Abstract
Chronic ethanol-mediated oxidative stress and lipid peroxidation increases the levels of various reactive lipid species including 4-hydroxynonenal (4-HNE), which can subsequently modify proteins in the liver. It has been proposed that 4-HNE modification adversely affects the structure and/or function of mitochondrial proteins, thereby impairing mitochondrial metabolism. To determine whether chronic ethanol consumption increases levels of 4-HNE modified proteins in mitochondria, male rats were fed control and ethanol-containing diets for 5 weeks and mitochondrial samples were analyzed using complementary proteomic methods. Five protein bands (approx. 35, 45, 50, 70, and 90 kDa) showed strong immunoreactivity for 4-HNE modified proteins in liver mitochondria from control and ethanol-fed rats when proteins were separated by standard 1D SDS-PAGE. Using high-resolution proteomic methods (2D IEF/SDS-PAGE and BN-PAGE) we identified several mitochondrial proteins immunoreactive for 4-HNE, which included mitofilin, dimethylglycine dehydrogenase, choline dehydrogenase, electron transfer flavoprotein α, cytochrome c1, enoyl CoA hydratase, and cytochrome c. The electron transfer flavoprotein α consistently showed increased 4-HNE immunoreactivity in mitochondria from ethanol-fed rats as compared to mitochondria from the control group. Increased 4-HNE reactivity was also detected for dimethylglycine dehydrogenase, enoyl CoA hydratase, and cytochrome c in ethanol samples when mitochondria were analyzed by BN-PAGE. In summary, this work identifies new targets of 4-HNE modification in mitochondria and provides useful information needed to better understand the molecular mechanisms underpinning chronic ethanol-induced mitochondrial dysfunction and liver injury.
Keywords: Alcohol, Liver, Proteome, Mitochondria, 4-Hydroxynonenal, Posttranslational modification
Graphical abstract
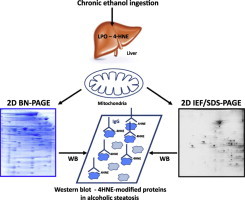
Highlights
-
•
Male rats were fed isocaloric Lieber–DeCarli control and ethanol-containing diets for 5 weeks.
-
•
Mitochondria were isolated and examined for 4-HNE modifications using proteomic methods.
-
•
A small number of mitochondrial proteins were found to be immunoreactive for 4-HNE.
-
•
Chronic ethanol consumption increased 4-HNE reactivity in the electron transfer flavoprotein-α.
Introduction
Chronic ethanol consumption causes liver disease through a complex process of metabolic and signaling alterations involving interactions of multiple liver cell types [1,2]. Early in the disease gut-derived endotoxin activates hepatic Kupffer cells, which release a variety of cytokines that can negatively affect hepatocyte biology [3]. Consequently, signaling events are triggered in hepatocytes causing increased production of reactive species that initiate widespread damage to intracellular organelles and macromolecules. One early and critical target of chronic alcohol-induced oxidative stress is the mitochondrion. Hepatic mitochondrial function, specifically bioenergetic function, is impaired in experimental rodent models of chronic ethanol consumption [4]. Depressed bioenergetics contributes to hepatocyte cell death, a key trigger for progression of alcoholic liver disease, as insufficient energy is produced to combat damage from secondary metabolic and environmental stressors (or ‘hits’) and for overall liver tissue repair [5,6].
While it is well accepted that hepatic mitochondrial function is compromised by chronic ethanol consumption, the molecular mechanisms responsible for these negative effects remain poorly understood. One potential mechanism underlying ethanol-dependent mitochondrial injury is increased production of reactive species including reactive oxygen, nitrogen, and lipid species (ROS, RNS, or RLS). Notably, chronic ethanol consumption increases mitochondrial ROS production [7,8], thus, mitochondria are both a source of reactive species and target of oxidative injury. We propose that chronic ethanol-mediated increases in reactive species contribute to increased oxidative modification of mitochondrial proteins. For example, one consequence of ethanol-mediated oxidative stress is the initiation of lipid peroxidation and generation of reactive electrophilic lipid species, which can adduct to proteins altering protein structure and function [9]. One electrophilic lipid species that has received significant attention is 4-hydroxynonenal or 4-HNE, a reactive α,β-unsaturated aldehyde species generated during conditions of oxidative stress and peroxidation of polyunsaturated fatty acids.
The biological effects of 4-HNE are dependent upon the location and amount of the species formed. Low levels of 4-HNE generated during normal cellular function are typically metabolized by reduction of the aldehyde functional group and/or conjugation to glutathione by cellular detoxification systems, e.g., glutathione S-transferase and aldehyde dehydrogenase [10]. However, during conditions of elevated oxidative stress and lipid peroxidation, 4-HNE is able to modify nucleophilic protein residues, specifically deprotonated side chains of cysteine, histidine, and lysine [11]. Interestingly, 4-HNE can form a variety of adducts with proteins due to both the aldehyde functional group and the adjacent α,β-unsaturated carbonyl moiety. Protein adducts with 4-HNE include Michael addition products mediated via the electrophilic β-carbon, and Schiff base formation via the aldehyde [12]. Michael adducts are primarily formed with cysteine, although histidine and lysine adducts have also been described [13]. Schiff base adducts occur by reaction of the aldehyde with the nitrogens of histidine or lysine (and less commonly arginine), resulting in the loss of a single molecule of water, or subsequent dehydration and cyclization to form a pentylpyrrole adduct (lysine only) [14]. It is important to note that the intrinsic reactivity of amino acids is dependent on the specific protein structure, cellular context, and local pH. Therefore, some proteins are inherently more susceptible to modification by reactive electrophiles such as 4-HNE. Thus, during oxidative stress, 4-HNE modification of a susceptible group of proteins increases, altering protein function and providing a footprint or biomarker of lipid peroxidation.
During conditions of chronic ethanol-induced oxidative stress, local concentrations of 4-HNE generated in the liver are sufficient for 4-HNE modification of proteins as increased adducts have been shown in liver using global immunohistochemistry (IMHC) approaches [15,16]. It is hypothesized that formation and retention of 4-HNE adducted proteins contributes to chronic ethanol-induced liver injury. Even though increased levels of 4-HNE adducts are consistently shown in liver from ethanol-exposed rodents via IMHC, few studies have identified specific 4-HNE modified proteins in ethanol models, especially mitochondrial proteins. Studies by Petersen and colleagues have been the most successful in identifying individual 4-HNE protein targets in liver from control and ethanol exposed rodents [17–21]. Therefore, to build on this work we performed a small focused study, using complementary proteomic approaches combined with immunoblotting, to identify 4-HNE modified proteins in rat liver mitochondria following chronic ethanol consumption. Using this experimental strategy, we identified several mitochondrial proteins immunoreactive for 4-HNE in mitochondria from control (ethanol-naïve) and ethanol-fed male rats. Modification of these proteins may contribute to mitochondrial dysfunction and liver injury in the chronic alcohol consumer.
