 on the heart.
(H) The conduction velocity of SHOX2-EB injected hearts was significantly slower than that from the control heart upon complete heart block. Data are represented as mean ± SEM. All experiments were performed on three independent biological replicates.
SHOX2-EBs Function as a Biological Pacemaker after Transplantation In Vivo
Motivated by the greater automaticity of the SHOX2-EBs, we tested whether SHOX2-EBs could provide biological pacemaker function when implanted in vivo. We and others have previously demonstrated that spontaneously beating human ESC-derived EBs could electrically couple with and drive the beating of host myocardium upon implantation in vivo (Kehat et al., 2004; Xue et al., 2005). In those studies, spontaneously beating areas of several EBs were microsurgically dissected prior to injection so as to remove nonbeating areas, thereby maximizing the proportion of beating cardiac tissue to nonbeating cells. Here, we tested whether the increased automaticity in SHOX2-EBs suffices to provide biological pacemaker function even without microsurgical removal of nonbeating cells. To this end, 10–12 spontaneously beating GFP-EBs or SHOX2-EBs were randomly selected at D6+4 and directly injected into the left ventricular apex of the rat heart in vivo. Two to 4 days after in vivo transplantation, the heart was isolated and retrogradely perfused so that its electrical activity could be optically mapped. In sinus rhythm, hearts injected with either GFP-EBs or SHOX2-EBs exhibited similar heart rates at room temperature (Figure 4E, “baseline”). Then complete heart block was implemented by surgically disrupting both the SA and atrioventricular nodes to unleash potential biological pacemaker activity. Afterward, control hearts injected with GFP-EBs exhibited slow junctional rhythms of 34 ± 2 beats per minute (bpm) (n = 4). In contrast, the SHOX2-EB-injected hearts exhibited a significantly higher ventricular rate (44 ± 3 bpm, n = 4, p < 0.05). Since the SHOX2-EBs were injected in the ventricular myocardium, heartbeats initiated from the injection site are expected to propagate more slowly than a junctional rhythm, which travels via the fast secondary conduction system. Under sinus rhythm, conduction velocities from hearts implanted with SHOX2-EBs were comparable to those from the hearts injected with GFP-EBs, with rapid spreading of the depolarization wave front. After the induction of complete heart block, conduction velocity slowed significantly in the SHOX2-EB-injected hearts, as anticipated for a wave front propagating through nonspecialized myocardium (Figures 4F and 4G; Movies S1 and S2). In contrast, the conduction velocity of GFP-EB-injected hearts did not change in complete heart block (Figure 4H, bar graphs). Electrical coupling between the donor and the host myocardium is evidenced by the presence of gap junction proteins at the interface between SHOX2-EBs and the rat ventricular myocardium (Figure S4). Taken together, the data indicate that SHOX2-EBs, but not GFP-EBs, function as biological pacemakers when the EBs were randomly selected and injected into the left ventricular apex of the rat heart.
Electrophysiology of Single Cells Isolated from SHOX2-EBs
In order to investigate the pacemaker phenotype of SHOX2-EBs at the single-cell level, we characterized APs from freshly isolated, spontaneously beating cells from SHOX2-EBs or GFP-EBs. The majority (62%) of single cells isolated from SHOX2-EBs exhibit a pacemaker-like AP profile, while 74% of the single cells isolated from GFP-EBs exhibit ventricular/atrial-like APs (Figures 5A and 5B). Single cells from SHOX2-EBs also fired APs at a higher rate than control cells (Figure 5C, left). The mean amplitude of spontaneous APs was lower in SHOX2-EB cells (Figure 5C, second from left), consistent with the lower INa density in genuine SAN pacemaker cells (Kapoor et al., 2013). Action potential duration (APD) was shorter, and each AP was preceded by faster phase 4 depolarization in SHOX2-EB cells compared with control (Figure 5C, right two panels). The density of If, a key contributor to spontaneous phase 4 depolarization (Lakatta and DiFrancesco, 2009), was significantly higher in SHOX2-EB cells compared with control (Figures 5D and 5E). Thus, SHOX2-EB cells more often display SAN-like AP parameters. Furthermore, the spontaneously beating single cells from both groups responded predictably to adrenergic stimulation and muscarinic challenge (Figures 5F and 5G), demonstrating their responsiveness to neurohumoral input.
Figure 5.
