Abstract
Genomic copy-number variations (CNVs) constitute an important cause of epilepsies and other human neurological disorders. Recent advancement of technologies integrating genome-wide CNV mapping and sequencing is rapidly expanding the molecular field of pediatric neurodevelopmental disorders. In a previous study, a novel epilepsy locus was identified on 6q16.3q22.31 by linkage analysis in a large pedigree. Subsequent array comparative genomic hybridization (array CGH) analysis of four unrelated cases narrowed this region to ∼5 Mb on 6q22.1q22.31. We sought to further narrow the critical region on chromosome 6q22. Array CGH analysis was used in genome-wide screen for CNVs of a large cohort of patients with neurological abnormalities. Long-range PCR and DNA sequencing were applied to precisely map chromosomal deletion breakpoints. Finally, real-time qPCR was used to estimate relative expression in the brain of the candidate genes. We identified six unrelated patients with overlapping microdeletions within 6q22.1q22.31 region, three of whom manifested seizures. Deletions were found to be de novo in 5/6 cases, including all subjects presenting with seizures. We sequenced the deletion breakpoints in four patients and narrowed the critical region to a ∼250-kb segment at 6q22.1 that includes NUS1, several expressed sequence tags (ESTs) that are highly expressed in the brain, and putative regulatory sequences of SLC35F1. Our findings indicate that dosage alteration in particular, of NUS1, EST AI858607, or SLC35F1 are important contributors to the neurodevelopmental phenotype associated with 6q22 deletion, including epilepsy and tremors.
Introduction
Despite a wealth of information on the developmental pathways that operate during neurogenesis, only a limited number of epileptogenic genes were identified before the advent of high-throughput technologies for studies of structural genomic variations. It has now become clear that copy-number variations (CNVs) frequently cause epilepsy and contribute to other neurodevelopmental disorders, including intellectual disability, neuropsychiatric disorders, autism spectrum disorder (ASD), and behavior abnormalities. Undoubtedly, the impact of recurrent genomic CNVs involving 14q12, 7q11.23, 15q13.3, 16p11.2, and 16p13.11 loci on pediatric epilepsy and other neurodevelopmental problems is substantial.1, 2, 3, 4, 5 Using linkage analysis in 15 subjects from a large family with a history of genetic epilepsy with febrile seizures plus (GEFS+), Poduri et al6 delineated an 18.1 Mb haplotype on 6q16.3q22.31 that defined a novel GEFS+8 locus (OMIM 613828). However, no mutations were found in any of the 16 protein-coding genes expressed in the brain within this interval in the affected subjects. In a recent study, Rosenfeld et al7 reported four individuals with seizures and overlapping microdeletions on 6q22.1q22.31, and narrowed the critical region to a ∼5-Mb genomic segment. Here, applying array comparative genomic hybridization (array CGH), we report overlapping microdeletions in six additional patients, three of them presenting with seizures, and narrow the locus to a ∼250 kb region on 6q22.1 including NUS1, brain-specific ESTs of unknown function, and putative cis-regulatory sequences of the SLC35F1 promoter. Our findings delineate a critical disease region within 6q22 and implicate novel genes in pediatric epilepsy and tremors within this region.
Materials and methods
Patient ascertainment
Individuals with 6q21q22.31 deletions reported here were identified after referral for chromosomal microarray analysis (CMA) to clinical laboratories, including Baylor College of Medicine (BCM) Medical Genetics Laboratories (MGL) (patients 1–4) and Signature Genomic Laboratories (SGL) (patients 5 and 6). Between June 2008 and June 2013, 13 208 children with neurological phenotypes were studied by array CGH at MGL. Of these, 8601 children were referred for developmental delay (65%), 2887 were studied for ASD (22%), and 1720 subjects were evaluated for seizures (13%). Informed consents via a protocol approved by the Institutional Review Board for Human Subject Research at BCM were obtained for patients with deletions within the 6q22 region. DNA samples were re-evaluated by DNA sequencing.
Subjects
Patient 1 was evaluated at 3 years of age with severe speech/language delay, ASDs, stereotypical repetitive behaviors, and staring episodes suggestive of atypical absence seizures. He walked independently at 15 months of age. His EEG background was diffusely slow, mostly in the theta range, with brief bursts of generalized high-amplitude spike and slow wave discharges of 1.5–2.5 hz. Plasma amino acids, urine organic acids, and cerebrospinal fluid (CSF) neurotransmitters were normal. The MRI of the brain was unremarkable. His hearing test was normal. No focal deficits were noted on his neurological exam.
