Abstract
Background
Actinomyces are a common part of the residential flora of the human intestinal tract, genitourinary system and skin. Isolation and identification of Actinomyces by conventional methods is often difficult and time consuming. In recent years, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) has become a rapid and simple method to identify bacteria.
Objective
The present study evaluated a new in-house algorithm using MALDI-TOF-MS for rapid identification of different species of oral Actinomyces cultivated from subgingival biofilm.
Design
Eleven reference strains and 674 clinical strains were used in this study. All the strains were preliminarily identified using biochemical methods and then subjected to MALDI-TOF-MS analysis using both similarity-based analysis and classification methods (support vector machine [SVM]). The genotype of the reference strains and of 232 clinical strains was identified by sequence analysis of the 16S ribosomal RNA (rRNA).
Results
The sequence analysis of the 16S rRNA gene of all references strains confirmed their previous identification. The MALDI-TOF-MS spectra obtained from the reference strains and the other clinical strains undoubtedly identified as Actinomyces by 16S rRNA sequencing were used to create the mass spectra reference database. Already a visual inspection of the mass spectra of different species reveals both similarities and differences. However, the differences between them are not large enough to allow a reliable differentiation by similarity analysis. Therefore, classification methods were applied as an alternative approach for differentiation and identification of Actinomyces at the species level. A cross-validation of the reference database representing 14 Actinomyces species yielded correct results for all species which were represented by more than two strains in the database.
Conclusions
Our results suggest that a combination of MALDI-TOF-MS with powerful classification algorithms, such as SVMs, provide a useful tool for the differentiation and identification of oral Actinomyces.
Keywords: MALDI-TOF-MS, oral Actinomyces, support vector machine
Actinomyces are a common part of the residential flora of the human intestinal tract as well as other habitats such as the genitourinary tract system and the skin. They are gram-positive, anaerobic, and aerotolerant, non-spore-forming, non-motile pleomorphic rods. Although the genus Actinomyces was already described in 1919, many new species were found quite recently. Although in 1986 only 10 species were recognized as Actinomyces (1), the number has increased to at least 36 by now (2), 20 of them being relevant for human medicine. Actinomyces species are mainly associated with cervicofacial actinomycosis, oral or cerebral abscesses, caries, and periodontitis (1, 3, 4). They seem to play a bigger role than expected in the pathogenesis of osteoradionecrosis- and bisphosphonate-related osteonecrosis of the jaw (5, 6), and can cause lethal infection such as mediastinitis (7). As a consequence, fast and reliable identification methods for Actinomyces species have become increasingly important.
Isolation and identification of Actinomyces by conventional methods is often difficult and time consuming. Many studies have been performed to characterize Actinomyces species using phenotypic (8–10) and molecular (11, 12) approaches. Most of the available commercial identification kits (Rapid ID 32 A, API Coryne, VITEK 2, ANC ID Card, bioMerieux, and VITEK-MS, bioMerieux) do not include the majority of newer species in their database and the sophisticated molecular methods, such as chromosomal DNA fingerprinting, arbitrarily primed PCR, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) (13), and 16S ribosomal RNA (rRNA) sequencing, are still available only in research and reference laboratories.
In recent years, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) has become a rapid and simple method to identify bacteria. However, this method can be used for routine detection only if high quality reference spectra databases are available (14, 15). Moreover, if phenotypically similar bacterial species are to be discriminated, powerful algorithms for spectra analysis are critical for success.
The present study aimed to evaluate a new in-house identification algorithm using MALDI-TOF-MS for rapid identification of different species of oral Actinomyces cultivated from subgingival biofilm.
Material and methods
Bacterial strains
In total, 685 bacterial strains were used in this study. Eleven were reference strains: Actinomyces dentalis (DSM 19115), Actinomyces georgiae (DSM 6843), Actinomyces gerencseriae (ATCC 23860), Actinomyces graevenitzii (DSM 15540), Actinomyces neuii (DSM 8576), Actinomyces odontolyticus (DSM 43331), Actinomyces radicidentis (DSM 15433), Actinomyces viscosus (DSM 43798), Actinomyces naeslundii (DSM 17233), Actinomyces oris (DSM 23056), and Actinomyces israelii ATCC 12107.