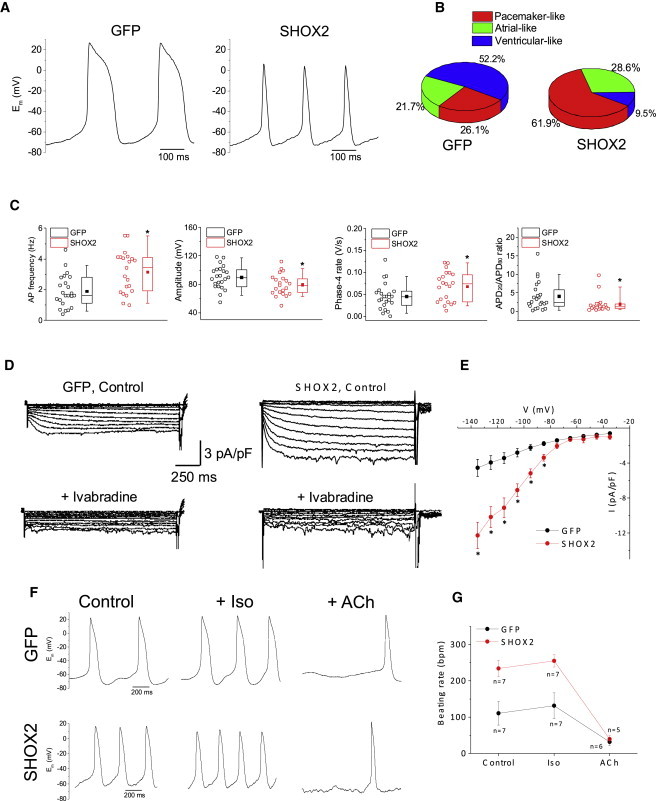
Single-Cell Electrophysiology of SHOX2-EBs Demonstrate SAN Pacemaker Cell-like Electrophysiology
(A) Representative raw traces of APs recorded from spontaneously-beating single cells from GFP-EBs (left) or SHOX2-EBs (right).
(B) Percentage of cells with pacemaker-like, atrial-like, or ventricular-like APs in GFP (n = 23) and SHOX2 (n = 21) groups.
(C) Comparison of AP parameters (from left to right): spontaneous beating rate, amplitude of the AP upstroke, spontaneous phase 4 diastolic depolarization rate, and ratio of AP duration at 20% repolarization (APD20) to 80% repolarization (APD80). ∗p < 0.05.
(D) Representative HCN ionic currents (If) recorded from a GFP-EB cell (left) and a SHOX2-EB cell (right). Lower panels show the inhibition of time-dependent currents upon addition of 10 μM ivabradine in the bath solution. HCN currents were recorded in normal Tyrode’s solution containing 1 mM BaCl2 in order to block contaminating inward rectifier K+ currents. The If currents were elicited with a family of voltage steps from −135 mV to +35 mV for 1.5 s with 10 mV increment from a holding potential of −35 mV.
(E) Current density-voltage relationships of GFP (black, n = 4) and SHOX2 (red, n = 5) groups. ∗p < 0.05.
(F) Representative APs of a GFP cell (top row) and a SHOX2 cell (bottom row) before (left) and after treatment with isoproterenol (Iso, 1 μM, middle) or acetylcholine (ACh, 50 nM, right).
(G) Summary of beating rate responses before and after treatment with isoproterenol or acetylcholine. Data are represented as mean ± SEM. All experiments were performed on three independent biological replicates.
Discussion
Shox2 is indispensable for proper formation and development of the SAN. Shox2 null/null mouse embryos exhibit severe hypoplasia of the SAN accompanied by an aberrant expression of chamber cardiomyocyte-specific markers in the SAN (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Mouse Shox2 is first detected at ED8.5 in the posterior region of the developing heart tube, and its expression terminates by ED 13.5, being restricted to the cardiac conduction system (Blaschke et al., 2007; Espinoza-Lewis et al., 2009). Genetic ablation of Shox2 results in slowed contraction rate in mESC-derived EBs (Hashem et al., 2013). Here, we demonstrate that transient and heterologous expression of SHOX2 greatly increases the percentage of spontaneously beating EBs, the number of beating foci in each EB, and the EB contraction rate compared with control (Figures 4A–4D). The enhanced automaticity correlates directly with more HCN4+ cells present in SHOX2-EBs compared with control (Figure 3A). Direct injection of SHOX2-EBs into the rat left ventricular apex created ectopic automaticity indicative of induced biological pacing at a rate faster than the junctional escape rhythm observed in hearts injected with GFP-EBs (Figure 4E). The enhanced automaticity is accompanied by increased expression of automaticity-promoting genes and gene products such as HCN4, NCX1, and CX45 (Figure 3).
As rationale for the timing of SHOX2 overexpression, it may have been more logical to base it when Shox2-positive progenitors exist. In this regard, Shox2 expression is detected as early as day 6 of differentiation (Hashem et al., 2013) in an mESC-derived EB system. This largely coincides with the time points of exogenous SHOX2 overexpression (D3, D6, D7) in our study. It is notable that the beating rates of the GFP-EBs rise and then recede between D6+9 and D6+11 (Figure 4C). The majority of embryonic cardiac myocytes have the tendency to beat spontaneously but become more quiescent as they mature electrophysiologically (Pelleg et al., 1980). Likewise, the rapid decrease in the beating rates of GFP-EBs may be due to changes in their electrophysiological components, similar to the swift increase in INa density in differentiating, mESC-derived EBs (Maltsev et al., 1994). In contrast, the spontaneous beating rates remain high in SHOX2-EBs (Figure 4C), further supporting the notion that the superior automaticity of SHOX2-EBs is due to enhanced differentiation to pacemaker cells rather than to nonspecific deterioration of chamber cardiomyocytes.