Patient 2 was born at term after an uneventful pregnancy. His prenatal and neonatal course was unremarkable. Mild hypotonia was noted by 6 months of age and physical therapy was initiated. He walked independently at 21 months of age. At 4 years of age, he had severe articulation problems, speaking mostly in two-word phrases. He continued to have difficulties with balance and coordination when evaluated. MRI of the brain was unremarkable with no structural abnormalities. Karyotype study, DNA for Fragile X, plasma amino acids, urine organic acids, and creatine and guanidinoacetate studies were normal. His physical examination at 4 years of age showed facial dysmorphisms including broad, flat nasal bridge, bilateral epicanthal folds, and a mildly high arched palate. His neurological exam revealed mild hypotonia and clumsy gait.
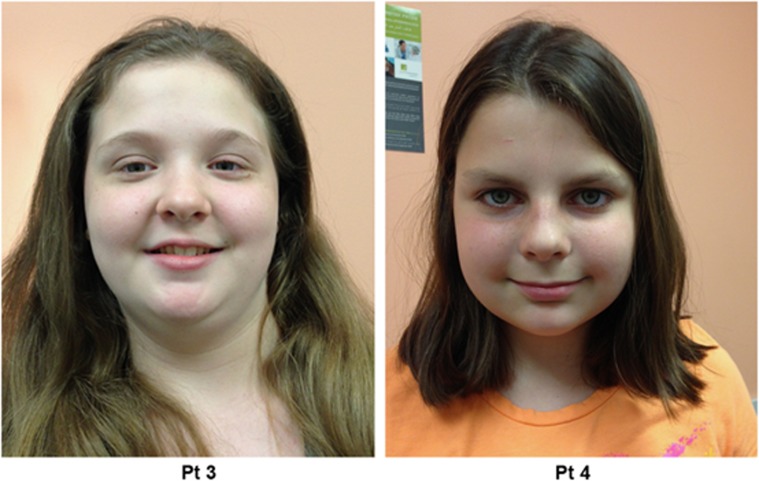
Patient 3 (Figure 1) was referred for evaluation of cognitive delay and epilepsy. She was the product of a twin pregnancy with complication of twin-twin transfusion syndrome, who was delivered by C-section at 33 weeks gestation. All developmental milestones, including those of language and motor, were attained within normal limits until 2 years of age, at which time she experienced the onset of seizures and had regression of previously acquired language and developmental skills. Seizures were characterized by eye rolling, staring, and eyelid fluttering or blinking. These lasted 3–10 s each with immediate return to baseline and occurred up to 100 times daily. She was diagnosed with generalized epilepsy with atypical absence seizures, refractory to multiple anticonvulsant medications, including phenobarbital, levetiracetam, valproic acid, topiramate, and lamotrigine. EEG showed appropriate background rhythms for age with frequent brief bursts of bifrontal-predominant, generalized spike-and-wave and polyspike-and-wave discharges that were rhythmic to semi-rhythmic at 3–3.5 Hz during wakefulness and sleep. She was also diagnosed with mild tremor. Neuropsychological evaluation (WISC-IV) at 8 years of age was consistent with a diagnosis of intellectual disability of moderate severity. Weaknesses were apparent in both receptive and expressive language, and there were relative weaknesses in visuomotor integration and visuospatial construction. Etiological testing including lactate, pyruvate, thyroid function, plasma amino acids, urine organic acids, acylcarnitine profile, CSF neurotransmitter levels, and febrile seizure panel (including SCN1A) were normal. MRI of the brain was also unremarkable.
Figure 1.
Patients 3 and 4 with de novo microdeletions within the 6q22.1 region, presenting with language delay, epilepsy, and tremors. No significant craniofacial abnormalities are noted.
Physical examination at 12 years of age revealed normal growth parameters including the head circumference. No dysmorphic features were noted on exam. She was able to follow simple commands well. Her neurological exam was generally unremarkable.
Patient 4 (Figure 1) was evaluated for cognitive delay and intractable cryptogenic generalized epilepsy. She was born at 39 weeks gestation and met all her developmental milestones in the first year of life. She walked independently at 14 months of age. At 2 years, she had surgical repair for spina bifida occulta cord tethering. She first began to experience recurrent seizures just after 2 years of age. The seizures were seemingly unprovoked and not associated with an intercurrent illness or sleep deprivation. She was diagnosed with cryptogenic generalized epilepsy with atypical absence and tonic/myoclonic seizures found to be refractory to multiple anticonvulsant medications. Her EEG showed background slowing and recurrent bursts of generalized 2–2.5 Hz spike and slow wave discharges. She also started having tremors, which worsened with intention. The tremor was constantly present, but varied considerably in intensity over the course of the day. The generalized mild tremulousness predominately affected her upper extremities. It was treated with Sinemet without improvement. Neuropsychological evaluation (Stanford-Binet) at 7 years of age revealed her intellectual functioning in the moderately deficient range with a Full Scale IQ of 50. She had poor visual motor integration skills. As part of her evaluation, she had DNA testing for MECP2 and for Huntington disease, all of which was normal. The ataxia evaluation for genes including SCA1, SCA2, SCA3, SCA5, SCA6, SCA7, SCA8, SCA10, SCA14, SCA17, and DRPLA was normal. Her brain imaging was essentially normal. Her neurological exam at the chronological age of 11 years showed mild axial hypotonia with good strength, prominent polyminimyoclonus-like isolated multifocal jerks and twitches, normal deep tendon reflexes, and mild intention tremor.