The other 674 strains were fresh clinical isolates from the subgingival biofilm of patients with chronic periodontitis. The presumptive identification of the clinical strains was performed by established biochemical methods: colony morphology, pigmentation, gram stain morphology, catalase test, CAMP test, and Rapid ID 32 A. Flowcharts for preliminary identification of Actinomyces species proposed from Sarkonen et al. were also used (9). The newly described species (Actinomyces timonensis, Actinomyces massiliensis, and A. dentalis) were preliminarily identified using the diagnostic traits of the type strains (16–18).
The genotypes of all the reference strains and of 232 clinical isolates were identified by sequence analysis of the 16S rRNA gene. The strain sequences were compared with sequences deposited in GenBank using the program BLAST through the NCBI server and with the sequences deposited in the Human Oral Microbiome Database (HOMD) using the program HOMD 16S rRNA Sequence Identification (19). We included in the reference database only the strains with percent identities of at least 99% in both programs.
The similarity in 16S rRNA gene sequence analysis was very similar for A. odontolyticus and Actinomyces meyeri in both programs that we used. Biochemical characteristics cannot separate them either, so we pooled the two species and attempted using MALDI-TOF-MS only to differentiate this group from other Actinomyces species.
MALDI-TOF-MS sample preparation
Individual colonies of each isolate or reference strain were subcultured on Columbia blood agar for 4 days at 37°C in an anaerobic chamber (Whitley MG1000 anaerobic workstation, Meintrup DWS Laborgeräte, GmbH, Germany). Colonies from the half surface of plates were suspended in 1 ml DNase-free water (SIGMA, Taufkirchen, Germany) and then centrifuged at 8,500 g for 15 min. The further processing of the samples was done according to Friedrichs et al. (15). For each strain, 10 consecutive spots were prepared. In order to demonstrate reproducibility, the sample preparation was repeated for each strain, starting with a new culture.
MALDI-TOF-MS analysis
The MALDI-TOF-MS procedures have been described in detail elsewhere (15). Briefly, we used a MALDI-TOF mass spectrometer, Autoflex II (Bruker Daltonics) with a nitrogen laser (337 nm) operated in positive linear mode (delay 150 ns, voltage 20 kV, mass range 3–20 kDa), and we used a Flexcontrol software version 2.4 (Bruker Daltonics). Each spectrum was automatically obtained (average of 500 laser shots). The spectra were calibrated externally using the Escherichia coli DH 5 alpha strain prepared the same way as the clinical samples. The data files were transferred to Flexanalysis version 2.4 (Bruker Daltonics) for automated peak extraction. Sixty peaks were automatically labeled in each spectrum according to their appearance above the background (threshold ratio 1.5). We controlled manually the correct labeling. Peak lists containing masses and intensities were exported as Excel files.
Cluster formation of the mass peaks
To refine spectra accuracy, peak lists were aligned for mass drift adjustment (15). Briefly, a mass-dependent size of the mass window was used according to window size=sizeabs+(sizerel * peak mass) with sizeabs=0.8 m/z and sizerel=0.001. Thus, for each bacterial species we arrived at a mean spectrum containing common m/z values. All spectra obtained for this species were aligned individually to the peaks of the mean spectrum by linear mass adjustment of the peaks (20). Subsequently, peak clusters were formed which contained all peaks originating from different individual spectra, however, occurring in the same window. All peaks assigned to one cluster are represented by the respective mean cluster mass. In this study, each sample was analyzed 10 times (see MALDI-TOF-MS sample preparation). The aligned m/z values of cluster peaks occurring in one sample and the respective mean peak intensities are used for further analysis and denoted sample centroid. To set up the mass spectra reference database all sample centroids of a species were combined to species centroids.
