Abstract
Psycho-social stress has been suggested to influence the development of cancer, but it remains poorly defined with regard to pancreatic cancer, a lethal malignancy with few effective treatment modalities. In this study, we sought to investigate the impacts of enriched environment (EE) housing, a rodent model of “eustress”, on the growth of mouse pancreatic cancer, and to explore the potential underlying mechanisms through gene expression profiling. The EE mice showed significantly reduced tumor weights in both subcutaneous (53%) and orthotopic (41%) models, while each single component of EE (inanimate stimulation, social stimulation or physical exercise) was not profound enough to achieve comparative anti-tumor effects as EE. The integrative transcriptomic and proteomic analysis revealed that in response to EE, a total of 129 genes in the tumors showed differential expression at both the mRNA and protein levels. The differentially expressed genes were mostly localized to the mitochondria and enriched in the citrate cycle and oxidative phosphorylation pathways. Interestingly, nearly all of the mitochondria-related genes were down-regulated by EE. Our data have provided experimental evidence in favor of the application of positive stress or of benign environmental stimulation in pancreatic cancer therapy.
Pancreatic cancer is one of the most deadly neoplastic diseases, with a 5-year survival rate of below 5%1. Pancreatic cancer is typically asymptomatic in its early stages and is profoundly resistant to conventional chemo- and radio-therapies. In addition, none of the novel approaches attempted over the past decades for the treatment of pancreatic cancer have been demonstrated to be clinically beneficial or to improve patient survival benefit2. Thus, there is an urgent need to identify new strategies to treat patients with this deadly disease.
Recently, there has been increasing interest in the effects of environmental factors, and specifically the physical living and social stimulation, on the development of peripheral cancer. Adverse psycho-social factors, including striking life events, high levels of depressive symptoms, and low levels of social support, have been related to higher cancer incidences, such as those of breast and colon cancer3,4,5. Psycho-social distress, which is a negative stress associated with exposure to severe aversive and hostile environments6, has also been related to poor survival in cancer patients3,7. Recently, a tumor-promoting effect of the distress response has been demonstrated in a pancreatic cancer xenograft mouse model8, indicating that psycho-social factors may be sufficiently profound to regulate the growth of pancreatic cancer.
The housing of laboratory rodents in an enriched environment (EE) is a classical and widely used model for studying environmental impacts in neuroscience. Compared to standard housing conditions, EEs consist of more complex housing with increased space and enhanced social interactions and physical activity9. The EE, which promotes “eustress” or positive psycho-social stress10, has been demonstrated to influence brain structure and function11. It elicits a number of beneficial effects on the central nervous system (CNS), such as reduced anxiety levels12,13, enhanced learning and memory14, induction of hippocampal neurogenesis and neural plasticity15,16, and improved recovery from brain injury and cerebral disorders9,17,18. Interestingly, EE has been associated with anti-tumor phenotypes and to significantly inhibit tumor growth in syngeneic melanoma, colon cancer10, and breast cancer models19. These intriguing results provide an experimental indication of the importance of eustress for tumor growth control20. Studies focusing on EEs and pancreatic cancer may provide insights to facilitate the improvement of pancreatic cancer interventions.
In the current study, we tested whether EE could influence the growth of pancreatic cancer in mouse subcutaneous and orthotopical xenograft models. Because EE is a complex stimulation composed of several environmental components, we also investigated the elements that may play roles in the regulation of cancer growth. Additionally, by comparing the global gene expression profiles of xenograft tumors from EE- and non EE-raised mice, we were able to demonstrate that mitochondrial metabolic genes were generally down-regulated by the EE.