Patient 5 was assessed for global delay with no history of seizures. He was born at 39 weeks after a normal pregnancy via C-section due to breech presentation. His early motor developmental milestones were appropriately attained. Speech and language delay was apparent by 3 years of age. He was additionally diagnosed with tremors and motor dyspraxia causing fine motor delays. He had psychometric testing around 7 years of age, which showed a full scale IQ of 60. The brain imaging study was normal. At the time of his evaluation at 8 ½ years, his head circumference was at the 2nd percentile. He has continued to make progress with no evidence of developmental regression.
Patient 6 was evaluated for hypotonia, motor delays, and dysmorphic facial features at 4 months of age. His cardiac evaluation was consistent with branch pulmonary stenosis and patent foramen ovale. There was no history of seizures by report. His physical examination was remarkable for an adducted left thumb and borderline microcephaly. His neurological exam was otherwise normal.
DNA and RNA isolation
Genomic DNA was extracted from peripheral blood using the Puregene DNA isolation kit (Gentra System, Minneapolis, MN, USA). Total RNA from the normal human brain was purchased from Ambion (Austin, TX, USA). Reference RNA was extracted from normal fetal lung fibroblasts, IMR-90 (ATCC CCL-186), using MasterPur Complete DNA and RNA Purification Kit (Epicentre, Madison, WI, USA). The RNA sample was treated with DNase using DNA-free Kit (Ambion).
Array CGH analysis
The four unrelated subjects referred for clinical CMA at the MGL at BCM were screened using custom-designed exon-targeted CGH oligonucleotide microarrays (patient 1 V7.1 OLIGO, 105 K; patient 2 V8.1 OLIGO, 180 K and patients 3 and 4 V8.2 OLIGO, 180 K) designed by MGL at BCM (http://www.bcm.edu/geneticlabs) and manufactured by Agilent Technologies (Santa Clara, CA, USA), as previously described.8, 9 Patients 1, 3, and 4 were also screened using custom-designed high-resolution 3 × 720 K microarrays covering 12 Mb in 6q21-6q22.31, designed and produced by Roche-NimbleGen (Madison, WI, USA). Labeling and hybridization of DNA samples were done according to the manufacturer's instructions (NimbleGen Array User's Guide v9.1 for array CGH). Patients 5 and 6 were screened using whole-genome oligonucleotide-based CGH microarrays custom-designed by SGL: 105 K SignatureChipOS v1.1 (manufactured by Agilent) and 135 K SignatureChipOS v2.0 (manufactured by Roche–NimbleGen), respectively, as previously described.10, 11
FISH analysis
Confirmatory and parental fluorescence in situ hybridization (FISH) analyses were performed using standard procedures. Deletions were verified by using the chromosome 6q-specific BAC clones RP11-61K10, RP11-282C5, RP11-141L18, or RP11-435B17.
Long-range PCR and DNA sequencing
Primers were designed using the Primer 3 software (http://frodo.wi.mit.edu/primer3). Amplification of junction fragments was performed using Takara LA Taq Polymerase (TaKaRa Bio USA, Madison, WI, USA) applying 30 cycles of 94 °C for 30 s and 68 °C for 5 min. PCR products were treated with ExoSAP-IT (USB, Cleveland, OH, USA) to remove unconsumed dNTPs and primers, and directly sequenced by the Sanger method using primers flanking deletion breakpoints (see Supplementary Table S1).
Real-time qPCR analysis of 6q22.1 transcripts
Total RNA from the normal human brain and lung fibroblasts was reverse-transcribed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Branchburg, NJ, USA). cDNA from piRNA, piR-56963, was synthesized using SuperScript First Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA, USA) and stem-loop primer 5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCCAG-3′. Primers to quantify piR-56963 were designed using miRNA Primer Design Tool. Primers to quantify transcripts of NUS1, SLC35F1, GAPDH (internal control), ESTs: CB111179, BI825833, AI858607, BQ429202, and a lncRNA, TCONS_00011550, were designed using PrimerQuest (http://www.idtdna.com/Primerquest/Home/Index) (see Supplementary Table S2). qPCR was repeated four times using Power SYBR Green PCR Master Mix (Applied Biosystems). qPCR conditions included 40 cycles of 95 °C for 15 s and 60 °C for 1 min. For relative quantification, the comparative CT method was used.