Similarity analysis
A hierarchical clustering procedure performed with the MatLab software (R2013b; the Math-Works Inc., Natick, MA) (21) was used for both characterization of the similarity relations between the species centroids of the mass spectra reference database and identification of samples by finding the species centroid of the data base which is most similar to the sample centroid. The similarity between centroids was determined by pairwise comparison. The Jaccard similarity measure was applied to the similarity of the sample centroids. From the Jaccard similarity coefficients distances were calculated (1-Jaccard similarity coefficient), which are the percentages of non-zero cluster mass peaks that differ between the sample centroids (21). The number of clusters to which the two centroids contributed was counted. By this procedure, a symmetric matrix of pairwise similarities (peak mass-based similarity matrix) was formed. Distance matrices were calculated from normalized similarity matrices and dendrograms were calculated on the basis of the distance matrices by using a complete linkage function.
Classification analysis
A machine learning method was used as an alternative to similarity analysis in order to better identify Actinomyces species. Sample centroids were classified using the support vector machine (SVM) tool implemented in the Bioinformatic toolbox of MatLab (21). The SVM algorithm was trained with the reference database sample centroids of bacteria of known identity using a ‘one against one’ approach. To estimate the class prediction a 10-fold cross-validation error was calculated for the training group. The training set was first divided into 10 subsets of equal size. Sequentially, one subset was tested by using the classifier trained on the remaining nine subsets. Consequently, each probe of the training set was predicted once. The cross-validation accuracy is the percentage of data which were correctly classified (15).
Results and discussion
Many scientific and clinical investigations have been hampered by problems in the identification of Actinomyces species. The laboratory growth of these organisms is challenging, so identification of Actinomyces species is often based on histopathology and biopsy material. This has led to an empirical treatment rather than a true diagnosis of actinomycotic infections. Therefore, efficient, reliable, and rapid methods for the identification of Actinomyces at the species level would be of considerable clinical value.
The sequence analysis of the 16S rRNA gene of all references strains confirmed their previous identification. One hundred and forty clinical strains were identified as Actinomyces species by sequence analysis of 16S rRNA genes showing percent identities of at least 99% in both the NCBI BLAST and the HOMD Blast. The sample centroids calculated from the MALDI-TOF-MS spectra obtained from the reference strains and 100 confirmed Actinomyces were used to create the Actinomyces mass spectra reference database (Table 1). The other 40 well-identified clinical strains (20 A. naeslundii and 20 A. oris) were used as a test group. For each Actinomyces species of the reference database a species centroid was calculated from all sample centroids available from strains of this species. The species centroids were used for quality assessment of the reference database and for identification of unknown clinical samples. Actinomyces johnsonii, formerly known as A. naeslundii serotype WVA 963, with only four entries proved to be very similar with A. naeslundii strains. That was the reason for pooling these two species in one group when we used the database to identify the unknown clinical strains.