Results
Establishment of the EE condition for rearing normal and pancreatic cancer-implanted C57BL/6 Mice
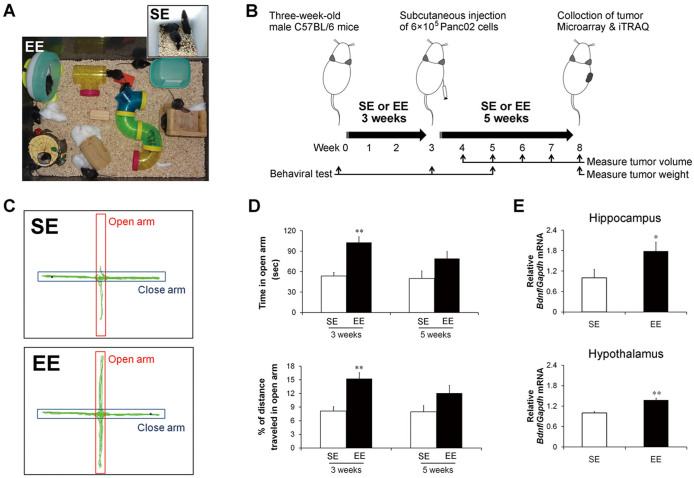
Figure 1A shows the housing conditions of EE and standard environment (SE), which are described in detail in the Methods section. The experimental protocol is schematically diagrammed in Figure 1B. Three-week-old C57BL/6 mice were randomly assigned to either EE or SE housing for 3 weeks. Both EE and SE mice were then given subcutaneous injections of Panc02 pancreatic cancer cells and returned to their respective homes for 5 additional weeks. Because one of the most important benefits of EE to the CNS is to reduce the anxiety level of residents12,13, we examined its effects on anxiety-like behaviors of mice by elevated plus maze (EPM), which is a behavioral test commonly used to assess anxiety levels in rodents. There were no differences in the behavioral parameters between the SE and EE groups upon study entry. However, mice housed in EE for 3 weeks (immediately before tumor implantation) showed significantly lower levels of anxiety-like behaviors, reflected by longer cumulative time spent in the open arms (SE = 53.7 ± 5.2 s, EE = 102.8 ± 5.2 s, P < 0.01) and higher percentages of distances travelled in the open arms (SE = 8.1 ± 1.0%, EE = 15.3 ± 1.3%, P < 0.01) (Figure 1C & D). The EPMs were repeated 2 weeks after tumor implantation (following a total of 5 weeks of EE housing). Consistently, the tumor-bearing mice living in the EE conditions showed decreased trend of anxiety-like behaviors with borderline significance (time in open arm: SE = 50.1 ± 10.8 s, EE = 79.2 ± 10.3 s, P = 0.06; percentage of distance travelled in open arm: SE = 7.9 ± 1.3%, EE = 12.1 ± 1.7%, P = 0.07) (Figure 1D).
Figure 1. Enriched environment confers beneficial effects to brain and behavior in mice.
(A) Enriched environment (EE) and standard environment (SE) housing conditions. (B) Schematic diagram of the experimental protocol. Mice were tested for anxiety-like behaviors with the elevated plus-maze (EPM) after 3 weeks (immediately before tumor implantation; C and D) or after 5 weeks (2 weeks after Panc02 tumor implantation; D) following housing in EE or SE. (C) Representative travel pathways of an SE and an EE mouse on the EPM. (D) Comparison of time spent in open arm (top) and distance traveled in open arm (bottom) between the SE and EE mice. More time or increased distance in open arm represents lower levels of anxiety-like behaviors. (E) Real-time analysis of brain BDNF mRNA levels in Panc02 tumor-bearing mice. BDNF mRNA expression significantly increased in the hippocampus (top) and hypothalamus (bottom) of the EE mice. (D, E) Data are presented as the mean ± SEM; n = 12 per group. * P < 0.05, ** P < 0.01 versus SE mice.
The expression of the brain-derived neurotrophic factor (BDNF) is considered to be an effector immediate-early gene responsive to EE10. To further confirm the CNS effects of EE in tumor-bearing mice, we measured brain BDNF levels and found that the decreased anxiety-like behaviors in the EE-housed mice were associated with significant increases in hippocampal BDNF mRNA levels (P < 0.05; Figure 1E). In addition to its influence on hippocampal BDNF, EE significantly increased hypothalamic BDNF mRNA levels in the tumor-bearing mice (P < 0.01; Figure 1E), which was consistent with previous observations10. Taken together, these results indicated the successful establishment of EE conditions for the Panc02 tumor-bearing mice.