Bioinformatics
Genomic sequences based on oligonucleotide coordinates obtained from the array CGH experiments were downloaded from the UCSC Genome Browser (NCBI build 37, May 2009, hg19, http://www.genome.ucsc.edu) and assembled using BLAT (UCSC genome browser), BLAST2 (NCBI, http://blast.ncbi.nlm.nih.gov), and the Sequencher v4.8 software (Gene Codes, Ann Arbor, MI, USA). Interspersed repeat sequences were identified using RepeatMasker (http://www.repeatmasker.org). GC content was analyzed using Cpgplot (http://www.ebi.ac.uk/Tools/seqstat/emboss_cpgplot).
Results
Further narrowing the 6q22 critical region
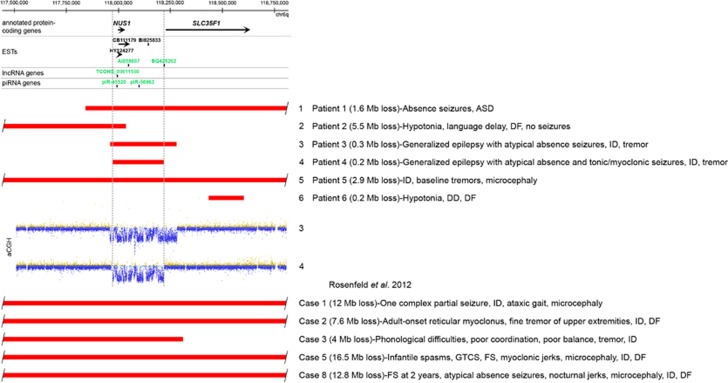
Applying array CGH methodology, we identified overlapping microdeletions within the 6q16.3q22.31 locus in five of six unrelated subjects (Figure 2, Table 1). The smallest region of deletion overlap that correlated with seizures (patients 1, 3 and 4) enabled narrowing the critical region to a ∼250-kb genomic segment, encompassing NUS1 and the promoter of SLC35F1 (Figure 2). This region is defined by the de novo deletion in patient 4 with intractable cryptogenic generalized epilepsy. Patient 3, with a similar clinical presentation, had an overlapping ∼300-kb deletion, also determined to be de novo, encompassing NUS1 as well as the 5′ portion of the SLC35F1 gene. Patient 1, with atypical absence seizures, had an ∼1.6-Mb deletion, completely overlapping the deletions found in patients 3 and 4. In 5/6 cases in which parental studies were performed, deletions were not detected in the parents (Table 1). Upon examination, only larger microdeletions, overlapping with this region were found in the DECIPHER and the ISCA (International Standards for Cytogenomic Arrays Consortium) databases. An ∼6-Mb deletion at 6q22 was found in a DECIPHER individual 250717 with infantile spasms and ASD.
Figure 2.
Schematic representation of the analyzed genomic region at 6q22.1q22.31. Genomic coordinates correspond to the hg19 build of the human genome. Red bars represent microdeletions. Dotted vertical lines mark the region of the microdeletion overlap in patients 1, 3, and 4 presenting with seizures. Protein-coding genes, ESTs, and non-protein-coding genes are shown. Black and green color depict opposite directions of transcription. Array CGH log ratio plots of microdeletions in patients 3 and 4 are shown. ASD, autism spectrum disorder; DD, developmental delay; DF, dysmorphic features; FS, febrile seizures; GTCS, generalized tonic-clonic seizures; ID, intellectual disability.
Table 1. Clinical features and analysis of 6q21q22.31 microdeletion breakpoints.
| Pt | Chromosomal location | Deletion size (Mb) | Breakpoint coordinates (hg19) | Proximal breakpoint | Distal breakpoint | Microhomology at the breakpoint | GC enrichment at the breakpoint | Parental origin | Seizures | Deleted annotated protein-coding genes |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 6q22.1q22.31 | 1.6 | 117 810 940–117 810 996/119 417 693–119 417 749 | SINE AluY | SINE AluYb8 | 55 bp | Present | De novo | Absence seizures | DCBLD1, GOPC, NUS1, SLC35F1, CEP85L, BRD7P3, PLN, MCM9, ASF1A, FAM184A |
| 2 | 6q21q22.1 | 5.5 | 112 511 751–112 511 752/118 037 595–118 037 596 | Unique sequence | LINE L1MA8 | Absent | Present | De novo | - | LAMA4, RFPL4B, MARCKS, LOC285758, HDAC2, HS3ST5, FRK, NT5DC1, COL10A1, TSPYL4, TSPYL1, DSE, FAM26F, TRAPPC3L, FAM26E, FAM26D, RWDD1, RSPH4A, ZUFSP, KPNA5, FAM162B, GPRC6A, RFX6, VGLL2, ROS1, DCBLD1, GOPC, NUS1 |
| 3 | 6q22.1 | 0.3 | 117 961 791–117 961 792/118 280 043–118 280 044 | Unique sequence | Unique sequence | Absent | Absent | De novo | Generalized epilepsy with atypical absence seizures | NUS1, SLC35F1 |
| 4 | 6q22.1 | 0.2 | 117 971 549–117 971 550/118 218 719–118 218 720 | LINE L1PA7 | LTR ERV1 MER4A1 | Absent | Present at the distal breakpoint | De novo | Cryptogenic generalized epilepsy with atypical absence and tonic/myoclonic seizures | NUS1 |
| 5 | 6q22.1q22.31 | 2.9 | 116 681 080–116 735 056/119 687 719–119 775 014 | Unique sequence | LTR ERVL LTR82A | N/A | N/A | De novo | — | DSE, FAM26F, TRAPPC3L, FAM26E, FAM26D, RWDD1, RSPH4A, ZUFSP, KPNA5, FAM162B, GPRC6A, RFX6, VGLL2, ROS1, DCBLD1, GOPC, NUS1, SLC35F1, CEP85L, BRD7P3, PLN, MCM9, ASF1A, FAM184A, MAN1A1 |
| 6a | 6q22.2q22.31 | 0.2 | 118 410 062–118 436 482/118 604 516–118 635 123 | Unique sequence | LINE L2b | N/A | N/A | Unknown | — | SLC35F1 |
Patient 6 additionally had duplications in Xp11.23 (46 768 761–47 033 527), Xq22.1 (99 441 716–99 949 649), and 16p11.2 (28 833 436–29 046 284).