Table 1.
Species of Actinomyces which were included in the reference database
| Species | No. of strains |
|---|---|
| Actinomyces dentalis | 6 |
| Actinomyces gerencseriae | 15 |
| Actinomyes georgiae | 2 |
| Actinomyces graevenitzii | 1 |
| Actinomyces urogenitalis | 1 |
| Actinomyces europaeus | 1 |
| Actinomyces israelii | 15 |
| Actinomyces meyeri/odontolyticus | 6 |
| Actinomyces oris | 20 |
| Actinomyces radicidentis | 1 |
| Actinomyces timonensis | 4 |
| Actinomyces naeslundii/johnsonii | 24 |
| Actinomyces neuii | 3 |
| Actinomyces massiliensis | 11 |
| Actinomyces viscosus | 1 |
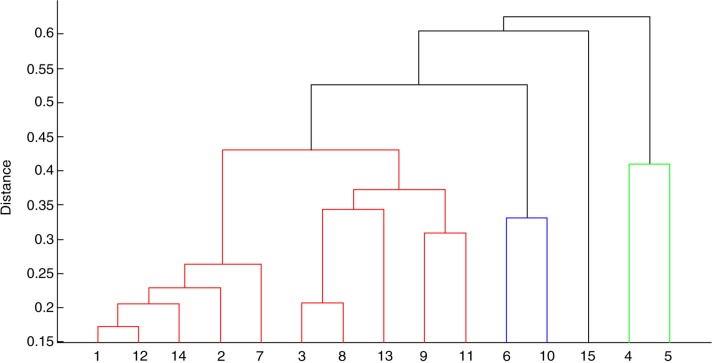
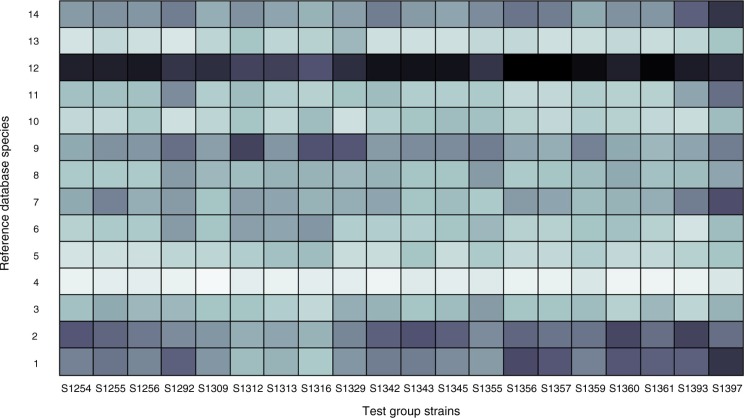
Already a visual inspection of the species centroids of different Actinomyces species reveals both similarities and differences. The results of a computational similarity analysis are shown in Fig. 1. The dendrogram consists of three significantly different main branches of Actinomyces. Remarkably, the one represented by A. graevenitzii and Actinomyces urogenitalis is more dissimilar to the other Actinomyces than is Streptococcus sanguinis, which was included as an external reference. With only one reference spectrum created, one could wonder if the strains are good representatives for these species. The species within each main branch show different degrees of similarity. However, the differences between them are not large enough to allow a reliable differentiation. This is mainly caused by the inhomogeneity of the sample centroids of each species due to biological variation of strains and in some cases also due to a small number of samples contributing to the respective species. Both effects hamper the species identification of unknown samples by similarity analysis. For illustration, the results of a similarity analysis of a test group of 20 confirmed A. naeslundii samples are shown in Fig. 2. The sample centroids of each sample were compared pairwise with the species centroids of the reference database. Black indicates identity, whereas white indicates maximum dissimilarity. For a reliable species identification, each sample should reveal a high degree of similarity to the reference database species centroid of A. naeslundii and much less similarity to all other species centroids. As can be seen, this is true only for some samples, for example, S1309, whereas other samples show comparable similarities to different species centroids, for example, S1397.
Fig. 1.
Phenotypic relation between the species centroids of the Actinomyces reference database. The dendrogram was generated by similarity analysis. 1. A. dentalis, 2. A. gerencseriae, 3. A. georgiae, 4. A. graevenitzii, 5. A. urogenitalis, 6. A. europaeus, 7. A. israelii, 8. A. meyeri/odontolyticus, 9. A. oris, 10. A. radicidentis, 11. A. timonensis, 12. A. naeslundii/johnsonii, 13. A. neuii, 14. A. massiliensis, 15. S. sanguinis.
Fig. 2.
Similarity of the test group of 20 A. naeslundii strains on the x-axis with the centroids of the Actinomyces reference database on the y-axis. Black indicates identity while white indicates maximum dissimilarity. 1. A. dentalis, 2. A. gerencseriae, 3. A. georgiae, 4. A. graevenitzii, 5. A. urogenitalis, 6. A. europaeus, 7. A. israelii, 8. A. meyeri/odontolyticus, 9. A. oris, 10. A. radicidentis, 11. A. timonensis, 12. A. naeslundii/johnsonii, 13. A. neuii, 14. A. massiliensis.
