Abstract
Metaphyseal anadysplasia (MANDP) is an uncommon chondrodysplasia characterized by early-onset metaphyseal dysplasia and short stature that improves with age. MANDP is caused by mutations in the matrix metalloproteinase (MMP) 9 and 13 genes. Autosomal dominant (AD) MANDP has been described as more severe, and has been associated with dominant-negative MMP13 mutations that suppress activity of both MMP9 and MMP13; autosomal recessive (AR) MANDP has been described as a milder form associated with AR missense mutations in MMP9 or MMP13. Here we describe the molecular characterization of skeletal dysplasia in two brothers who presented with short stature and mixed epiphyseal and metaphyseal dysplasia. Whole-exome sequencing (WES) identified a homozygous C>T transition mutation in exon 2 of MMP13 (c.325C>T) on chromosome 11q22.2 resulting in a premature stop codon p.(R109*) that is predicted to abolish MMP13 activity. This report extends the MANDP phenotype by illustrating that AR nonsense mutations in MMP13 can lead to short stature that persists beyond childhood.
Introduction
Metaphyseal anadysplasia (MANDP) types 1 and 2 are caused by mutations in the matrix metalloproteinase 13 (MMP13; MIM*600108) and 9 (MMP9; MIM*120361) genes.1 These conditions share a similar phenotype marked by short stature and metaphyseal dysplasia of early onset that improves with age.2, 3, 4, 5, 6 Lausch et al1 proposed a clinical classification scheme that is based on phenotype and inheritance pattern, wherein MANDP1 is more severe and associated with autosomal dominant (AD) mutations in MMP13, whereas MANDP2 is milder and associated with autosomal recessive (AR) mutations in MMP13 or MMP9. More recently, a preliminary report described recessive mutations in MMP13 as the basis for metaphyseal chondrodysplasia, Spahr Type (MDST; MIM 250400).7 This finding challenges the clinical classification scheme proposed by Lausch et al, wherein AR MANDP is viewed as a milder condition, as patients with MDST can have severe short stature that persists into adulthood.8 Here we report the use of whole-exome sequencing (WES) to identify homozygosity for a nonsense mutation in MMP13 as the basis for AR MANDP in two brothers presenting with short stature, mixed epiphyseal and metaphyseal dysplasia, and joint pain.
Methods
Study participants
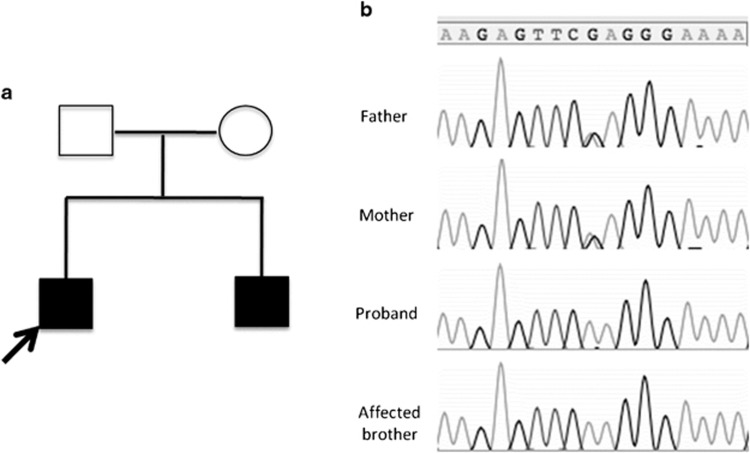
The study included two brothers and their unaffected parents (Figure 1a), all of whom provided written informed consent for participation in the study. The study was approved by the institutional review board at The Children's Hospital of Philadelphia.
Figure 1.
(a) Pedigree of family showing two affected brothers and unaffected parents. (b) Sanger sequencing confirmed the identified mutation in affected brothers and parents.
The proband, patient 1, presented at age 15 years for evaluation of chronic knee pain and skeletal dysplasia. He was 149.8 cm (<1st percentile, −2.6 SD) tall and weighed 49.3 kg (17th percentile, −1.0 SD); arm span to standing height ratio was 1.06; upper to lower segment ratio was 1.09. There was mesomelic shortening of the lower limbs and mild genu varum. The bone age at chronological age of 14 years and 9 months was 15 years (1 SD±10.4 months). Skeletal survey revealed areas of sclerosis and irregularity in the metaphyses and epiphyses of the distal femurs, in the proximal tibiae and the distal tibiae, and in the fibulae (Supplementary Figure 1–4). Narrowing of the joint space was seen at the knee. Metaphyses and epiphyses of the upper extremities were normal. The spine was normal except for mild reversal of cervical lordosis.