EE-induced growth inhibition of mouse pancreatic cancer
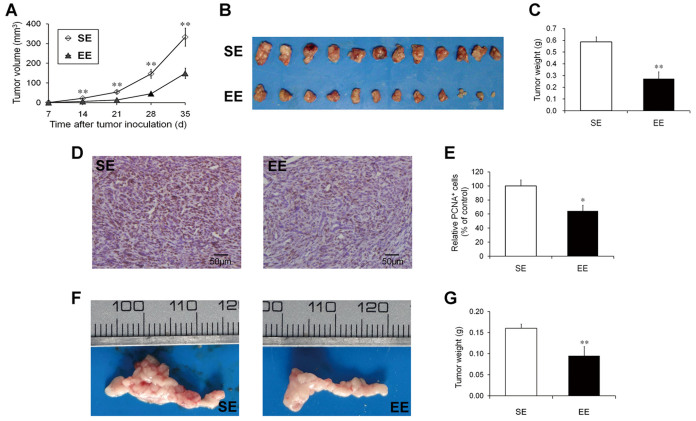
The volumes of the subcutaneous tumors were monitored once per week after the Panc02 cell injections (Figure 1B). The growth curve showed that the rate of tumor growth was substantially delayed in the EE mice (Figure 2A). At the time of sacrifice (35 days post-inoculation), the mean tumor volume for the EE-housed mice was significantly smaller than that of the SE-housed mice (148.9 ± 26.1 mm3 versus 332.8 ± 45.6 mm3, P < 0.01; Figure 2A and B). Similar to the tumor volume, the tumor weight at sacrifice was lower in EE mice by ~53.4% (P < 0.01) (Figure 2C). We further analyzed tumor cell proliferation with proliferating cell nuclear antigen (PCNA) staining. As predicted, the reduced tumor size in the EE mice was associated with a significant decrease in cell proliferation (35.7%, P < 0.05; Figure 2D and E).
Figure 2. Enriched environment reduced tumor growth in syngeneic mouse models of pancreatic cancer.
Mice were housed in the enriched environment (EE) or the standard environment (SE) for 3 weeks and then subcutaneously injected with Panc02 cells (6 × 105 cells per mouse). (A) Growth curve based on the tumor volumes monitored once a week after implantation. (B) Subcutaneous Panc02 tumors dissected at the time of sacrifice. (C) Comparison of tumor weights between the SE and EE mice. EE housing significantly inhibited the growth of the subcutaneous Panc02 tumors. Reduced tumor sizes in EE were associated with the decreased proliferating cell nuclear antigen (PCNA) staining of the Panc02 tumors. (D) Representative immunohistochemical staining of PCNA in the Panc02 tumors from the SE and EE mice. Scale bars: 50 μm. (E) Quantitative analysis of PCNA+ tumor cells. EE- and SE-housed mice were also orthotopically injected with Panc02 cells (1 × 105 cells per mouse). (F) Orthotopic Panc02 tumors dissected at the time of sacrifice. (G) Comparison of orthotopic tumor weights between the SE and EE mice. There were significantly reductions in the tumor weights in the EE-housed mice. (A, C, E, G) Data are presented as the mean ± SEM; n = 12 per group. * P < 0.05, ** P < 0.01 versus SE mice.
To further confirm the inhibitory effects of EE on the growth of pancreatic cancer, we established an orthotopic pancreatic cancer model in EE- and SE-housed mice. The experimental protocol was similar to that described above with the exception that the Panc02 cells were orthotopically implanted into the mouse pancreases. The mice were sacrificed at three weeks after tumor implantation. As shown in Figure 2F, the orthotopic tumors of the EE-housed mice were substantially smaller than those of the SE-housed mice. The tumor weight was lower in the EE-housed mice by 40.6% (P < 0.01) (Figure 2G). Taken together, our results provided evidence that EE can lead to considerable growth inhibition of pancreatic cancer in mice.