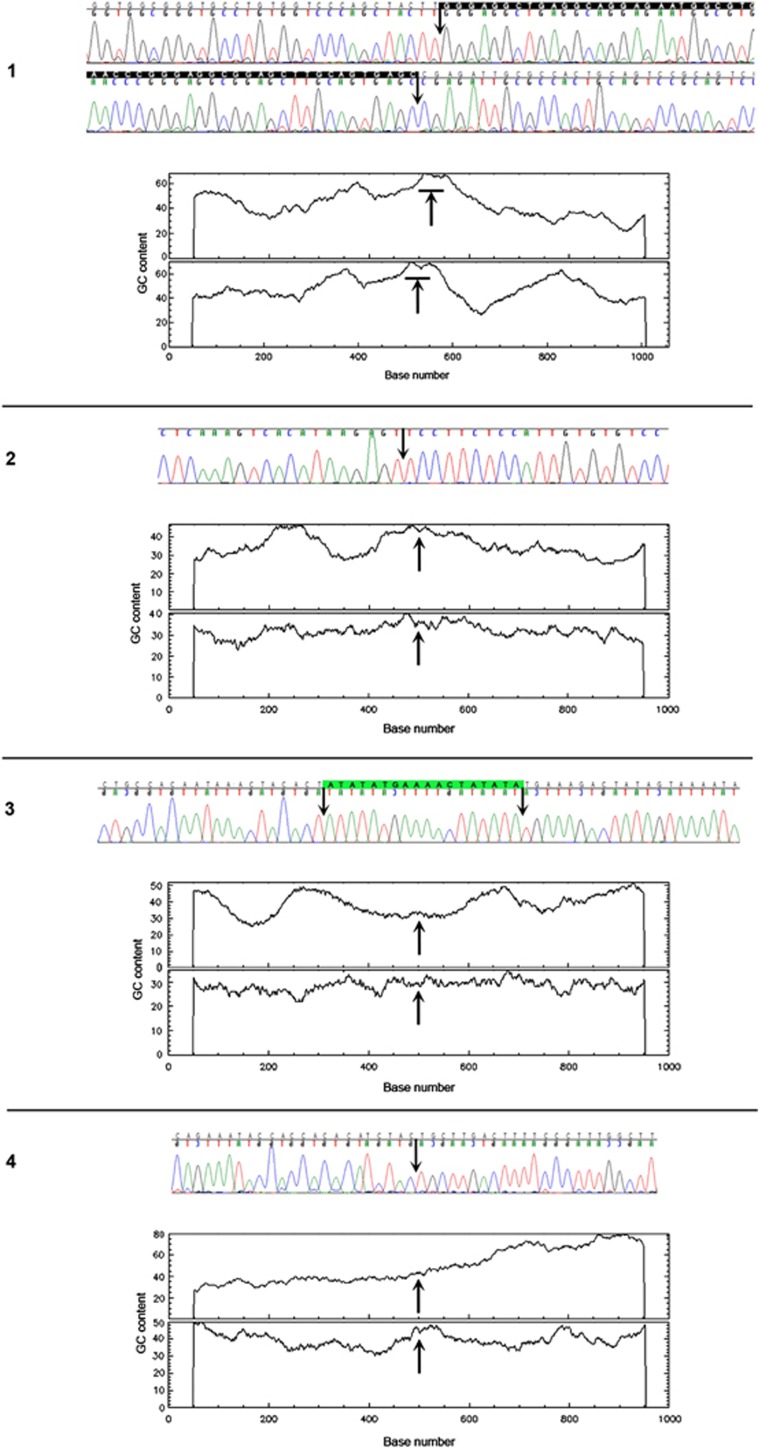
Identification and analysis of microdeletion breakpoints
We sequenced the 6q21q22.31 microdeletion breakpoints of patients 1–4. Five out of eight breakpoints were located within repetitive elements (Table 1). Microhomology was identified in patient 1, consistent with a template switching replicative error mechanism such as FoSTeS/MMBIR.12 We also found the recombination motif 5′-CCTCCCT-3′ and its inverted copy ∼300 bases downstream and upstream of the proximal and distal breakpoints, respectively, in patient 4. Of interest, the chromosomal location of five out of eight breakpoints correlated with a significant local enrichment of GC content (Figure 3, Table 1). The genomic position of the 6q breakpoints in each individual was determined as follows: Patient 1: chr6.hg19:g.(117,810,940_117,810,996)_(119,417,693_119,417,749)del; patient 2: chr6.hg19:g.(112,511,751_112,511,752)_(118,037,595_118,037,596)del; patient 3: chr6.hg19:g.(117,961,791_117,961,792)_(118,280,043_118,280,044)del; patient 4: chr6.hg19:g.(117,971,549_117,971,550)_(118,218,719_118,218,720)del; patient 5: chr6.hg19:g.(116,681,080_116,735,056)_(119,687,719_119,775,014)del; patient 6: chr6.hg19:g.(118,410,062_118,436,482)_(118,604,516–118,635,123)del. The genomic location of all 6q deletions has been submitted to ClinVar database.
Figure 3.
Chromatopherograms of the sequences across the breakpoint junctions and GC content around the breakpoints. Arrows indicate location of the breakpoints. Microhomology is shown in white letters on black background. Junction sequence of unknown origin is highlighted in green.
Expression of the 6q22.1 genes in the brain
The 6q22.1 delineated region includes only one annotated protein-coding gene, NUS1 (Figure 2), and putative regulatory sequences of SLC35F1 (see Supplementary Figure S1). This region also contains several unique ESTs and groups of overlapping ESTs. Two groups of ESTs overlap with NUS1 and may include its