Methods
Animals and feeding protocol
Male Sprague-Dawley rats (200–225 g) obtained from Charles River Laboratories were individually housed under a 12 h light–12 h dark cycle and fed nutritionally adequate control and ethanol-containing liquid diets (Bio-Serv, Frenchtown, NJ) for 5 weeks as described previously [22]. The ethanol-containing diet provides 36% of total daily calories as ethanol, 35% as fat, 11% of carbohydrate, and 18% as protein [23]. Control rats were pair-fed identical diets except that ethanol calories were replaced isocalorically with carbohydrate (maltose–dextrin). These studies were approved by the institutional animal care committee and conducting in accordance to The Guide for the Care and Use of Laboratory Animals (USDHHS, NIH publication no. 86-23, 1996).
Isolation of mitochondria
Liver mitochondria were isolated according to standard differential centrifugation procedures using an ice-cold mitochondrial isolation medium consisting of 0.25 M sucrose, 1 mM EDTA, and 5 mM Tris–HCl and protease inhibitors were added to the isolation medium to prevent protein degradation for proteomic analyses [24]. Mitochondria were carefully washed three times and the final mitochondrial pellet was re-suspended at a concentration of 30–40 mg protein/mL for respiration studies. Mitochondrial quality (i.e., tightness of coupling) was assessed by determining the respiratory control ratio (state 3 respiration/state 4 respiration) using succinate as substrate and are reported in [22]. Equal amounts of total mitochondrial protein were isolated from the livers of ethanol and control-fed animals [22]. In a separate experiment, freshly isolated mitochondria (0.5 mg) were incubated with varying amounts of 4-HNE (0, 10, or 50 µM dissolved in ethanol) under conditions used for respiration studies. Incubations with 4-HNE were done for 10 min and samples were used for 1D SDS-PAGE and 2D IEF/SDS-PAGE experiments (see description of these methods in subsequent sections). These experiments were done to help show specificity of the 4-HNE antibody. Mitochondrial samples for all proteomic analyses were divided into aliquots, frozen in liquid N2, and stored at −80 °C until their use in proteomic analyses.
Standard one-dimensional (1D) SDS-PAGE and western blotting
For the detection of 4-HNE modified proteins, equal amounts of mitochondrial and cytosolic protein (100 µg) were subjected to SDS-PAGE using large format 8–20% gradient gels to separate proteins. Separated proteins were then transferred to nitrocellulose membranes (0.2 µM pore size) according to standard immunoblotting procedures [25]. After transfer, membranes were stained with Ponceau S dye to verify equal loading and transfer of proteins and then incubated for 1 h with 5% (w/v) non-fat milk. Levels of 4-HNE modified proteins were detected following an overnight incubation with a 1:10,000 dilution of a polyclonal antibody made against 4-HNE-modified keyhole limpet hemocyanin (KLH) as described in [26]. Characterization of the antibody has shown that it recognizes cysteine, lysine, and histidine 4-HNE protein adducts [26] and that it is highly specific to 4-HNE derived protein adducts as the antibody does not cross react with proteins treated with other aldehydes like malondialdehyde [27]. Blots were then incubated with a 1:5000 dilution of goat anti-rabbit IgG HRP conjugate for 1 h before immunoreactive bands were detected using enhanced chemiluminescence (SuperSignal West Pico Reagent, Thermo Scientific Inc., Rockford, IL). Analysis of liver cytosolic proteins from control and ethanol-fed rats revealed no strong 4-HNE immunoreactive protein adducts, thus cytosolic proteins were not studied further (data not shown). Levels of the electron transfer flavoprotein in rat liver mitochondria were detected using a polyclonal antibody raised in rabbits against the porcine protein [28]. Electron transfer flavoprotein was detected following an overnight incubation with a 1:10,000 dilution of antibody. Scanned TIFF images for 1D gels and western blots were analyzed by standard densitometry methods using Scion Image Beta 4.02 software (Scion Corporation, MD) as described in [22].
Two-dimensional (2D) IEF–SDS-PAGE
For 2D IEF/SDS-PAGE, mitochondria were suspended in isoelectric buffer consisting of 9.5 M urea, 2.0% CHAPS, 1.0% DTT, and 0.8% (v/v) of each of pH 3–10, 5–8, and 4–6 ampholines (Sigma Aldrich, St. Louis, MO). After 1 h incubation to solubilize samples, 100 µg of protein were loaded onto IEF gels (pH 4–8.5) and focused for 16 h at 400 V and 800 V for 1 h. IEF gels were equilibrated and loaded onto 8–20% gradient gels for SDS-PAGE as described previously [22]. For each pair of control and ethanol samples, gels were run in duplicate with one gel stained with Coomassie Blue (visualize proteins) and the other gel used for immunoblotting with the 4-HNE antiserum as described above, with the exception that the 4-HNE antibody was diluted 1:5000 for 2D blots.
Image analysis for the 4-HNE blots generated from 2D IEF/SDS-PAGE gels was done using methods similar to those described in our previous publication [29]. To facilitate comparisons and image analysis, four pairs of control and ethanol 2D gels and blots were processed and developed together. The 2D total protein gel and western blot images (films) were saved as TIFF images and analyzed using PDQuest Image Analysis software (Bio-Rad Laboratories, Inc., Hercules, CA). Protein spots were detected and a match-set containing all four control blots with their corresponding ethanol blots was created. To compare protein spot density across the different blots, a reference blot (i.e., master image) was selected. To increase the likelihood that the highest possible number of protein spots in each blot could be matched to the corresponding protein spots in the master image, the selection of the master image was based on the highest number of detected proteins spots and the best protein spot resolution across the entire blot image. Thus, the ethanol blot from pair # 4 met all the requirements and was used as the master image for spot matching purposes. Due to the low number of spots detected on 4-HNE blots (4 protein spot clusters), matching was performed manually. Quantities from the individual spots that were part of spot clusters (Fig. 3C and D, spots #2 and #3) were combined using the spot combine tool. Spot densities were transferred to Microsoft Excel for statistical analyses. Because spot # 4 was detected on only 2 of the 4 control blots, but was present in all of the ethanol blots (Fig. 4A), we also choose to express results as a percentage of the total HNE spot density (or reactivity) on blots. Image analysis for protein gels has been reported previously from our laboratories [22].
Fig 3.
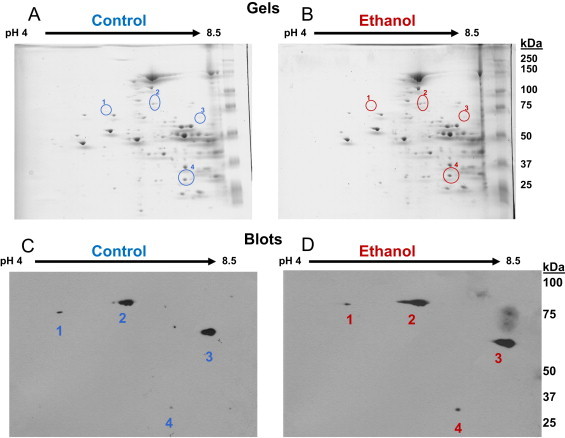
High-resolution separation of liver mitochondrial proteins from control and ethanol fed rats using 2D IEF/SDS-PAGE and 4-HNE detection by western blot. Panels A and B: For 2D IEF/SDS-PAGE, 100 µg of liver mitochondrial protein was loaded onto IEF gels (pH gradient 4–8.5) and focused overnight with the 2D separation performed using an 8–20% gradient gel. Representative gels are shown for one pair of control (A) and ethanol (B) fed rats. Panels C and D: Corresponding 2D western blots for gels. Four mitochondrial protein spot clusters were immunoreactive for 4-HNE adducts in mitochondrial samples from control (C) and ethanol (D) fed rats. Each of these 4-HNE modified proteins was identified using mass spectrometry (Table 1).