Cardiac derivatives from ESCs consist of cardiomyocytes of atrial and ventricular phenotypes as well as pacemaker-like cells (Wobus et al., 1995). Because of the absence of specific extracellular epitopes, no live cell sorting method exists to purify pacemaker, ventricular, or atrial myocytes without genetic tagging. As such, the inherent heterogeneity of pluripotent stem cell-derived cardiomyocytes poses a critical impediment to therapeutic/diagnostic applications. For biological pacemaker applications, contaminating ventricular/atrial myocytes in the newly derived cardiac derivatives may compromise pacemaking activity from the implanted cells. Conversely, the presence of pacemaker cells may be arrhythmogenic if ESC-derived cardiomyocytes were to replace a large mass of damaged myocardium. Although implantation of human ESC-derived cardiomyocytes in the guinea pig hearts was shown to be antiarrhythmic (Shiba et al., 2012), delivery of the same cells in nonhuman primates resulted in premature ventricular contractions and ventricular tachycardia in all experimental animals (Chong et al., 2014).
As an in vitro diagnostic tool, induced pluripotent stem cell-derived cardiac myocytes have been proposed as an in vitro platform for drug screen for arrhythmic diseases such as Long QT syndrome (Itzhaki et al., 2011). However, the inherent heterogeneity of these cells, complicated by the fact that the derived cardiomyocytes are largely immature (Mummery et al., 2010; Mummery et al., 2012), may result in erroneous index of how a putative drug may react in atrial or ventricular myocardium. These concerns underscore the need for attaining subtype-specific populations of cardiomyocytes. Previous work by others has tried to address this by inducing pluripotent stem cells to cardiac pacemaker-like cells with various pharmacological agents (Kleger et al., 2010; Müller et al., 2012; Wiese et al., 2011). Here, we demonstrate that SHOX2-mediated pacemaker cell potentiation leads to heightened automaticity in the EBs and that the SHOX2-EBs provide biological pacemaker function upon transplantation into the rat myocardium in vivo. Implantation of the SHOX2-EBs was performed without microsurgical excision of nonbeating areas of the EBs, further highlighting the intensified automaticity in the SHOX2-EBs. The long-term potential of ESC-derived EBs as durable biological pacemakers is an important step toward clinical realization (Jung et al., 2014). However, we do not offer the present findings as a direct prelude to long-term biological pacing tools, but rather as insights into the progression of pacemaker cell development as they are reflected in the EB system.
Because embryonic development of the SAN occurs concurrently with the looping of the linear heart tube (Christoffels et al., 2004), major signaling motifs may impart a unique signature on the developing SAN from the general myocardium. Wnt signaling is indispensable for embryonic heart development (Schneider and Mercola, 2001) and can be manipulated to enrich the cardiomyocyte population from human ESCs (Lian et al., 2012; Willems et al., 2011). Since canonical Wnt signaling is necessary for maintaining mesenchymal precursors that later form the sinus horn (Norden et al., 2011), we looked for differential expression of Wnt ligands and regulators in SHOX2-EBs. On day 6+4 expression of noncanonical Wnt, ligands are mostly suppressed, while that of Wnt inhibitors are higher in SHOX2-EBs compared with control (Figure S5A), but the expression pattern reverses later at D6+14 (Figures S5B and S5C). Inhibition of endogenous Wnt generally potentiates cardiac differentiation of ESCs (Kattman et al., 2011; Paige et al., 2010). In contrast, the expression of classical Wnt inhibitors such as Dkk1, Sfrp4, and Wif1 is significantly downregulated in SHOX2-EBs at D6+4 (Figure S5C). The data point to antagonistic effects of Wnt signaling on cardiac myocyte subtype specification, manipulation of which may steer the ESCs to differentiate into a specific cardiac cell type.
Experimental Procedures
Mouse ESC Culture
The mESC line, R1 (ATCC) was cultured in KnockOut Dulbecco’s modified Eagle’s medium supplemented with 15% KnockOut Serum Replacement, 2 mM l-glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, 1% MEM nonessential amino acids, and β-mercaptoethanol. The mESCs were kept undifferentiated by culturing them on confluent monolayers of mitomycin C-treated primary mouse embryonic fibroblasts (Millipore) and by adding purified recombinant mouse leukemia inhibitory factor (ESGRO, 1,000 units/ml; Millipore). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Molecular Cloning and Adenovirus Purification
The human SHOX2 gene with a C-terminal myc/FLAG tag was excised from pCMV6-Tbx18 (Origene) by digestion with FseI and SalI and then subcloned into a NotI- and XhoI-digested lentiviral expression vector with the desired reporter gene, pLVX-IRES-ZsGreen1 (Clontech) to create pLV-Tbx18-IRES-ZsGreen. The recombinant target gene was then introduced to an adenovirus vector backbone by Gateway recombination cloning using Gateway-adapted vectors (Invitrogen). LR recombination reaction was performed between the entry clone and the destination vector, pAd/CMV/V5-DEST, to generate the desired adenoviral expression construct, pAd-CMV-SHOX2-IRES-ZsGreen. Positive constructs were verified to have the correct target gene by DNA sequencing (Laragen). The expression constructs were digested with PacI to expose inverted terminal repeats before transfecting into 293A cells to produce adenoviral stocks. Adenoviruses were amplified and affinity-column-purified using Adenopure kit (Puresyn) and stored at −80°C.