Roles of different enrichment elements in tumor inhibition
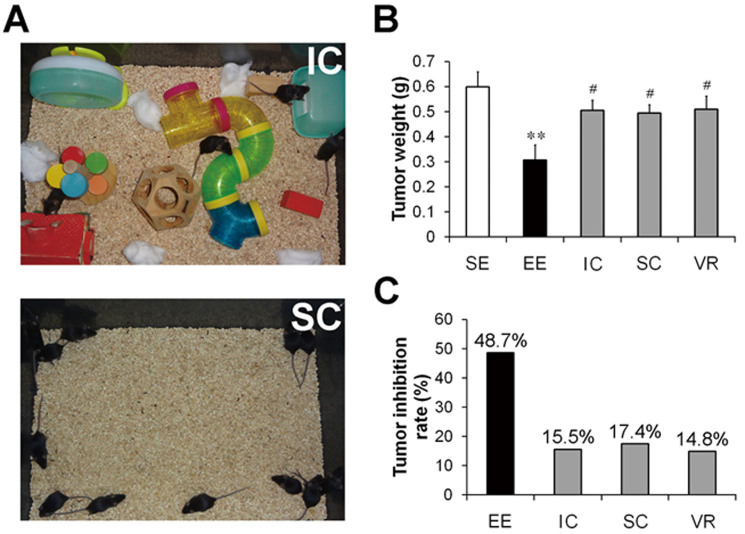
An EE is classically defined as a combination of two critical environmental stimulations: 1) complex inanimate stimulations such as toys, tunnels, etc. and 2) social stimulation21. To determine whether inanimate or social stimulation alone could achieve tumor inhibitory effects, we subjected mice either to a complex inanimate condition (IC, providing inanimate stimulation only) or to a social condition (SC, providing social stimulation only) (Figure 3A). Unlike the significant inhibition of tumor growth caused by the EE, both the IC and SC showed only slight, non-significant inhibitory effects on the growth of the subcutaneous Panc02 tumors as compared to SE (Figure 3B). As shown in Figure 3C, the tumor inhibition rates of the IC and SC were 15.5% and 17.4%, respectively, which were much lower than that of EE (48.7%).
Figure 3. A combination of multiple enrichment factors is essential for enriched environment-induced Panc02 tumor inhibition.
(A) Inanimate condition (IC) and social condition (SC). The inanimate housing condition (top), in which the mice were housed in groups of 4 per cage in the enriched environment (EE) cages supplemented with a variety of objects, only provided inanimate stimulation. The social housing condition (bottom), in which the mice were housed in groups of 12 per cage in the EE cages without any supplements, only provided social stimulation. We also assessed a voluntary running (VR) condition, in which the mice were housed with running wheels under the standard environment (SE) conditions, to investigate the impacts of physical exercise on the Panc02 tumors. The mice were housed under different conditions for 3 weeks and then were subcutaneously injected with Panc02 cells (6 × 105 cells per mouse). Thirty-five days after tumor implantation, the mice were sacrificed and the tumors were harvested and weighed. (B) Tumor weights determined at the time of sacrifice. Data are presented as the mean ± SEM; n = 12 per group. ** P < 0.01 versus SE mice; # P < 0.05 versus EE mice. (C) Tumor inhibition calculated from the tumor weight data. The values on each column indicate the tumor inhibition rates for each group. Inanimate stimulation, social stimulation or physical exercise alone is insufficient to account for EE-induced tumor inhibition.
We also subjected the mice to voluntary wheel running (VR) to investigate whether the EE-induced inhibition of the Panc02 tumors was largely attributed to physical exercise, which has been proposed to be an important component of EE22. The results showed that exercise only exhibited weak inhibitory effects on tumor growth, the extent of which was similar to those obtained with IC or SC (Figure 3B & C). These data suggest that inanimate stimulation, social stimulation or physical exercise alone is not sufficient to induce significant inhibition of tumor growth.