Fig 4.
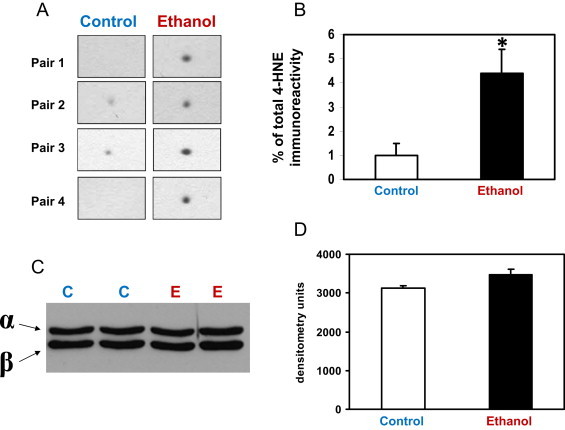
Chronic ethanol consumption increases 4-HNE immunoreactivity of the electron transfer flavoprotein, alpha (ETFα) subunit without a change in total ETFα protein. Panel A illustrates the increase in 4-HNE immunoreactivity for the ETFα subunit in four pairs of control and ethanol fed rats. Note that 4-HNE immunoreactivity for the ETFα subunit is absent for two controls but is consistently present and elevated in ethanol mitochondria. Panel B: Quantification of the density of the 4-HNE immunoblots for ETFα (n=4, *p=0.03). Panels C and D illustrate that chronic ethanol consumption has no effect on the total amount of ETFα subunit protein. Panel C shows representative immunoblots for both the α- and β-subunits of ETF in liver mitochondria from two pairs of control and ethanol fed rats. The ETF antiserum recognizes both subunits. Panel D: Quantification of ETFα subunit protein in liver mitochondria from control and ethanol fed rats (n=4, p=0.11).
Two-dimensional (2D) blue native PAGE (BN-PAGE)
Separation of mitochondrial proteins by BN-PAGE was essentially done as described in our previous publications [22,30]. BN-PAGE is a special type of gel electrophoresis technique that is largely used to separate the proteins that make up the oxidative phosphorylation (OxPhos) system; however, other mitochondrial proteins associated with the OxPhos complexes can be resolved with BN-PAGE as witnessed herein (Fig. 5 and Table 2) and in other papers [30]. Briefly, frozen mitochondrial protein (stored as 1.0 mg pellets) were treated with 10% (w/v) lauryl maltoside in 0.75 M aminocaproic acid, 50 mM BisTris, pH 7.0, to gently release the OxPhos complexes intact from the mitochondrial inner membrane. The protein extracts were then mixed with 6.3 µL of a 5% (w/v) suspension of Coomassie Brilliant Blue G-250 in 0.5 M aminocaproic acid and applied to 5–12% non-denaturing gradient mini-gels to separate the five OxPhos complexes in their intact form. After native gel electrophoresis, the vertical gel lanes for each individual sample were each cut from the gel and then laid on top of a 10% denaturing Tris/Tricine/SDS-PAGE gel to resolve the individual proteins that comprise each OxPhos complex. Again, samples were run in duplicate so that one gel could be stained with Coomassie Blue to visualize total protein and the other gel subjected to immunoblotting to visualize 4-HNE adducted proteins. Conditions for westerns blots were the same as those used for 2D IEF/SDS-PAGE. Images of the Coomassie Blue stained protein gels were obtained using a Bio-Rad Fluor-S Imager (Bio-Rad, Hercules, CA). Due to the irregular shape of the protein spots in the 2D BN-PAGE/4-HNE western blots, protein densities were determined using the freehand drawing tool present in Quantity One Image Analysis software (Bio-Rad Laboratories, Inc., Hercules, CA). Immunoblotting for cytochrome c was done to aid in the identification of one of the 4-HNE positive adducted proteins that could not be identified by mass spectrometry [31]. The cytochrome c antibody was used at a 1:1000 dilution (BD Biosciences, San Jose, CA).
Fig 5.
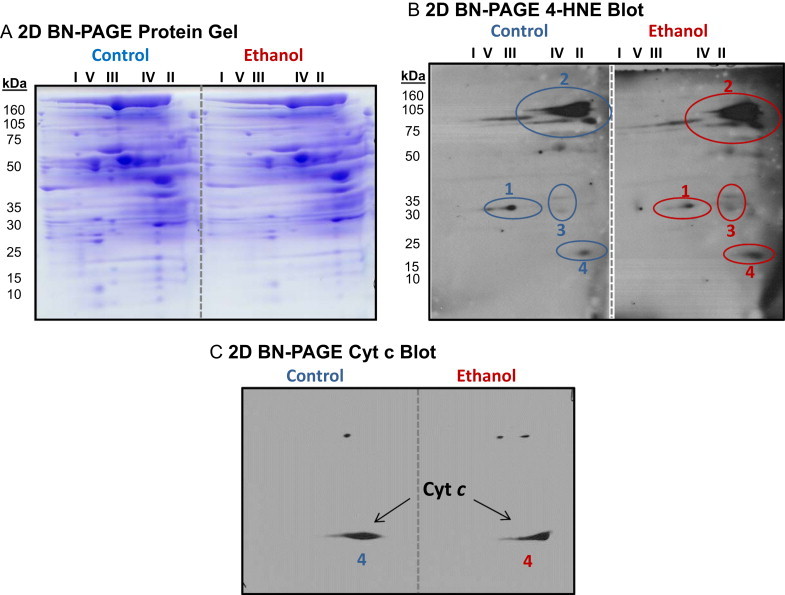
High-resolution separation of liver mitochondrial proteins from control and ethanol fed rats using 2D BN-PAGE and 4-HNE detection by western blot. Panel A: Representative 2D BN-PAGE gels of liver mitochondrial proteins from one pair of control and ethanol-fed rats. BN-PAGE was performed as described in methods sections. Panel B: Corresponding 2D 4-HNE western blots are shown for gels in panel A. Four protein spot clusters showed immunoreactivity for 4-HNE in mitochondria isolated from livers of control (C) and ethanol (E) fed rats. Three of the four protein spots (1–3) were identified by mass spectrometry (Table 2). Note that for protein spot cluster no. 3 the bottom spot is enoyl CoA hydratase and the top spot is most likely β-hydroxybutyrate dehydrogenase; however, MOWSE score was too low for positive ID. panel C: Immunoblot for cytochrome c from control and ethanol fed mitochondrial samples separated using 2D BN-PAGE. The protein spot immunoreactive for cytochrome c co-localizes to protein spot 4 in panel B. Previous studies from our laboratory show no difference in the amount of total cytochrome c protein in liver mitochondria isolated from control and ethanol-fed rats [8].
Table 2.