Differentiation of mESCs and Adenoviral Transduction
The cardiac differentiation media are composed of Iscove’s modified Dulbecco’s medium with Glutamax, ESC qualified fetal bovine serum (Embryomax, 20%), minimal essential media nonessential amino acids (1%), penicillin (50 units/ml), streptomycin (50 μg/ml), and β-mercaptoethanol (0.1 mM). On day 1, mESCs were dissociated with 0.05% trypsin-EDTA. Single ESCs were cultured in hanging drops (at 400 cells per 20 μl) to form EBs for 2 days. On day 3, the EBs were transferred to ultra-low-attachment dishes (Corning) and transduced either with an adenoviral vector expressing SHOX2 (Ad-SHOX2-IRES-GFP) or a control vector (Ad-GFP) at a multiplicity of infection (MOI) of 500. At day 6, control and treated EBs were plated onto gelatin-coated culture dishes. The EBs were transduced again at day 6 and D6+1 with the same MOI as at day 3 in order to maximize somatic gene transfer.
Scoring the Beating EBs
The numbers of spontaneously beating EBs and of beating foci per EB were counted under a bright-light microscope in a temperature- and CO2-controlled chamber. The position of each beating cluster was marked to avoid double counting.
RNA Extraction, Quantitative Real-Time PCR, and Wnt PCR Array
Total RNA was extracted from the embryonic mouse heart tissues with RNeasy Mini Kit (QIAGEN) followed by DNase treatment. RNA concentration was assessed by spectrophotometry (NanoDrop; Thermo Scientific). Real-time quantitative PCR was performed with the Taqman One Step PCR kit (Applied Biosystems) according to the manufacture’s recommended protocol. Primers used for quantitative RT-PCR were all from Applied Biosystems. Housekeeping genes such as glyceraldeyde-3 phosphate dehydrogenase and Hydroxymethylbilane synthase (HMBS) were employed as internal control. For PCR array analyses, total RNA (1 μg) was reverse transcribed into cDNA using the RT2 First Strand Kit (SA Biosciences). The resulting cDNA was analyzed using the Wnt Pathway PCR Array (catalog number PAMM-043; SA Biosciences). All quantitative PCR reactions were performed on an ABI 7900 HT fast Real-Time PCR System (Applied Biosystems). All experiments were performed in triplicates with three biological replicates. All primer sets used for quantitative RT-PCR (qRT-PCR) were purchased from Applied Biosystems (Table 1).
Table 1.
Primer Sets for Quantitative PCR
| Catalog Identification | Interrogated Sequence (RefSeq) | NCBI Chromosome Location | Exon Boundary | Amplicon Length | |
|---|---|---|---|---|---|
| Human SHOX2 | Hs00243203_m1 | NM_001163678.1 | chromosome 3: 157813800–157823952 | 2–3 | 129 |
| Mouse Shox2 | Mm00443183_m1 | NM_013665.1 | chromosome 3: 66973266–66981771 | 1–2 | 97 |
| Mouse Tbx3 | Mm01195726_m1 | NM_011535.3 | chromosome 5: 119670669–119684601 | 7–8 | 65 |
| Mouse Tbx18 | Mm00470177_m1 | NM_023814.4 | chromosome 9: 87702800–87731260 | 6–7 | 64 |
| Mouse Nkx2.5 | Mm00657783_m1 | NM_008700.2 | chromosome 17: 26838665–26841565 | 1–2 | 117 |
| Mouse Actc1 | Mm01333821_m1 | NM_009608.3 | chromosome 2: 114047289–114052811 | 3–4 | 62 |
| Mouse TnnT2 | Mm01290256_m1 | NM_001130174.2 | chromosome 1: 135836386–135852261 | 8–9 | 110 |
| Mouse Hmbs | Mm01143545_m1 | NM_001110251.1 | chromosome 9: 44336348–44344228 | 6–7 | 81 |
Western Blot Analysis
Whole-cell lysates were harvested with RIPA Buffer (Thermo Scientific) containing protease inhibitors (HALT Protease Cocktail; Pierce). Protein concentrations were determined with bicinchoninic acid protein assay (MicroBCA Kit; Pierce). Samples were loaded onto 12% SDS-PAGE, and the separated proteins were transferred to a polyvinylidine fluoride membrane. Blots were incubated overnight at 4°C with the primary antibodies against HCN4 (Abcam), Connexin-45(CX45; Invitrogen), Connexin-43 (CX43; Sigma-Aldrich), and Na+/Ca2+ exchanger (NCX1; Abcam). The primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies raised in appropriate species, followed by chemiluminescence detection (Pico Chemiluminescence Substrate; Pierce). Protein loading was controlled by reprobing the membranes with monoclonal anti-β-actin antibody (Sigma-Aldrich).