Patient 2 presented at age 7 years and 5 months for evaluation of abnormal gait and skeletal dysplasia. He was 114 cm tall (3rd percentile, −1.9 SD) and weighed 26.6 kg (73rd percentile, 0.6 SD); arm span to standing height ratio was 1.09 and upper to lower segment ratio was 1.05. Physical examination was notable for coxa vara, genu vara, pes planus and a waddling gait. Bone age obtained at 6 years and 11 months was 6 years (SD of 8.9 months). Skeletal survey revealed mild bilateral femoral bowing and shortening, as well as irregular metaphyseal changes and flaring at the distal femurs. The spine was normal (Supplementary Figures 5–8).
Patients 1 and 2 were full brothers born to parents of Albanian nationality who denied consanguinity. The father was 170 cm tall (17th percentile, −1.0 SD) and the mother was 152 cm tall (4th percentile, −1.7 SD), resulting in a predicted mid-parental height of 168 cm (10th percentile, −1.3 SD). Past medical history was unremarkable for both patients. Both boys had normal renal, hepatic and endocrine function and age-appropriate markers of bone metabolism (Supplementary Table 1). Family history was notable only for constitutional delay of growth and development in the father and short stature (height<152 cm) in the maternal grandmother and a paternal aunt. There was no known history of skeletal abnormalities. MANDP was not initially considered because patient 1 had more severe short stature compared with his younger brother.
Whole-exome sequence analysis
The exome was captured and sequenced for patients 1 and 2 using standard protocols using genomic DNA from peripheral blood mononuclear cells. The raw reads were aligned to the reference human genome using the BWA,9 and single-nucleotide variants (SNVs) and small insertions/deletions (indels) were identified using the GATK.10 Annovar11 and SnpEff (http://snpeff.sourceforge.net/) were used to functionally annotate the variants.
Based on family history, an AR mode of inheritance was assumed and genes carrying either rare homozygous or compound heterozygous variants were considered. We excluded coding variants that (1) were synonymous, (2) had a minor allele frequency (MAF) of >0.01 in either 1000 Genomes Project or NHLBI Exome Sequencing Project (ESP6500SI; http://evs.gs.washington.edu/EVS/) or (3) had been previously identified in our in-house database (Supplementary Table 2). There is a high rate of identity by descent (IBD) among Albanians;12 therefore, to shorten the list of candidate variants, we applied a Hidden–Markov Model Algorithm13 to the WES data to infer the loci in which both patients shared either homozygosity or heterozygosity (here termed as IBD=2). Validation of the mutation candidate was performed by Sanger sequencing in all family members.
Results
Exome sequencing and bioinformatics
An average coverage of 56 × was obtained. A total of 43 296 SNVs and 3476 indels were identified in the two samples that underwent WES, of which 42 431 (98.0%) and 2300 (66.2%) were reported in dbSNP135. Analysis revealed a homozygous substitution (c.325C>T) in exon 2 of MMP13 (exons are numbered like in GenBank: NG_021404.1 covering MMP13 gene transcript NM_002427.3) as the likely disease-causing candidate. The brothers were compound heterozygous for mutations in PRRC2A (NM_080686.2) and SYT15 (NM_031912.4); this was excluded as a disease candidate because these genes are not known to affect skeletal development.
The MMP13 mutation was confirmed by Sanger sequencing (Figure 1b), and both parents were heterozygous. The variant was present in the ESP6500SI data set with a MAF of 0.000077 and was reported in dbSNP138 (rs369083541) without any clinical significance, but analysis of the 1000 Genomes Project, the catalog of somatic mutations in cancer (COSMIC v.67) and existing sequence data from 1200 WES samples in our database did not reveal other occurrences of this mutation. Analyses of SNPs in and near the MMP13 genes in the affected siblings indicated that the c.325C>T mutation arose in a common founder and that the two mutant alleles were identical by descent (Supplementary Figure 9). The c.325C>T mutation inserts a nonsense codon p.(Arg109Ter) in the transcribed mRNA, which if translated is predicted to result in a markedly truncated, nonfunctional protein. Alternatively, nonsense-mediated decay of abnormal transcripts would be expected to prevent translation of protein. The variant was submitted to the Leiden Open Variation Database (LOVD, http://MMP13.lovd.nl).