Identification and quantification of 4-HNE modified proteins in liver mitochondria from control and ethanol-fed rats: results from 2D BN-PAGE.
| Spot no. | Protein | MOWSE | Control | Ethanol | P value |
|---|---|---|---|---|---|
| 1 | Cytochrome c1, Complex III | 142 | 2538±459 | 1654 ± 318 | 0.027 |
| 2 | Dimethylglycine dehydrogenase | 114 | 3828±461 | 5567 ± 480 | 0.013 |
| 3 | Enoyl CoA hydratase | 118 | 3068 | 5577 | − |
| 4 | Cytochrome c | − | 477 | 879 | − |
Protein spot cluster nos. 1–3 were identified by MALDI–TOF as described in the methods section, whereas protein spot no. 4 was identified by immunoblotting. 4-HNE immunoreactivity for enoyl CoA hydratase (spot cluster no. 3, bottom spot) and cytochrome c (spot no. 4) in both control and ethanol samples is reported for two of six pairs of samples run on BN-PAGE gels. Statistics are provided for cytochrome c1 (spot no. 1) and dimethylglycine dehydrogenase (spot cluster no. 2), as these protein spots immunoreactive for 4-HNE were detected in all six pairs of control and ethanol samples. Data represent the mean±SEM for spot cluster densities for 1 and 2, and the mean densities only for spot clusters 3 and 4.
Mass spectrometry identification
Films and scanned images of 4-HNE blots were aligned with their duplicate stained total protein gels so that the corresponding 4-HNE adducted proteins could be identified by mass spectrometry techniques. Matrix assisted laser desorption ionization/time-of-flight (MALDI–TOF) mass spectrometry was done to identify proteins with the exception that the electron transfer flavoprotein was identified by quadropole time-of-flight (Q-TOF) mass spectrometry. MALDI–TOF was done using a Voyager Elite Instrument (Applied Biosystems, Inc. Foster, City, CA) equipped with a nitrogen laser set at 337 nm and operated in a delayed extraction mode. Identification of electron transfer flavoprotein, alpha was performed using a Q-TOF 2 mass spectrometer with instrument operation, data acquisition, and analyses performed with MassLynx software (Micromass, Manchester, UK). An electrospray interface was used for tandem mass spectrometry. Peptide masses and amino acid sequences were entered into Mascot database for protein identification (http://www.matrixscience.com). Details regarding mass spectrometry protocols used for protein identification are provided in our papers [22,29] and on the UAB mass spectrometry shared facility website (http://www.uab.edu/proteomics).
Statistics
Results were analyzed using Student's t-test with significance set at p<0.05. The sample size for most measures was n=3–6 for control and ethanol groups.
Results
Rats fed the Lieber–DeCarli ethanol-containing diet for four or more weeks develop steatosis (i.e., fatty liver), thus, this well-accepted feeding paradigm is a good preclinical model for the early stage of alcoholic liver disease. In addition to histopathology, chronic ethanol feeding induces oxidative stress, alterations in redox signaling pathways, and mitochondrial dysfunction. The bioenergetic parameters of the mitochondrial preparations used in the current study have been reported previously [22] and show that chronic ethanol feeding decreases mitochondrial respiratory function and the activities of several electron transport system components. It is important to point out that even though respiration is decreased by chronic ethanol feeding, mitochondria from ethanol-fed rats are functionally viable upon isolation. This is important as poor preparation of mitochondrial fractions (e.g., high state 4 respiration due to damage to the inner membrane) negatively impacts proteomic analyses especially those conducted using the BN-PAGE method.
In this first set of experiments, the extent of 4-HNE modification in mitochondrial proteins was assessed using a low resolution approach, i.e., 1D SDS-PAGE followed by western blotting. Five bands with molecular weights of approximately 90, 70, 50, 45, and 35 kDa showed the strongest immunoreactivity for 4-HNE modifications in mitochondria from control and ethanol-fed rats (Fig. 1). Results are shown for two pairs of control and ethanol-fed rats from a total of six pair-fed groups (Fig. 1A). Image analysis showed that there was no statistically significant difference in the density of the five major bands resolved on the 1D gel with respect to 4-HNE immunoreactivity (Fig. 1B). When mitochondria were incubated with 4-HNE ex vivo for a short period of time (10 min) a significant increase in 4-HNE modified proteins was detected using the antibody (Fig. 2A). The extent of protein modifications was dose-dependent as greater 4-HNE immunoreactivity (i.e., more protein bands and stronger signal) was observed in mitochondria incubated with 50 µM 4-HNE (blot # 2) versus 10 µM 4-HNE (blot # 1) for the same length of time (Fig. 2A). Thus, the antibody was able to detect this key oxidative stress-linked posttranslational modification.
Fig 1.
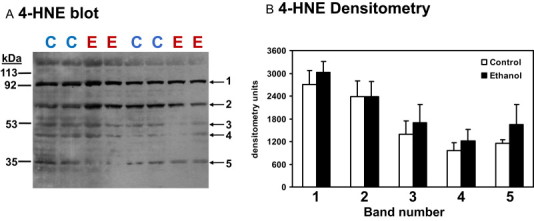
Immunoblot of 4-HNE modified proteins in liver mitochondria from control and ethanol fed rats. Mitochondrial protein (100 µg) was separated on a large format 8–20% gradient gel by SDS-PAGE. After electrophoresis, proteins were transferred to nitrocellulose membranes and probed for 4-HNE modifications using the conditions described in the methods section. Panel A: Five major protein bands with molecular weights of approximately 90, 70, 50, 45, and 35 kDa showed strong immunoreactivity for 4-HNE modifications in mitochondria from control (C) and ethanol (E) fed rats. Representative blots are shown for two pairs of control and ethanol fed rats with samples run in duplicate. Panel B: Quantification of the density of the 5 major protein bands showing 4-HNE immunoreactivity in mitochondria from control and ethanol fed rats. Total number of samples run was n=6 pairs of control and ethanol samples.
Fig 2.
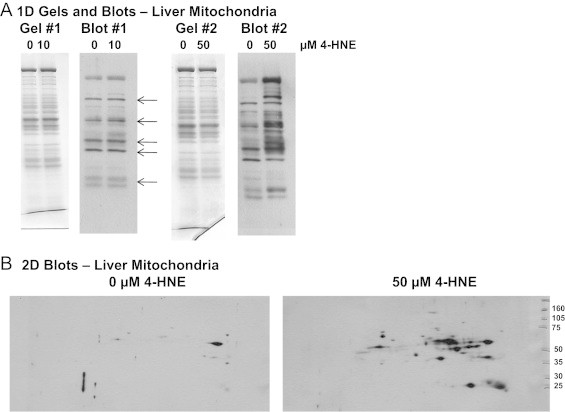
Immunoblots of 4-HNE modified proteins in liver mitochondria incubated with 4-HNE. Freshly isolated liver mitochondria (0.5 mg) were incubated with 0, 10, or 50 µM 4-HNE for 10 min to generate samples containing 4-HNE adducted proteins. After incubations, samples were flash frozen in liquid N2 and stored at −80 °C until used in electrophoresis experiments. Panel A: Mitochondrial protein (20 µg) was separated on 10% SDS-PAGE gels and immunoblotted to detect 4-HNE modified proteins. Results show increased in 4-HNE modified proteins in mitochondria incubated with 50 µM 4-HNE. A small increase in 4-HNE labeling is observed at 10 µM 4-HNE. The five 4-HNE modified proteins detected in mitochondria from control and ethanol-fed rats (Fig. 1) are highlighted here by the arrows. A slightly different pattern in separation is observed here as samples were separated using 10% mini-gels compared to an 8–20% large-format gradient gels (Fig. 1). Panel B: Protein (100 µg) from mitochondria treated with 0 and 50 µM 4-HNE was separated by 2D IEF SDS-PAGE and immunoblotted to visualize 4-HNE modified proteins. Results show an increase in the number of 4-HNE modified proteins in mitochondria exogenously treated with 50 µM 4-HNE as compared to untreated mitochondria.