Immunocytochemistry
Whole EBs were fixed using 4% paraformaldehyde, washed with PBS, and permeabilized with 0.1% Triton X-100. Fixed cells were blocked with 5% normal donkey serum overnight at 4°C. The EBs were then incubated with primary antibodies for HCN4 (Abcam) and a secondary antibody conjugated with Alexa Fluor 488 at 1:500 dilution for 1 hr at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
In Vivo Injection and Ex Vivo Optical Mapping
All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee of the Cedars-Sinai Medical Center. For testing of biological pacemaker function in animals implanted with the EBs in vivo, 10–12 whole SHOX2-EBs or GFP-EBs were randomly selected and injected with a 24G needle subepicardially into the left ventricular anterior wall of adult Sprague Dawley female rats upon thoracotomy. At 24 to 48 hr after injection, heart was harvested and retrogradely perfused with normal Tyrode’s solution containing 10 μM blebbistatin for 30 min in order to uncouple excitation-contraction, thereby minimizing motion artifacts (Fedorov et al., 2007). The heart was then perfused with di-4-ANEPPS (2 μM; Molecular Probes) for 15 min. Perfusion was made through the aorta, and temperature was continuously maintained at 37°C ± 1°C. During mapping, the heart was positioned in a transparent chamber filled with Tyrode’s solution with the injected site facing the optical light path, and the optical signals were recorded from a 5 × 5 mm area at baseline (sinus rhythm) and after complete heart block. Heart block was achieved by mechanically excising the SAN and the AV node. The fluorescence changes were recorded on a 496-diode array mapping system (WuTech).
Single-Cell Electrophysiology
To examine the electrophysiological properties of individual cardiomyocytes derived from mESC, EBs at D6+14 were dispersed into single cells with a protocol modified from a previous report (Zhang et al., 2002). EBs were digested with a nominally calcium-free solution (in mM: NaCl 140, KCl 5.4, KH2PO4 1.2, MgCl2 0.5, HEPES 5, taurine 50, glucose 5.5 [pH = 6.9] with NaOH) containing collagenase B (Roche, 0.5 mg/ml) at 37°C for 50 min followed by digestion with Liberase (Roche, 75 μg/ml) at 35°C for 20 min. Single, whole-cell AP and ionic current measurements were carried out using standard microelectrode whole-cell, patch-clamp technique with an Axopatch 200B amplifier (Molecular Devices) with a sampling rate of 20 kHz and low-pass Bessel-filtered at 5 kHz. Experiments were performed at 33°C ± 1°C. Cells were perfused with a normal Tyrode’s solution containing (mM: NaCl 138, KCl 5, CaCl2 1.8, MgCl2 0.5, glucose 10, and HEPES 10 [pH = 7.4]) with NaOH. Pipette solution contained (mmol/l) K-glutamate 98, KCl 50, MgCl2 1.0, HEPES 10, EGTA 2, and K-ATP 5 (pH = 7.2) with KOH. Microelectrodes had tip resistances of 2–4 MΩ when filled with the internal recording solution. Spontaneous APs were recorded with I = 0 mode. Effects of adrenergic and cholinergic agonists on AP firing rate were tested by perfusing cells with Tyrode solution containing isoproterenol (1 μM, Sigma) or acetylcholine (50 nM; Sigma). Data were corrected for the estimated liquid junction potentials (−12.5 mV). The diastolic depolarization rate was determined from the slope of a 20-ms segment after the maximum diastolic potential. Funny currents (If) were recorded in a voltage-clamp mode with cells bathed in normal Tyrode’s solution containing 1 mM BaCl2 in order to block contaminating inward rectifier K+ currents (IK1). Holding potential was set at −35 mV, and a family of voltage steps from −135 mV to +35 mV for 1.5 s with 10 mV increment was applied to elicit If. The If was identified by their time-dependent activation and their sensitivity to ivabradine (10 μM; Sigma-Aldrich).
Similar to the criteria adopted in the human ESC-derived cardiomyocytes (He et al., 2003), the APs were designated ventricular-, atrial- and pacemaker-like cells based on the AP duration and the slope of spontaneous phase 4 depolarization rate. Ordinary ventricular cardiomyocytes exhibit prominent plateau phase, reflected by their slow early repolarization rate (i.e., long APD20) and fast late repolarization rate (short APD80). Accordingly, cells with APD20/APD80 >3.0 were categorized to ventricular-like cells. Pacemaker-like APs were designated to cells with fast phase 4 diastolic depolarization rate (>0.04 V/s) and short APD (APD20/APD80 ≤3.0). Atrial-like APs were designated to cells with short APD (APD20/APD80 ≤3.0) and slow phase 4 diastolic depolarization (≤0.04 V/s).