alternative spliceoforms, encoding the N-terminal or C-terminal half of nogo-B receptor, NgBR (see Supplementary Figure S2). The unique ESTs include AI858607, BI825833, and BQ429202. In addition, the 6q22.1 region includes the gene for a lncRNA, TCONS_00011550, and two genes for piRNAs, piR-45520 and piR-56963.
Besides NUS1 and SLC35F1, the expression of the ESTs and the non-protein-coding genes located at 6q22.1 is unknown. To determine whether any of them could be considered as an additional candidate gene for epilepsy, we measured by qPCR the abundance of their transcripts in total RNA from the normal human brain and lung fibroblasts as a reference (Table 2). We confirmed enrichment of the expression of NUS1 and SLC35F1 in the brain and found that ESTs CB111179, AI858607, and BQ429202 also exhibited significantly higher expression in the brain compared with control lung fibroblasts, with EST AI858607 having the highest expression. Expression of the lncRNA and piR-56963 genes was very low both in the brain and the lung fibroblasts (data not shown).
Table 2. Expression levels of epilepsy candidate genes/ESTs located at 6q22.1.
| Relative expression (qPCR) | |||||
|---|---|---|---|---|---|
| Gene/EST | Strand | Genomic coordinates (chr6, hg19) | Lung fibroblasts (IMR-90) | Brain | P-value |
| NUS1 | + | 117 996 617–118 031 886 | 0.46±0.14 | 1.06±0.12 | 0.005 |
| EST CB111179 | + | 117 997 225–118 055 519 | 0.074±0.021 | 0.14±0.01 | 0.001 |
| EST AI858607 | − | 118 048 105–118 048 621 | 59.0±6.0 | 269.1±18.0 | 0.0001 |
| EST BI825833 | + | 118 144 355–118 145 047 | 1.6E−05±1.2E−07 | 3.2E−05±2.7E−05 | 0.3 |
| EST BQ429202 | − | 118 220 513–118 221 286 | 1.6E−06±6.6E−07 | 0.52±0.44 | 0.05 |
| SLC35F1 | + | 118 228 689–118 638 839 | 10.3±13.1 | 65.7±15.2 | 0.002 |
The highest expressed gene/EST are highlighted in gray. SD and unpaired t test two-tailed P-values for the difference between expression in the control lung fibroblasts and in the brain are shown.