In light of the fact that modification of individual proteins may be masked due to the co-migration of proteins with very similar molecular weights when using low resolution 1D SDS-PAGE, we repeated these measurements using 2D proteomics techniques in the hope to identify more 4-HNE targets in liver mitochondria from control and ethanol-fed rats. Thus, a higher resolution separation of mitochondrial proteins was done by separating proteins in the first dimension (1D) by isoelectric focusing (IEF) followed by separating proteins in the second dimension (2D) based on their respective molecular weights. Proteins separated using this method show a very characteristic and similar pattern or ‘map’ of mitochondria proteins isolated from liver of both control and ethanol-fed rats (Fig. 3A and B). There is no difference in the global protein density (staining) between the gels generated from control and ethanol mitochondrial samples [22,29]; however, as reported previously we do observe ethanol-mediated changes in the abundances of approximately two dozen individual mitochondrial proteins [22]. Notably, the proteins we identified here as being modified by 4-HNE were not found to be altered in abundance in response to chronic ethanol [22]. Even though there was significantly increased resolution of mitochondrial proteins using the 2D approach (Fig. 3A and B), we only observed four discrete protein spot clusters as being immunoreactive for 4-HNE in mitochondria from control and ethanol-fed rats (Fig. 3C and D). Again, when we performed 2D IEF/SDS-PAGE on mitochondria incubated with 4-HNE ex vivo many more proteins showed immunoreactivity for 4-HNE (Fig. 2B). Thus, our results show a low number of 4-HNE modified proteins in liver mitochondria of control and ethanol-fed rats in this experimental model system.
The protein spots found to be immunoreactive for 4-HNE (Fig. 3C and D) were matched to their corresponding proteins spots in the protein gels (Fig. 3A and B) and proteins were identified by mass spectrometry. The protein spot numbers in Fig. 3C and D correspond to the identified proteins listed in Table 1. All four protein spots were identified and include mitofilin (MF, spot no. 1), dimethylglycine dehydrogenase (DMGDH, spot no. 2), choline dehydrogenase (CDH, spot no. 3), and electron transfer flavoprotein, alpha (ETFα, spot no. 4). While chronic ethanol consumption increased 4-HNE immunoreactivity in all four proteins when raw protein spot densities were compared, ETFα was the only protein that showed a statistically significant increase in 4-HNE immunoreactivity (Table 1). This was also the case when results were analyzed based on the percentage of total 4-HNE immunoreactivity (Table 1). As stated in the Methods section, it is preferable to report these results as a percent of total 4-HNE immunoreactivity as the spot positive for 4-HNE modified ETFα was not detected in two of the four control samples, but easily detected in all four ethanol samples analyzed (Fig. 4A and B). To confirm that the increase in 4-HNE modified ETFα (Fig. 4A) was simply not due to increased abundance of this protein in mitochondria from ethanol-fed rats, we obtained an antibody for ETFα and performed western blotting. Results showed there is no significant difference between the amount of ETFα (and β) immunoreactive protein in mitochondria from control and ethanol-fed rats (Fig. 4C and D). This finding supports the contention that chronic ethanol feeding increased 4-HNE modification of the ETFα protein (Fig. 4A).
Table 1.
Identification and quantification of 4-HNE modified proteins in liver mitochondria from control and ethanol-fed rats: results from 2D IEF/SDS-PAGE.
| Spot no. | Protein | MOWSE | Raw spot densities |
P value | % of total 4-HNE reactivity |
P value | ||
|---|---|---|---|---|---|---|---|---|
| Control | Ethanol | Control | Ethanol | |||||
| 1 | Mitofilin | 135 | 16,508±3222 | 21,162±536 | 0.20 | 3.04±1.3 | 3.27±1.0 | 0.40 |
| 2 | Dimethylglycine dehydrogenase | 233 | 212,830±20,687 | 231,017±19,455 | 0.40 | 58.2±9.9 | 56.9±6.3 | 0.30 |
| 3 | Choline dehydrogenase | 133 | 164,343±53,099 | 170,536±45,409 | 0.90 | 37.9±8.7 | 36.5±6.2 | 0.40 |
| 4 | Electron transfer flavoprotein, alpha | – | 3988±2202 | 17,282±2358 | 0.01 | 1.0±0.5 | 4.4±1.0 | 0.03 |
The total 4-HNE immunoreactivity per control and ethanol blot was 393,611±69,256 and 434,862±66,732 densitometry units, respectively (n=4, p=0.46). Data analysis was performed on either the raw spot densities or as a % of the total 4-HNE reactivity. Similar results were observed using both methods. Electron transfer flavoprotein, alpha was identified by Q-TOF with the other proteins (spot nos. 1–3) identified by MALDI–TOF. The MOWSE score is an algorithmic calculation used to assign statistical weight to peptides match. A higher MOWSE indicates a higher statistical likelihood of the match being correct. All scores were >40 indicating significant homology and matches (p≤0.05).
Because many of the proteins that comprise the OxPhos system are poorly resolved on conventional 2D IEF/SDS-PAGE gels [32], we extended our studies using BN-PAGE to see if additional protein targets of 4-HNE could be detected in mitochondria from control and ethanol-fed rats. Representative 2D BN-PAGE gels for control and ethanol groups are presented in Fig. 5A, and show that chronic ethanol feeding decreases the amount of OxPhos proteins. These results are in agreement with previous studies from our laboratory [22]. To determine the extent of 4-HNE modification using this electrophoresis approach, 2D BN-PAGE gels were subjected to western blotting and membranes were probed with 4-HNE antisera. We observed four major protein spot clusters with 4-HNE immunoreactivity on the 2D BN-PAGE blots (Fig. 5B). These 4-HNE immunoreactive spots (Fig. 5B) were matched to their corresponding proteins spots in gels (Fig. 5A) and proteins (nos. 1–3) were identified by mass spectrometry. The protein spot numbers in Fig. 5B correspond to the identified proteins listed in Table 2. The proteins identified were cytochrome c1 (Cyt c1, spot no. 1), dimethylglycine dehydrogenase (DMGDH, spot no. 2), enoyl CoA hydratase (ECH, spot no. 3), and cytochrome c (Cyt c, spot no. 4). Protein spot no. 4 was identified by co-localization of western blotting for cytochrome c (Fig. 5C), which agrees with previous studies [31]. Using the 2D BN-PAGE approach, DMDH showed a significant increase in 4-HNE immunoreactivity in mitochondria from ethanol-fed rats (Table 2). Ethanol feeding also increased 4-HNE reactivity in ECH and Cyt c (Table 2); however, we were only able to detect these protein spot clusters in two out of six pairs of control and ethanol mitochondrial samples run. In contrast, 4-HNE reactivity for Cyt c1 was decreased in mitochondria from ethanol-fed rats (Table 2); however, this is most likely due to the fact that Cyt c1 protein is significantly decreased by chronic ethanol feeding [22].