Curve Fitting
The time course of endogenous Shox2 expression data was processed by nonlinear curve fitting (Figure 2A) with amplitude version of Gaussian peak function with an equation, y = y0 + A∗exp(−0.5 × ((x − xc)/w)∧2). Levenberg Marquardt algorithm was used for iteration. The final values for each parameter at the end of the iteration are provided in Table 2.
Table 2.
Parameters for the Nonlinear Curve Fitting of the Data in Figure 2A
| Value | SE | |
|---|---|---|
| y0 | 1.88621 | 0.12281 |
| xc | 5.0783 | 0.23033 |
| w | 3.34034 | 0.3499 |
| A | −1.904 | 0.13304 |
| Full width at half maximum | 7.86591 | 0.82396 |
| Area | −15.9422 | 2.2425 |
| Number of points | 10 | |
| Degrees of freedom | 6 | |
| Reduced chi squared | 1.28641 | |
| Adjusted R square | 0.96031 |
Statistical Analysis
All data are represented as mean ± SEM. Statistical significance was calculated with an unpaired Student’s t test. A confidence of p ≤ 0.05 was considered significant.
Author Contributions
V.I. designed and carried out all experiments except the single-cell electrophysiology work. W.L. carried out all experiments that relate to the single-cell electrophysiology.
Acknowledgments
We thank Giselle Galang for adenoviral vector construction, amplification, and purification. Jordan Mak aided in mESC maintenance. The study was supported by the Cedars-Sinai Board of Governors Heart Stem Cell Center, Canadian Institutes of Health Research Fellowship to W.L. and the American Heart Association (12SDG9020030) and National Heart, Lung, and Blood Institute (1R01HL111646-01A1) to H.C.C.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
A nylon suture was tied to the heart adjacent to the EB injection site at the time of surgery in order to locate the site of EB implantation during optical mapping.
A nylon suture was tied to the heart adjacent to the EB injection site at the time of surgery in order to locate the site of EB implantation during optical mapping.
References
- Bakker M.L., Boukens B.J., Mommersteeg M.T., Brons J.F., Wakker V., Moorman A.F., Christoffels V.M. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Bakker M.L., Boink G.J., Boukens B.J., Verkerk A.O., van den Boogaard M., den Haan A.D., Hoogaars W.M., Buermans H.P., de Bakker J.M., Seppen J. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc. Res. 2012;94:439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- Blaschke R.J., Hahurij N.D., Kuijper S., Just S., Wisse L.J., Deissler K., Maxelon T., Anastassiadis K., Spitzer J., Hardt S.E. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- Bleeker W.K., Mackaay A.J., Masson-Pévet M., Bouman L.N., Becker A.E. Functional and morphological organization of the rabbit sinus node. Circ. Res. 1980;46:11–22. doi: 10.1161/01.res.46.1.11. [DOI] [PubMed] [Google Scholar]
- Boheler K.R., Czyz J., Tweedie D., Yang H.T., Anisimov S.V., Wobus A.M. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- Bonke F.I. Electronic spread in the sinoatrial node of the rabbit heart. Pflugers Arch. 1973;339:17–23. doi: 10.1007/BF00586978. [DOI] [PubMed] [Google Scholar]
- Bressan M., Liu G., Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340:744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C.L., Martin J.C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.C., Marbán E. Biological therapies for cardiac arrhythmias: can genes and cells replace drugs and devices? Circ. Res. 2010;106:674–685. doi: 10.1161/CIRCRESAHA.109.212936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels V.M., Burch J.B., Moorman A.F. Architectural plan for the heart: early patterning and delineation of the chambers and the nodes. Trends Cardiovasc. Med. 2004;14:301–307. doi: 10.1016/j.tcm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Christoffels V.M., Mommersteeg M.T., Trowe M.O., Prall O.W., de Gier-de Vries C., Soufan A.T., Bussen M., Schuster-Gossler K., Harvey R.P., Moorman A.F., Kispert A. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ. Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- Christoffels V.M., Grieskamp T., Norden J., Mommersteeg M.T., Rudat C., Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–E9. doi: 10.1038/nature07916. discussion E9–E10. [DOI] [PubMed] [Google Scholar]
- Christoffels V.M., Smits G.J., Kispert A., Moorman A.F. Development of the pacemaker tissues of the heart. Circ. Res. 2010;106:240–254. doi: 10.1161/CIRCRESAHA.109.205419. [DOI] [PubMed] [Google Scholar]
- DiFrancesco D. The role of the funny current in pacemaker activity. Circ. Res. 2010;106:434–446. doi: 10.1161/CIRCRESAHA.109.208041. [DOI] [PubMed] [Google Scholar]
- Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G., Gramolini A., Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza-Lewis R.A., Yu L., He F., Liu H., Tang R., Shi J., Sun X., Martin J.F., Wang D., Yang J., Chen Y. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 2009;327:376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov V.V., Lozinsky I.T., Sosunov E.A., Anyukhovsky E.P., Rosen M.R., Balke C.W., Efimov I.R. Application of blebbistatin as an excitation-contraction uncoupler for electrophysiologic study of rat and rabbit hearts. Heart Rhythm. 2007;4:619–626. doi: 10.1016/j.hrthm.2006.12.047. [DOI] [PubMed] [Google Scholar]
- Hashem S.I., Lam M.L., Mihardja S.S., White S.M., Lee R.J., Claycomb W.C. Shox2 regulates the pacemaker gene program in embryoid bodies. Stem Cells Dev. 2013;22:2915–2926. doi: 10.1089/scd.2013.0123. [DOI] [PubMed] [Google Scholar]
- Hattori F., Chen H., Yamashita H., Tohyama S., Satoh Y.S., Yuasa S., Li W., Yamakawa H., Tanaka T., Onitsuka T. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- He J.Q., Ma Y., Lee Y., Thomson J.A., Kamp T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- Itzhaki I., Maizels L., Huber I., Zwi-Dantsis L., Caspi O., Winterstern A., Feldman O., Gepstein A., Arbel G., Hammerman H. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- Jansen J.A., van Veen T.A., de Bakker J.M., van Rijen H.V. Cardiac connexins and impulse propagation. J. Mol. Cell. Cardiol. 2010;48:76–82. doi: 10.1016/j.yjmcc.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Joyner R.W., van Capelle F.J. Propagation through electrically coupled cells. How a small SA node drives a large atrium. Biophys. J. 1986;50:1157–1164. doi: 10.1016/S0006-3495(86)83559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.J., Husse B., Rimmbach C., Krebs S., Stieber J., Steinhoff G., Dendorfer A., Franz W.M., David R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Rep. 2014;2:592–605. doi: 10.1016/j.stemcr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor N., Liang W., Marbán E., Cho H.C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A., Ellis J., Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kehat I., Kenyagin-Karsenti D., Snir M., Segev H., Amit M., Gepstein A., Livne E., Binah O., Itskovitz-Eldor J., Gepstein L. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J. Clin. Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehat I., Khimovich L., Caspi O., Gepstein A., Shofti R., Arbel G., Huber I., Satin J., Itskovitz-Eldor J., Gepstein L. Electromechanical integration of cardiomyocytes derived from human embryonic stem cells. Nat. Biotechnol. 2004;22:1282–1289. doi: 10.1038/nbt1014. [DOI] [PubMed] [Google Scholar]
- Kleger A., Seufferlein T., Malan D., Tischendorf M., Storch A., Wolheim A., Latz S., Protze S., Porzner M., Proepper C. Modulation of calcium-activated potassium channels induces cardiogenesis of pluripotent stem cells and enrichment of pacemaker-like cells. Circulation. 2010;122:1823–1836. doi: 10.1161/CIRCULATIONAHA.110.971721. [DOI] [PubMed] [Google Scholar]
- Kolossov E., Lu Z., Drobinskaya I., Gassanov N., Duan Y., Sauer H., Manzke O., Bloch W., Bohlen H., Hescheler J., Fleischmann B.K. Identification and characterization of embryonic stem cell-derived pacemaker and atrial cardiomyocytes. FASEB J. 2005;19:577–579. doi: 10.1096/fj.03-1451fje. [DOI] [PubMed] [Google Scholar]
- Lakatta E.G., DiFrancesco D. What keeps us ticking: a funny current, a calcium clock, or both? J. Mol. Cell. Cardiol. 2009;47:157–170. doi: 10.1016/j.yjmcc.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev V.A., Rohwedel J., Hescheler J., Wobus A.M. Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech. Dev. 1993;44:41–50. doi: 10.1016/0925-4773(93)90015-p. [DOI] [PubMed] [Google Scholar]
- Maltsev V.A., Wobus A.M., Rohwedel J., Bader M., Hescheler J. Cardiomyocytes differentiated in vitro from embryonic stem cells developmentally express cardiac-specific genes and ionic currents. Circ. Res. 1994;75:233–244. doi: 10.1161/01.res.75.2.233. [DOI] [PubMed] [Google Scholar]
- Mandel Y., Weissman A., Schick R., Barad L., Novak A., Meiry G., Goldberg S., Lorber A., Rosen M.R., Itskovitz-Eldor J., Binah O. Human embryonic and induced pluripotent stem cell-derived cardiomyocytes exhibit beat rate variability and power-law behavior. Circulation. 2012;125:883–893. doi: 10.1161/CIRCULATIONAHA.111.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michou A.I., Santoro L., Christ M., Julliard V., Pavirani A., Mehtali M. Adenovirus-mediated gene transfer: influence of transgene, mouse strain and type of immune response on persistence of transgene expression. Gene Ther. 1997;4:473–482. doi: 10.1038/sj.gt.3300412. [DOI] [PubMed] [Google Scholar]
- Mommersteeg M.T., Hoogaars W.M., Prall O.W., de Gier-de Vries C., Wiese C., Clout D.E., Papaioannou V.E., Brown N.A., Harvey R.P., Moorman A.F., Christoffels V.M. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- Müller M., Stockmann M., Malan D., Wolheim A., Tischendorf M., Linta L., Katz S.F., Lin Q., Latz S., Brunner C. Ca2+ activated K channels-new tools to induce cardiac commitment from pluripotent stem cells in mice and men. Stem Cell Rep. 2012;8:720–740. doi: 10.1007/s12015-011-9324-9. [DOI] [PubMed] [Google Scholar]