Therefore, classification based on a SVM algorithm was applied as an alternative approach for differentiation and identification of Actinomyces at the species level. A cross-validation of the reference database represented by the sample centroids of the 14 Actinomyces species yielded correct results (accuracy = 100%) for all species which were represented by more than two strains in the database. This provides a sound basis for an assignment of unknown Actinomyces strains to those species which are represented in the reference database by a sufficient number of different strains to reflect the biological heterogeneity within a species. Identification results pointing to species which are represented by less than 10 samples (A. georgiae, A. radicidentis, A. urogenitalis, Actinomyces europaeus, A. neuii, and A. graevenitzii) may be correct but have to be interpreted with caution and should be verified by 16S rRNA sequencing. The performance of the classification analysis was tested by two test groups of 20 samples each of confirmed A. naeslundii and A. oris. All of them were correctly identified (correct rate=100%, sensitivity=1, specificity=1). Taken into account that A. oris, formerly known as A. naeslundii genotype 2, is closely related to A. naeslundii, this result is encouraging.
The results of the identification of the unknown clinical samples by classification analysis are given in Table 2.
Table 2.
Identification of unknown Actinomyces species using classification by pattern recognition
| Bacterial strains | No. of clinical strains |
|---|---|
| Actinomyces dentalis | 44 |
| Actinomyces gerencseriae | 58 |
| Actinomyces georgiae | 16 |
| Actinomyces graevenitzii | 1 |
| Actinomyces urogenitalis | 17 |
| Actinomyces europaeus | 0 |
| Actinomyces israelii | 57 |
| Actinomyces meyeri/odontolyticus | 8 |
| Actinomyces oris | 58 |
| Actinomyces radicidentis | 0 |
| Actinomyces timonensis | 7 |
| Actinomyces naeslundii/johnsonii | 291 |
| Actinomyces neuii | 3 |
| Actinomyces massiliensis | 14 |
The quality and reliability of the identification depends on the quality and the amount of reference spectra present in the database (22). A higher number of entries for the same species will better reflect the diversity within the species. Although obvious differences between spectra were observed, only tentative identification can be made for the species which were not very well represented in our collection: A. georgiae, A. radicidentis, A. urogenitalis, A. europaeus, A. neuii, and A. graevenitzii.
Conclusions
Our results suggest that a combination of MALDI-TOF-MS with powerful classification algorithms, such as SVMs, provide a useful tool for the differentiation and identification of oral Actinomyces, provided that the database contains enough entries for one species to reflect biological intra-species heterogeneity.
Acknowledgements
The authors appreciate the laboratory work of Angela Pöschel and Annett Hennig-Rolle.
Footnotes
Both authors had contributed equally to this work.
Conflict of interest and finding
The authors declare no conflict of interest in this study.
References
- 1.Funke G. Actinomyces spp. und verwandte fakultativ anaerobe grampositive Stäbchen. In: Neumeister B, Geiss HK, Braun R, Kimmig P, editors. Mikrobiologische diagnostik. Stuttgart: Georg Thieme Verlag KG; 2009. pp. 533–8. [Google Scholar]
- 2.Schaal KP, Yassin AF, Stackebrandt E. The family Actinomycetaceae: the Genera Actinomyces, Actinobaculum, Arcanobacterium, Varibaculum, and Mobiluncus . In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes – a handbook on the biology of bacteria. 3rd ed. New York: Springer Verlag; 2006. pp. 430–537. [Google Scholar]
- 3.Marsh P, Martin MV. Oral microbiology. fourth edn. Oxford: Reed Educational and Professional Publishing; 1999. [Google Scholar]
- 4.Brailsford SR, Tregaskis RB, Leftwich HS, Beighton D. The predominant Actinomyces spp. isolated from infected dentin of active root canal lesions. J Dent Res. 1999;78:1525–34. doi: 10.1177/00220345990780090701. [DOI] [PubMed] [Google Scholar]