Discussion
Previous studies from our laboratory showed that chronic ethanol feeding significantly alters the liver mitochondrial proteome [22,29]. We have shown that chronic ethanol feeding altered the abundance of approximately two dozen mitochondrial matrix proteins and significantly decreased the levels of many mitochondrial- and nuclear-encoded polypeptides that comprise the OxPhos system [22]. In addition, we have reported that chronic ethanol altered the redox state of protein thiol groups in several mitochondrial proteins [29], presumably via increased oxidative and/or nitrosative stress. Herein, we extended this work and examined the effect chronic ethanol feeding has on 4-HNE modification of mitochondrial proteins using a proteomics approach. For this, we used three complementary gel electrophoresis approaches in combination with a well-characterized 4-HNE antibody [26]. Interestingly, we detected only a small number of mitochondrial proteins immunoreactive for 4-HNE in our experimental model of alcoholic steatosis. However, this may not be too surprising as in our previous study we also observed only a small handful of proteins with modified protein thiols following chronic ethanol consumption[29]. These proteins included aldehyde dehydrogenase 2, GRP78, glutamate dehydrogenase, pyruvate carboxylase, and acetyl CoA acyl transferase 2. These results support the concept that there is likely specificity to posttranslational modifications in vivo as modifications are dependent on protein structure and protein microenvironment conditions. Therefore, while exposure to high concentrations of 4-HNE ex vivo leads to a large number of adducted proteins (Fig. 2), only a small number of adducted proteins are present under conditions of a chronic low-level oxidative stress in vivo (Figs. 3 and 5).
Our combined 2D proteomics analyses were successful in identifying seven mitochondrial proteins immunoreactive for 4-HNE in samples from both control and ethanol-fed rats. These proteins include mitofilin (MF), dimethylglycine dehydrogenase (DMGDH), choline dehydrogenase (CDH), electron transfer flavoprotein, alpha (ETFα), cytochrome c1 (Cyt c1), enoyl CoA hydratase (ECH), and cytochrome c (Cyt c). While several proteins had increased 4-HNE immunoreactivity in ethanol samples analyzed by 2D IEF/SDS-PAGE, 4-HNE reactivity was only statistically increased in the ETFα protein in response to chronic ethanol feeding. ETF is comprised of two subunits, α and β, and functions as the electron acceptor for various flavoprotein dehydrogenases of fatty acid β-oxidation, as well as some dehydrogenases involved in amino acid and choline metabolism, e.g., DMGDH and CDH. In the mitochondrion, reducing equivalents from these dehydrogenases are transferred sequentially to ETF, ETF–ubiquinone oxidoreductase complex (ETF–QO), and then to ubiquinone, thus entering the electron transport system at the level of Complex III. Because ETF, together with the ETF–QO, are essential components of β-oxidation, a defect in the functioning of the ETFα as a consequence of 4-HNE mediated modification, might contribute, in part, to accumulation of liver fat as a result in the inability to metabolize fatty acyl-CoAs. Our finding may be significant as defects in fatty acid oxidation occur in response to chronic ethanol consumption [33]. With this said, however, future studies are required to identify the sites of 4-HNE modification in ETFα and whether modification alters activity.
In addition, BN-PAGE proteomics showed statistically increased 4-HNE reactivity in DMGDH in ethanol samples. Interestingly, DMGDH immunoreactivity in ethanol samples was not different from control when mitochondrial samples were analyzed by 2D IEF/SDS-PAGE. While the reason for this discrepancy is not known, it may be related to differences in the separation techniques whereby BN-PAGE may have enhanced the accessibility of the 4-HNE modified sites. DMGDH is an abundant mitochondrial matrix enzyme that participates in the interconnected cycles of methionine, betaine, glycine, and choline metabolism [34]. DMGDH itself plays a key role in usage of methyl groups from choline. Specifically, DMGDH, a flavin-containing enzyme, catalyzes the oxidative demethylation of dimethylglycine to sarcosine, which in turn is converted to glycine by the enzyme sarcosine dehydrogenase. Again, while we do not know whether the increase in 4-HNE modification affected the activity of the enzyme, it is known that chronic ethanol perturbs function of the metabolic cycles critical for one-carbon metabolism and that these derangements contribute to steatosis [35]. Importantly, betaine homocysteine methyltransferase (BHMT) is inhibited by accumulation of its product dimethylglycine [36]. As Ji and colleagues have shown that BHMT is highly protective in models of alcoholic steatosis [37], we propose that DMGDH inactivation may participate in the disease process through impairment of BHMT. However, until the physiological roles and importance of dimethylglycine and sarcosine and associated enzymes are better understood, the significance of this result remains unclear.
We also observed increased 4-HNE reactivity in ECH and Cyt c from ethanol samples. Unfortunately, we were only able to detect these 4-HNE immunoreactive protein spots in two out of six pairs of control and ethanol samples. Therefore, it is premature to speculate as to how 4-HNE modification of these two proteins might impact mitochondrial function. However, studies by Darley-Usmar and colleagues [14] reported significant modification of Cyt c by 4-HNE treatment in vitro. Notably, mapping these modifications on to the X-ray crystal structure of Cyt c revealed that 4-HNE modification of lysine residues could have detrimental effects on electron transport efficiency, a parameter known to be impaired by chronic ethanol consumption [38]. These initial observations open a potential new line of inquiry for examining ethanol-mediated defects on mitochondrial bioenergetic function.
Finally, we would also like to point out that usage of different 4-HNE antibodies in studies many times lead to different results. For example, Patel et al. [39] demonstrated that chronic ethanol consumption elevated 4-HNE immunoreactivitity to one 55 kDa mitochondrial protein in 1D SDS-PAGE/immunoblots using the 4-HNE antibody from Alpha Diagnostics (San Antonio, TX). We found the same result when using this antibody (data not shown). Their lab went on to identify this protein as 3-hydroxy-3-methylglutaryl (HMG-CoA) synthase, the rate-limiting enzyme in ketone body formation. When we stripped and re-probed our 2D IEF/SDS-PAGE blots with this antibody we detected three additional 4-HNE targets at the basic end of gels. Two of which we were able to identify as acyl CoA dehydrogenase, very long chain and HMG-CoA synthase (data not shown). Furthermore, using the monoclonal antibody specific for the 4-HNE histidine adduct (generated by Esterbauer [40]), Henderson and colleagues [41] showed that short-term ethanol exposure enhanced formation of 4-HNE adduct in cytochrome c oxidase, specifically subunit IV. And, recently Frtiz et al. [15] using a 4-HNE antibody developed in their laboratory [13] have shown that the mitochondrially localized sirtuin 3 is a target for 4-HNE adduction. Together, these studies highlight the diversity in different 4-HNE antibody preparations and suggest that multiple antibodies be employed to maximize detection of adducted proteins in biological samples.