- Mummery C.L., Davis R.P., Krieger J.E. Challenges in using stem cells for cardiac repair. Sci. Transl. Med. 2010;2:27ps17. doi: 10.1126/scitranslmed.3000558. [DOI] [PubMed] [Google Scholar]
- Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden J., Greulich F., Rudat C., Taketo M.M., Kispert A. Wnt/β-catenin signaling maintains the mesenchymal precursor pool for murine sinus horn formation. Circ. Res. 2011;109:e42–e50. doi: 10.1161/CIRCRESAHA.111.245340. [DOI] [PubMed] [Google Scholar]
- Nunes S.S., Miklas J.W., Liu J., Aschar-Sobbi R., Xiao Y., Zhang B., Jiang J., Massé S., Gagliardi M., Hsieh A. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat. Methods. 2013;10:781–787. doi: 10.1038/nmeth.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige S.L., Osugi T., Afanasiev O.K., Pabon L., Reinecke H., Murry C.E. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS ONE. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelleg A., Vogel S., Belardinelli L., Sperelakis N. Overdrive suppression of automaticity in cultured chick myocardial cells. Am. J. Physiol. 1980;238:H24–H30. doi: 10.1152/ajpheart.1980.238.1.H24. [DOI] [PubMed] [Google Scholar]
- Schneider V.A., Mercola M. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 2001;15:304–315. doi: 10.1101/gad.855601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y., Fernandes S., Zhu W.Z., Filice D., Muskheli V., Kim J., Palpant N.J., Gantz J., Moyes K.W., Reinecke H. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tripathy S.K., Black H.B., Goldwasser E., Leiden J.M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat. Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- Van Vliet P., Wu S.M., Zaffran S., Pucéat M. Early cardiac development: a view from stem cells to embryos. Cardiovasc. Res. 2012;96:352–362. doi: 10.1093/cvr/cvs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C., Grieskamp T., Airik R., Mommersteeg M.T., Gardiwal A., de Gier-de Vries C., Schuster-Gossler K., Moorman A.F., Kispert A., Christoffels V.M. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- Wiese C., Nikolova T., Zahanich I., Sulzbacher S., Fuchs J., Yamanaka S., Graf E., Ravens U., Boheler K.R., Wobus A.M. Differentiation induction of mouse embryonic stem cells into sinus node-like cells by suramin. Int. J. Cardiol. 2011;147:95–111. doi: 10.1016/j.ijcard.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E., Spiering S., Davidovics H., Lanier M., Xia Z., Dawson M., Cashman J., Mercola M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ. Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus A.M., Wallukat G., Hescheler J. Pluripotent mouse embryonic stem cells are able to differentiate into cardiomyocytes expressing chronotropic responses to adrenergic and cholinergic agents and Ca2+ channel blockers. Differentiation. 1991;48:173–182. doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- Wobus A.M., Rohwedel J., Maltsev V., Hescheler J. Development of cardiomyocytes expressing cardiac-specific genes, action potentials, and ionic channels during embryonic stem cell-derived cardiogenesis. Ann. N Y Acad. Sci. 1995;752:460–469. doi: 10.1111/j.1749-6632.1995.tb17456.x. [DOI] [PubMed] [Google Scholar]
- Xue T., Cho H.C., Akar F.G., Tsang S.Y., Jones S.P., Marbán E., Tomaselli G.F., Li R.A. Functional integration of electrically active cardiac derivatives from genetically engineered human embryonic stem cells with quiescent recipient ventricular cardiomyocytes: insights into the development of cell-based pacemakers. Circulation. 2005;111:11–20. doi: 10.1161/01.CIR.0000151313.18547.A2. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Dobrzynski H., Tellez J., Niwa R., Billeter R., Honjo H., Kodama I., Boyett M.R. Extended atrial conduction system characterised by the expression of the HCN4 channel and connexin45. Cardiovasc. Res. 2006;72:271–281. doi: 10.1016/j.cardiores.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Zhang Y.M., Hartzell C., Narlow M., Dudley S.C., Jr. Stem cell-derived cardiomyocytes demonstrate arrhythmic potential. Circulation. 2002;106:1294–1299. doi: 10.1161/01.cir.0000027585.05868.67. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wilson G.F., Soerens A.G., Koonce C.H., Yu J., Palecek S.P., Thomson J.A., Kamp T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A nylon suture was tied to the heart adjacent to the EB injection site at the time of surgery in order to locate the site of EB implantation during optical mapping.
A nylon suture was tied to the heart adjacent to the EB injection site at the time of surgery in order to locate the site of EB implantation during optical mapping.