Discussion
To our knowledge this is the first report of the c.325C>T nonsense mutation in MMP13 in an AR form of MANDP. Lausch et al1 described the molecular basis of MANDP, identifying mutations in MMP13 and MMP9 as underlying the diagnosis of MANDP. There have been three families with AD MANDP in which two different dominant-negative missense mutations have been identified in exon 2 of MMP13. These mutations disrupt the pro-domain of MMP13 and resulted in decreased secretion of both MMP9 and MMP13 proteins.1 An AD missense mutation in the pro-domain of MMP13 has also been identified as the cause of spondyloepimetaphyseal dysplasia Missouri type (SEMDMO).14
The previously described case of AR MANDP due to MMP13 was ascribed to a homozygous missense mutation in exon 5 (c.722C>A). The affected individual was described to have short stature during childhood; however, adult height was not reported.1 Similarly, adult height was not reported in the abstract detailing AR MMP13 mutations as the cause of MDST.7
Notably, the two patients reported by us have not demonstrated catch-up growth, and the older brother has a more significant height deficit than his younger brother (height SD of −2.6 SD at 15 years vs −1.9 at 7 years). Moreover, both heterozygous parents have modest height deficits, raising the possibility of a gene dosage effect. The contribution of MMP9 and MMP13 function to height appears complex. Mouse models revealed that the loss of both MMP9 and MMP13 proteins in the Mmp9−/−; Mmp13−/− double knockout mice led to more severe growth plate abnormalities compared with loss of MMP13 alone.15, 16 It is not clear that this is the case in humans, however, as there is considerable variability and overlap in the degree of short stature reported in cases of AD MANDP (loss of MMP9 and MMP13) and MDST (loss of MMP13 alone).1, 8, 17
Also notable is the chronic joint pain described by patient 1, a finding not classically described in MANDP but that has been reported in association with MDST.8 This may be related to epiphyseal sclerosis and narrowing of the joint space (Supplementary Figure 1), although the absence of serial radiographs makes it unclear whether the epiphyseal involvement is directly related to MMP13 dysfunction or is a complication of long-standing varus deformity.
In summary, we have identified a founder mutation in exon 2 of the MMP13 gene as a new cause of AR MANDP in two Albanian brothers. These cases add to the reported phenotype of AR MANDP, suggesting that this condition is associated with deleterious skeletal effects including short stature that can persist into adulthood. This finding may have important clinical implications in the counseling and treatment strategies employed in the care of children affected with this disease.
Acknowledgments
We acknowledge Sara Pinney for referring the patients to the Bone Health Center, MaryJane Benton, who helped in the attainment of participant samples, and Cuiping Hou, Maizan Lai and Evan Opas, who provided technical assistance. The research presented in this manuscript was supported by an Institutional Clinical and Translational Science Award: NIH/NCATS, Grant UL1TR000003.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Author Contributions
Study design: DL, HH, ML. Study conduct: DL, HH, ML. Data analysis: DL, HH, ML. Data interpretation: DL, DW, MD, HH, ML. Drafting manuscript: DL and DW. Revising manuscript content: DL, DW, MD, HH, ML. Approving the final version of the manuscript: DL, DW, MD, HH, ML. ML takes responsibility for the integrity of the data analysis.
Supplementary Material
References
- Lausch E, Keppler R, Hilbert K, et al. Mutations in MMP9 and MMP13 determine the mode of inheritance and the clinical spectrum of metaphyseal anadysplasia. Am J Hum Genet. 2009;85:168–178. doi: 10.1016/j.ajhg.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura G, Ikegawa S, Saga T, Nagai T, Aya M, Kawano T. Metaphyseal anadysplasia: evidence of genetic heterogeneity. Am J Med Genet. 1999;82:43–48. doi: 10.1002/(sici)1096-8628(19990101)82:1<43::aid-ajmg9>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Wiedemann HR, Spranger J. Chondrodysplasia metaphysaria (dysostosis metaphysaria) – ein neur typ. Z Kinderheilk. 1970;108:171–186. [PubMed] [Google Scholar]
- Maroteaux P, Verloes A, Stanescu V, Stanescu R. Metaphyseal anadysplasia: a metaphyseal dysplasia of early onset with radiological regression and benign course. Am J Med Genet. 1991;39:4–10. doi: 10.1002/ajmg.1320390103. [DOI] [PubMed] [Google Scholar]
- Le Merrer M, Maroteaux P. Metaphyseal anadysplasia type II: a new regressive metaphyseal dysplasia. Pediatr Radiol. 1998;28:771–775. doi: 10.1007/s002470050463. [DOI] [PubMed] [Google Scholar]
- Slama M, Mathieu M, Dehouck I, et al. Metaphyseal anadysplasia in two sisters. Pediatr Radiol. 1999;29:372–375. doi: 10.1007/s002470050610. [DOI] [PubMed] [Google Scholar]
- Gorna M, Liang J, Campos-Xavier AB, et al. 2013. Available at http://isdsbologna2013.org/blog/presentation/mmp2013-mutations-are-the-cause-of-recessive-metaphyseal-dysplasia-spahr-type/ .
- Megarbane A, Chouery E, Ghanem I. A multiplex family with possible metaphyseal Spahr-type dysplasia and exclusion of RMRP and COL10A1 as candidate genes. Am J Med Genet. 2008;146a:1865–1870. doi: 10.1002/ajmg.a.32390. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 2007;96:644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Rodelsperger C, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Kennedy AM, Inada M, Krane SM, et al. MMP13 mutation causes spondyloepimetaphyseal dysplasia, Missouri type (SEMD(MO) J Clin Invest. 2005;115:2832–2842. doi: 10.1172/JCI22900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickens D, Behonick DJ, Ortega N, et al. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada M, Wang Y, Byrne MH, et al. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci USA. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag TI, Teebi AS. The second family with Spahr-type metaphyseal chondrodysplasia: autosomal recessive inheritance confirmed. Clin Genet. 1990;38:237–239. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.