In summary, we provide new results showing chronic ethanol-mediated alterations to the mitochondrial proteome at the posttranslational level. We identified a small number of proteins that are modified by the electrophilic lipid 4-HNE in rat liver mitochondria. Interestingly, some of the proteins identified as having increased 4-HNE modification in response to chronic ethanol consumption are linked to one-carbon metabolism. Specifically, we observed that CDH, DMGDH, and ETF were modified by 4-HNE, with chronic ethanol enhancing 4-HNE modification in the latter two proteins. Thus, it is possible that within this small study we have identified a previously unknown focal point involved in alcohol-mediated alterations to the mitochondrial proteome in liver. Identification of these proteins provides a new potential mechanism whereby modification of proteins in vivo by chronic ethanol consumption may contribute, in part, to ethanol-mediated mitochondrial dysfunction and toxicity.
Conflict of interest
The authors declare that there are no conflicts of interest.
Funding
This work was supported by the National Institute of Health grants to SMB (R01 AA15172 and R01 AA18841).
Acknowledgements
The authors would like to thank Ashley Davis, Anita Pinner, and Gloria Robinson for technical assistance with gels and western blotting, and Marion Kirk and Landon Wilson for mass spectrometry analyses done in the UAB mass spectrometry Core that is under the direction of Dr. Stephen Barnes. The authors thank Dr. Luke I. Szweda, Oklahoma Medical Research Foundation, Oklahoma City, OK, for kindly providing the 4-HNE antibody. The antibody was provided to our laboratory when Dr. Szweda was faculty at Case Western University, Cleveland, OH. The ETF antiserum was kindly provided by Dr. Frank E. Frerman, University of Colorado Anschutz Medical Campus, Denver, CO. The authors would also like to thank Dr. Victor Darley-Usmar, University of Alabama at Birmingham, for guidance in data analysis.
References
- 1.An L., Wang X., Cederbaum A.I. Cytokines in alcoholic liver disease. Archives of Toxicology. 2012;86:1337–1348. doi: 10.1007/s00204-012-0814-6. 22367091 [DOI] [PubMed] [Google Scholar]
- 2.Feldstein A.E., Bailey S.M. Emerging role of redox dysregulation in alcoholic and nonalcoholic fatty liver disease. Antioxidants & Redox Signaling. 2011;15:421–424. doi: 10.1089/ars.2011.3897. 21254858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo G., Bala S. Alcoholic liver disease and the gut-liver axis. World Journal of Gastroenterology: WJG. 2010;16:1321–1329. doi: 10.3748/wjg.v16.i11.1321. 20238398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham C.C., Bailey S.M. Ethanol consumption and liver mitochondria function. Neurosignals. 2001;10:271–282. doi: 10.1159/000046892. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham C.C., Van Horn C.G. Energy availability and alcohol-related liver pathology. Alcohol Research & Health: Journal of the National Institute on Alcohol Abuse and Alcoholism. 2003;27:291–299. 15540800 [PMC free article] [PubMed] [Google Scholar]
- 6.Young T.A., Bailey S.M., Van Horn C.G., Cunningham C.C. Chronic ethanol consumption decreases mitochondrial and glycolytic production of ATP in liver. Alcohol and Alcoholism. 2006;41:254–260. doi: 10.1093/alcalc/agl017. 16571619 [DOI] [PubMed] [Google Scholar]
- 7.Bailey S.M., Cunningham C.C. Acute and chronic ethanol increases reactive oxygen species generation and decreases viability in fresh, isolated rat hepatocytes. Hepatology. 1998;28:1318–1326. doi: 10.1002/hep.510280521. 9794917 [DOI] [PubMed] [Google Scholar]
- 8.Bailey S.M., Robinson G., Pinner A., Chamlee L., Ulasova E., Pompilius M., Page G.P., Chhieng D., Jhala N., Landar A., Kharbanda K.K., Ballinger S., Darley-Usmar V. S-adenosylmethionine prevents chronic alcohol-induced mitochondrial dysfunction in the rat liver. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2006;291:G857–G867. doi: 10.1152/ajpgi.00044.2006. 16825707 [DOI] [PubMed] [Google Scholar]
- 9.Fritz K.S., Petersen D.R. An overview of the chemistry and biology of reactive aldehydes. Free Radical Biology & Medicine. 2013;59:85–91. doi: 10.1016/j.freeradbiomed.2012.06.025. 22750507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartley D.P., Ruth J.A., Petersen D.R. The hepatocellular metabolism of 4-hydroxynonenal by alcohol dehydrogenase, aldehyde dehydrogenase, and glutathione S-transferase. Archives of Biochemistry and Biophysics. 1995;316:197–205. doi: 10.1006/abbi.1995.1028. 7840616 [DOI] [PubMed] [Google Scholar]
- 11.Wall S.B., Smith M.R., Ricart K., Zhou F., Vayalil P.K., Oh J.Y., Landar A. Detection of electrophile-sensitive proteins. Biochimica et Biophysica Acta. 2014;1840:913–922. doi: 10.1016/j.bbagen.2013.09.003. 24021887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasil’ev Y.V., Tzeng S.C., Huang L., Maier C.S. Protein modifications by electrophilic lipoxidation products: adduct formation, chemical strategies and tandem mass spectrometry for their detection and identification. Mass Spectrometry Reviews. 2014;33:157–182. doi: 10.1002/mas.21389. 24818247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doorn J.A., Petersen D.R. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chemical Research in Toxicology. 2002;15:1445–1450. doi: 10.1021/tx025590o. 12437335 [DOI] [PubMed] [Google Scholar]
- 14.Isom A.L., Barnes S., Wilson L., Kirk M., Coward L., Darley-Usmar V. Modification of cytochrome c by 4-hydroxy- 2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. Journal of the American Society for Mass Spectrometry. 2004;15:1136–1147. doi: 10.1016/j.jasms.2004.03.013. 15276160 [DOI] [PubMed] [Google Scholar]
- 15.Fritz K.S., Galligan J.J., Smathers R.L., Roede J.R., Shearn C.T., Reigan P., Petersen D.R. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chemical Research in Toxicology. 2011;24:651–662. doi: 10.1021/tx100355a. 21449565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chacko B.K., Srivastava A., Johnson M.S., Benavides G.A., Chang M.J., Ye Y.Z., Jhala N., Murphy M.P., Kalyanaraman B., Darley-Usmar V.M. Mitochondria-targeted ubiquinone (mitoq) decreases ethanol-dependent micro and macro hepatosteatosis. Hepatology. 2011;54:153–163. doi: 10.1002/hep.24377. 21520201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone D.L., Doorn J.A., Kiebler Z., Ickes B.R., Petersen D.R. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. Journal of Pharmacology and Experimental Therapeutics. 2005;315:8–15. doi: 10.1124/jpet.105.088088. 15951401 [DOI] [PubMed] [Google Scholar]