- 5.Hansen T, Kunkel M, Kirkpatrick CJ, Weber A. Actinomyces in infected osteoradionecrosis – underestimated? Hum Pathol. 2006;37:61–7. doi: 10.1016/j.humpath.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 6.De Ceulaer J, Tacconelli E, Vandecasteele SJ. Actinomyces osteomyelitis in bisphosphonate-related osteonecrosis of the jaw (BRONJ): the missing link? Eur J Clin Microbiol Infect Dis. 2014;33:1873–80. doi: 10.1007/s10096-014-2160-5. [DOI] [PubMed] [Google Scholar]
- 7.Branquinho DF, Andrade DR, Almeida N, Sofia C. Mediastinitis by Actinomyces meyeri after oesophageal stent placement. BMJ Case Rep. 2014 doi: 10.1136/bcr-2014-204499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brander MA, Jousimies-Somer HR. Evaluation of the RapID ANA II and API ZYM systems for identification of Actinomyces species from clinical specimens. J Clin Microbiol. 1992;30:3112–16. doi: 10.1128/jcm.30.12.3112-3116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkonen N, Könönen E, Summanen P, Könönen M, Jousimies-Somer H. Phenotypic identification of Actinomyces and related species isolated from human sources. J Clin Microbiol. 2001;39:3955–61. doi: 10.1128/JCM.39.11.3955-3961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerttula AM, Carlson P, Sarkonen N, Hall V, Könönen E. Enzymatic/biochemical analysis of Actinomyces with commercial test kits with an emphasis on newly described species. Anaerobe. 2005;11:99–108. doi: 10.1016/j.anaerobe.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Hall V. Actinomyces – gathering evidence of human colonization and infection. Anaerobe. 2008;14:1–7. doi: 10.1016/j.anaerobe.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Henssge U, Do T, Radford DR, Gilbert SC, Clark D, Beighton D. Emended description of Actinomyces naeslundii and descriptions of Actinomyces oris sp. nov. and Actinomyces johnsonii sp. nov., previously identified as Actinomyces naeslundii genospecies 1, 2 and WVA 963. Int J Syst Evol Microbiol. 2009;59:509–16. doi: 10.1099/ijs.0.000950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruby JD, Li Y, Luo Y, Caufield PW. Genetic characterization of the oral Actinomyces . Arch Oral Biol. 2002;47:457–63. doi: 10.1016/s0003-9969(02)00023-7. [DOI] [PubMed] [Google Scholar]
- 14.Stingu CS, Eschrich K, Rodloff AC, Schaumann R, Jentsch H. Periodontitis is associated with a loss of colonization by Streptococcus sanguinis . J Med Microbiol. 2008;57:495–9. doi: 10.1099/jmm.0.47649-0. [DOI] [PubMed] [Google Scholar]
- 15.Friedrichs C, Rodloff AC, Chhatwal GS, Schellenberger W, Eschrich K. Rapid identification of viridans streptococci by mass spectrometric discrimination. J Clin Microbiol. 2007;45:2392–7. doi: 10.1128/JCM.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renvoise A, Raoult D, Roux V. Actinomyces timonensis sp. nov., isolated from a human clinical osteo-articular sample. Int J Syst Evol Microbiol. 2010;60:1516–21. doi: 10.1099/ijs.0.012914-0. [DOI] [PubMed] [Google Scholar]
- 17.Renvoise A, Raoult D, Roux V. Actinomyces massiliensis sp. nov., isolated from a patient blood culture. Int J Syst Evol Microbiol. 2009;59:540–4. doi: 10.1099/ijs.0.001503-0. [DOI] [PubMed] [Google Scholar]
- 18.Hall V, Collins MD, Lawson PA, Falsen E, Duerden BI. Actinomyces dentalis sp. nov., from a human dental abscess. Int J Syst Evol Microbiol. 2005;55:427–31. doi: 10.1099/ijs.0.63376-0. [DOI] [PubMed] [Google Scholar]
- 19.Chen T, Yu WH, Izard J, Baranova O, Lakshmanan A, Dewhirst FE. The human oral microbiome database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:1764–75. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 21.MATLAB and Statistics Toolbox. Natick, MA: The Mathworks, Inc; 2013. [Google Scholar]
- 22.Veloo AC, Knoester M, Degener JE, Kuijper EJ. Comparison of two matrix-assisted laser desorption ionisation-time of flight mass spectrometry methods for the identification of clinically relevant anaerobic bacteria. Clin Microbiol Infect. 2011;17:1501–6. doi: 10.1111/j.1469-0691.2011.03467.x. [DOI] [PubMed] [Google Scholar]