Discussion
In this report, we identified six unrelated patients with non-recurrent deletions within 6q21q22.31 and established that the loss of this region results in a neurodevelopmental disorder, characterized by language delay, cognitive deficits, epilepsy, and tremors with variable expressivity. Language delay was ascertained in all patients after 2 years of age. Seizure disorder and tremors were observed in three out of six individuals. The incomplete penetrance for the seizure phenotype in this study is consistent with the published data, which suggest that epilepsy is observed in about half of cases when 6q22 is deleted.6 At least two out of six patients with non-overlapping deletions were reported to have mild dysmorphic features. The variability in the phenotype could be indicative of effects due to contiguous gene deletion, rather than loss of a single dosage sensitive gene. Alternatively, the variability may result from the deletion of different loci, as seen in patient 6 with hypotonia, motor delays and dysmorphic facial features. In addition to the 200 kb loss on 6q22.2q22.31 involving SLC35F1, patient 6 had additional CNVs including 265-kb duplication in Xp11.23, 508-kb duplication in Xq22.1, and 213-kb gain of 16p11.2. Parental samples were unavailable for further evaluation in the case of patient 6 and it is unclear whether the small CNV on chromosome 6 is truly pathogenic in this individual. The deletions arose de novo in the remaining five out of six cases.
Despite increasing interest in the genetic bases of pediatric epilepsy, only a few genome-wide studies of CNVs have been performed in individuals with seizure disorder.2, 13, 14, 15, 16, 17, 18 In this study, we narrowed the critical region to the ∼250-kb segment at 6q22.1, harboring only one protein-coding annotated gene, NUS1, several ESTs, two piRNA genes, and a lncRNA gene. Quantitative analyses of the transcripts of these genes and ESTs by qPCR showed that NUS1, ESTs AI858607, CB111179, BQ429202, and adjacent to the deletion overlapping region, SLC35F1 gene are relatively highly expressed in the brain when compared with their expression in cultured lung fibroblasts used as a control.
NUS1 encodes a precursor of the membrane receptor (NgBR) for the N terminus of nogo-B.19 Nogo proteins A, B, and C are important regulators of neuronal and endothelial cell motility and growth.20 NgBR also interacts with and stabilizes NPC2 protein. Of interest, mutations in NPC2 result in Niemann-Pick Type C,21 an autosomal recessive disease characterized by inability to metabolize cholesterol by the cell, and also associated with seizures and ataxia. NUS1 overlaps with two groups of ESTs, some of which likely correspond to its alternative spliceoforms. One of these groups, represented by EST HY124277, encodes the N-terminal half of NgBR, including its transmembrane helix (see Supplementary Figure S2). The other group, represented by EST CB111179, includes part of NUS1 exon 1, the entire exon 2, almost the entire exon 3 and an additional exon not present in NUS1. It encodes the C-terminal part of the NgBR cytoplasmic domain and also includes eight amino-acid residues not present in NgBR (see Supplementary Figure S2). The functional significance of these isoforms is unknown. At this time, we can only speculate that they could function as competitive inhibitors of NgBR and NgB or have novel functions. We found that the level of the expression of the cytoplasmic domain-coding spliceoform corresponding to the EST CB111179 is lower than that of NUS1, although it is still relatively high and brain-specific.
SLC35F1 encodes putative solute transporter that is also mainly expressed in the brain.22 No part of SLC35F1 is deleted in patient 4, who had epilepsy. Nevertheless, the 6q22.1 deletion in this patient removes putative upstream regulatory sequences of SLC35F1. Poduri et al6 sequenced exons of the 16 candidate genes in the 6q16.3q22.31 region, including NUS1 and SLC35F1 from the large pedigree with GEFS+, but no sequence alterations or CNVs were found in the affected subjects. However, testing of promoters, distant and intronic enhancers, and other regulatory regions of these genes has not been reported.
Brain-specific non-coding EST BQ429202 (not deleted in patient 4) does not likely encode any polypeptide, and thus may correspond to an antisense lncRNA that, based on the proximity to SLC35F1, may regulate expression of SLC35F1. EST AI858607 exhibits the highest brain expression of all genes and ESTs at 6q22.1. It is transcribed in the opposite direction than NUS1 or SLC35F1 and encodes a polypeptide that shows limited amino-acid sequence similarity to an olfactory receptor (see Supplementary Figure S3). Because of its relatively high brain-specific expression, we propose considering this EST for future structural and functional studies.
Of note, the critical region at 6q22.1 also harbors two piRNAs and a lncRNA gene. The lncRNA, TCONS_00011550, is an antisense RNA that may regulate expression of the neighboring NUS1 gene. The brain expression of the piRNA gene, piR-56963, is very low in comparison with the annotated protein-coding genes and the majority of ESTs. This is consistent with the fact that the majority of piRNAs have germline-restricted expression where they epigenetically regulate transposition events.23
In summary, we conclude that the most likely candidate genes for the neurodevelopmental phenotype, including epilepsy and tremors, on 6q21 locus involve NUS1 and EST AI858607. It is also possible that mutations or CNVs in SLC35F1 or its regulatory regions contribute to epilepsy. In addition, supporting our previous reports,1, 9, 24, 25 we continue to observe increased GC content locally at the DNA-breakpoint junctions of microdeletions. We believe that this may be a more general phenomenon with broader implications for understanding mechanisms of CNVs on a genome-wide scale.