- 18.Sampey B.P., Carbone D.L., Doorn J.A., Drechsel D.A., Petersen D.R. 4-Hydroxy-2-nonenal adduction of extracellular signal-regulated kinase (Erk) and the inhibition of hepatocyte Erk-Est-like protein-1-activating protein-1 signal transduction. Molecular Pharmacology. 2007;71:871–883. doi: 10.1124/mol.106.029686. 17164404 [DOI] [PubMed] [Google Scholar]
- 19.Sampey B.P., Korourian S., Ronis M.J., Badger T.M., Petersen D.R. Immunohistochemical characterization of hepatic malondialdehyde and 4-hydroxynonenal modified proteins during early stages of ethanol-induced liver injury. Alcoholism, Clinical and Experimental Research. 2003;27:1015–1022. doi: 10.1097/01.ALC.0000071928.16732.26. 12824824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smathers R.L., Galligan J.J., Shearn C.T., Fritz K.S., Mercer K., Ronis M., Orlicky D.J., Davidson N.O., Petersen D.R. Susceptibility of L-FABP−/− mice to oxidative stress in early-stage alcoholic liver. Journal of Lipid Research. 2013;54:1335–1345. doi: 10.1194/jlr.M034892. 23359610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smathers R.L., Galligan J.J., Stewart B.J., Petersen D.R. Overview of lipid peroxidation products and hepatic protein modification in alcoholic liver disease. Chemico-Biological Interactions. 2011;192:107–112. doi: 10.1016/j.cbi.2011.02.021. 21354120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatraman A., Landar A., Davis A.J., Chamlee L., Sanderson T., Kim H., Page G., Pompilius M., Ballinger S., Darley-Usmar V., Bailey S.M. Modification of the mitochondrial proteome in response to the stress of ethanol-dependent hepatotoxicity. Journal of Biological Chemistry. 2004;279:22092–22101. doi: 10.1074/jbc.M402245200. 15033988 [DOI] [PubMed] [Google Scholar]
- 23.Lieber C.S., DeCarli L.M. The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcoholism, Clinical and Experimental Research. 1982;6:523–531. doi: 10.1111/j.1530-0277.1982.tb05017.x. 6758624 [DOI] [PubMed] [Google Scholar]
- 24.Bailey S.M., Patel V.B., Young T.A., Asayama K., Cunningham C.C. Chronic ethanol consumption alters the glutathione/glutathione peroxidase-1 system and protein oxidation status in rat liver. Alcoholism, Clinical and Experimental Research. 2001;25:726–733. 11371722 [PubMed] [Google Scholar]
- 25.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences of the United of States of America. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida K., Szweda L.I., Chae H.Z., Stadtman E.R. Immunochemical detection of 4-hydroxynonenal protein adducts in oxidized hepatocytes. Proceedings of the National Academy of Sciences of the United of States of America. 1993;90:8742–8746. doi: 10.1073/pnas.90.18.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida K., Itakura K., Kawakishi S., Hiai H., Toyokuni S., Stadtman E.R. Characterization of epitopes recognized by 4-hydroxy-2-nonenal specific antibodies. Archives of Biochemistry and Biophysics. 1995;324:241–248. doi: 10.1006/abbi.1995.0036. 8554315 [DOI] [PubMed] [Google Scholar]
- 28.Loehr J.P., Goodman S.I., Frerman F.E. Glutaric acidemia type ii: Heterogeneity of clinical and biochemical phenotypes. Pediatric Research. 1990;27:311–315. doi: 10.1203/00006450-199003000-00024. 2320399 [DOI] [PubMed] [Google Scholar]
- 29.Venkatraman A., Landar A., Davis A.J., Ulasova E., Page G., Murphy M.P., Darley-Usmar V., Bailey S.M. Oxidative modification of hepatic mitochondria protein thiols: Effect of chronic alcohol consumption. American Journal of Physiology. Gastrointestinal and Liver Physiology. 2004;286:G521–G527. doi: 10.1152/ajpgi.00399.2003. 14670822 [DOI] [PubMed] [Google Scholar]
- 30.Andringa K., King A., Bailey S. Blue native-gel electrophoresis proteomics. Methods in Molecular Biology. 2009;519:241–258. doi: 10.1007/978-1-59745-281-6_15. 19381587 [DOI] [PubMed] [Google Scholar]
- 31.Brookes P.S., Pinner A., Ramachandran A., Coward L., Barnes S., Kim H., Darley-Usmar V.M. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics. 2002;2:969–977. doi: 10.1002/1615-9861(200208)2:8<969::AID-PROT969>3.0.CO;2-3. 12203892 [DOI] [PubMed] [Google Scholar]
- 32.Bailey S.M., Landar A., Darley-Usmar V. Mitochondrial proteomics in free radical research. Free Radical Biology & Medicine. 2005;38:175–188. doi: 10.1016/j.freeradbiomed.2004.10.011. 15607901 [DOI] [PubMed] [Google Scholar]
- 33.Fischer M., You M., Matsumoto M., Crabb D.W. Peroxisome proliferator-activated receptor alpha (PPARalpha) agonist treatment reverses PPARalpha dysfunction and abnormalities in hepatic lipid metabolism in ethanol-fed mice. Journal of Biological Chemistry. 2003;278:27997–28004. doi: 10.1074/jbc.M302140200. 12791698 [DOI] [PubMed] [Google Scholar]
- 34.Mato J.M., Martínez-Chantar M.L., Lu S.C. Methionine metabolism and liver disease. Annual Review of Nutrition. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. 18331185 [DOI] [PubMed] [Google Scholar]
- 35.Kharbanda K.K. Alcoholic liver disease and methionine metabolism. Seminars in Liver Disease. 2009;29:155–165. doi: 10.1055/s-0029-1214371. [DOI] [PubMed] [Google Scholar]
- 36.Allen R.H., Stabler S.P., Lindenbaum J. Serum betaine, N,N-dimethylglycine and N-methylglycine levels in patients with cobalamin and folate deficiency and related inborn errors of metabolism. Metabolism: Clinical and Experimental. 1993;42:1448–1460. doi: 10.1016/0026-0495(93)90198-w. 7694037 [DOI] [PubMed] [Google Scholar]
- 37.Ji C., Shinohara M., Vance D., Than T.A., Ookhtens M., Chan C., Kaplowitz N. Effect of transgenic extrahepatic expression of betaine-homocysteine methyltransferase on alcohol or homocysteine-induced fatty liver. Alcoholism, Clinical and Experimental Research. 2008;32:1049–1058. doi: 10.1111/j.1530-0277.2008.00666.x. 18498552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bailey S.M., Cunningham C.C. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radical Biology & Medicine. 2002;32:11–16. doi: 10.1016/s0891-5849(01)00769-9. 11755312 [DOI] [PubMed] [Google Scholar]
- 39.Patel V.B., Spencer C.H., Young T.A., Lively M.O., Cunningham C.C. Effects of 4-hydroxynonenal on mitochondrial 3-hydroxy-3-methylglutaryl (HMG-CoA) synthase. Free Radical Biology & Medicine. 2007;43:1499–1507. doi: 10.1016/j.freeradbiomed.2007.08.004. 17964421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waeg G., Dimsity G., Esterbauer H. Monoclonal antibodies for detection of 4-hydroxynonenal modified proteins. Free Radical Research. 1996;25:149–159. doi: 10.3109/10715769609149920. 8885333 [DOI] [PubMed] [Google Scholar]
- 41.Chen J.J., Robinson N.C., Schenker S., Frosto T.A., Henderson G.I. Formation of 4-hydroxynonenal adducts with cytochrome c oxidase in rats following short-term ethanol intake. Hepatology. 1999;29:1792–1798. doi: 10.1002/hep.510290611. 10347122 [DOI] [PubMed] [Google Scholar]