Acknowledgments
We thank the families for participating in the study. Dr Seema R Lalani is supported by grants from the Doris Duke Charitable Foundation (DDCF) and Gillson Longenbaugh Foundation.
DISCLAIMER
We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
The Department of Molecular and Human Genetics at Baylor College of Medicine offers extensive genetic laboratory testing, including chromosomal microarray analysis, and derives revenue from this activity. Jill A Rosenfeld is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer Inc. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Ramocki MB, Bartnik M, Szafranski P, et al. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am J Hum Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorkowski AR, Thio LL, Rosenfeld JA, et al. Copy number variants and infantile spasms: evidence for abnormalities in ventral forebrain development and pathways of synaptic function. Eur J Hum Genet. 2011;19:1238–1245. doi: 10.1038/ejhg.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassuk AG, Geraghty E, Wu S, et al. Deletions of 16p11.2 and 19p13.2 in a family with intellectual disability and generalized epilepsy. Am J Med Genet A. 2013;161A:1722–1725. doi: 10.1002/ajmg.a.35946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig I, Hartmann C, Mefford HC. The unexpected role of copy number variations in juvenile myoclonic epilepsy. Epilepsy Behav. 2013;28 (Suppl 1:S66–S68. doi: 10.1016/j.yebeh.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Mullen SA, Carvill GL, Bellows S, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013;81:1507–1514. doi: 10.1212/WNL.0b013e3182a95829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Wang Y, Gordon D, et al. Novel susceptibility locus at chromosome 6q16.3-22.31 in a family with GEFS+ Neurology. 2009;73:1264–1272. doi: 10.1212/WNL.0b013e3181bd10d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld JA, Amrom D, Andermann E, et al. Genotype-phenotype correlation in interstitial 6q deletions: a report of 12 new cases. Neurogenetics. 2012;13:31–47. doi: 10.1007/s10048-011-0306-5. [DOI] [PubMed] [Google Scholar]
- Boone PM, Bacino CA, Shaw CA, et al. Detection of clinically relevant exonic copy-number changes by array-CGH. Hum Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski P, Schaaf CP, Person RE, et al. Structures and molecular mechanisms for common 15q13.3 microduplications involving CHRNA7: benign or pathological. Hum Mutat. 2010;31:840–850. doi: 10.1002/humu.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, McDonald-McGinn DM, et al. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet. 2008;74:469–475. doi: 10.1111/j.1399-0004.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- Duker AL, Ballif BC, Bawle EV, et al. Paternally inherited microdeletion at 15q11.2 confirms a significant role for the SNORD116 C/D box snoRNA cluster in Prader-Willi syndrome. Eur J Hum Genet. 2010;18:1196–1201. doi: 10.1038/ejhg.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Guerrini R. Chromosomal disorders associated with epilepsy. Epileptic Disord. 2005;7:181–192. [PubMed] [Google Scholar]
- Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Ann Neurol. 2011;70:974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodiya SM, Mefford HC. Genetic contribution to common epilepsies. Curr Opin Neurol. 2011;24:140–145. doi: 10.1097/WCO.0b013e328344062f. [DOI] [PubMed] [Google Scholar]
- Bartnik M, Szczepanik E, Derwińska K, et al. Application of array comparative genomic hybridization in 102 patients with epilepsy and additional neurodevelopmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:760–771. doi: 10.1002/ajmg.b.32081. [DOI] [PubMed] [Google Scholar]
- Miao RQ, Gao Y, Harrison KD, et al. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc Natl Acad Sci USA. 2006;103:10997–11002. doi: 10.1073/pnas.0602427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab ME. Functions of Nogo proteins and their receptors in the nervous system. Nat Rev Neurosci. 2010;11:799–811. doi: 10.1038/nrn2936. [DOI] [PubMed] [Google Scholar]
- Harrison KD, Miao RQ, Fernandez-Hernándo C, Suárez Y, Dávalos A, Sessa WC. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 2009;10:208–218. doi: 10.1016/j.cmet.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Suzuki S, Satoh T, Naito S. Tissue-specific mRNA expression profiles of human solute carrier 35 transporters. Drug Metab Pharmacokinet. 2009;24:91–99. doi: 10.2133/dmpk.24.91. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Dharmadhikari AV, Kang SH, Szafranski P, et al. Small rare recurrent deletions and reciprocal duplications in 2q21.1, including brain-specific ARHGEF4 and GPR148. Hum Mol Genet. 2012;21:3345–3355. doi: 10.1093/hmg/dds166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittwald P, Gambin T, Szafranski P, et al. NAHR-mediated copy-number variants in a clinical population: mechanistic insights into both genomic disorders and Mendelizing traits. Genome Res. 2013;23:1395–1409. doi: 10.1101/gr.152454.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.