Identification of differentially expressed genes caused by EE in mouse pancreatic tumors
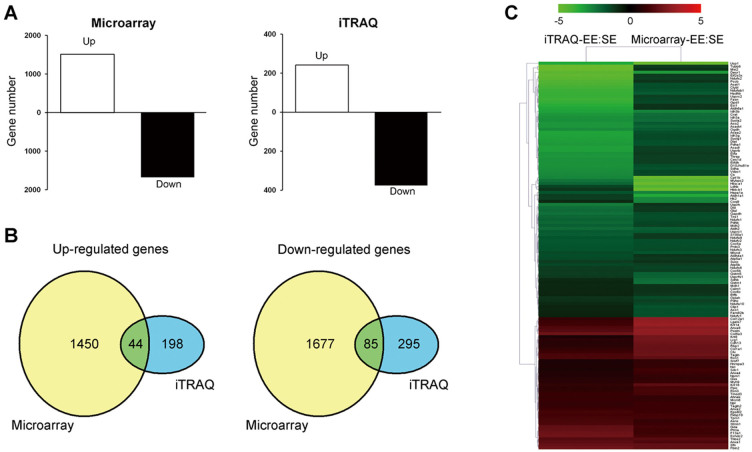
Pooled RNA from the subcutaneous Panc02 tumors of EE or of SE mice were subjected to genome-wide comparative analysis using a microarray approach. After obtaining normalized signal data, 30,319 probes were qualified for differential expression analysis. A total of 3,256 genes were found to be differentially expressed (fold change of > 2.0 or < 0.5) between the EE and SE groups, among which approximately 55% (1762 genes) were down-regulated by the EE (Figure 4A).
Figure 4. Integrative analyses of differentially expressed genes/proteins in Panc02 tumor caused by enriched environment exposure.
The pooled RNA and protein samples from the Panc02 tumors of the standard environment (SE)- or the enriched environment (EE)-housed mice were subjected to microarrays and isobaric tags for relative and absolute quantification (iTRAQ) assays, respectively. (A) Numbers of mRNAs (left) and proteins (right) differentially expressed in microarray (fold change > 2.0 or < 0.5) and iTRAQ (fold change > 1.5 or < 0.67) assays in the comparison of EE versus SE. (B) Venn diagrams of the numbers of up-regulated (left) and down-regulated (right) mRNAs and proteins identified by the microarray and iTRAQ assays. A total of 44 genes were up-regulated, and 85 were down-regulated at both the mRNA and protein levels. (C) Hierarchical clustering of the 129 genes, for which both the mRNA and protein expression levels changed in the same directions. The fold changes for all genes were log2 transformed prior to performing the clustering analysis. The colored images are presented as described; the green and red colors indicate decreased and increased levels of expression, respectively.
In parallel, a comparative proteomic Isobaric Tags for Relative and Absolute Quantification (iTRAQ) analysis was performed using samples obtained from subcutaneous Panc02 tumors from a different set of experiments. Using the cut-offs for the unused protein score of > 1.3 and the number of peptides of ≥ 2, a total of 1657 proteins were identified, and 1625 proteins were quantified. In total, we found 242 up-regulated proteins (fold change > 1.5) and 380 down-regulated proteins (fold change < 0.67) in the Panc02 tumors from the EE mice as compared with SE mice (Figure 4A & Supplementary Table S1).
A combined analysis of the above two experiments revealed that 129 genes exhibited expression changes at both the mRNA and protein levels. Among the 129 genes, 44 were up-regulated, and 85 were down-regulated (Figure 4B). The expression patterns of these 129 genes were subjected to hierarchical clustering analysis, and the heat map is shown in Figure 4C. The extents of the mRNA expression changes did not always parallel those of the protein expression changes, although the directions were the same23, suggesting the possible role of post-translational modifications in contributing to these discrepancies.
Validation of differentially expressed genes by quantitative RT-PCR
We performed quantitative RT-PCR to validate the gene expression changes in the Panc02 tumors caused by EE in an independent set of samples (n = 12 for each group). Twelve genes (4 up-regulated and 8 down-regulated) were randomly selected to validate the 129 genes showing expression changes. As shown in Figure 5, the results of the quantitative RT-PCR confirmed the directions of changes for all selected genes. Ten of the 12 genes still showed significant expression changes (P < 0.05). These data supported the validity of the criteria used in our analysis.
Figure 5. Validations of dysregulated genes by quantitative real-time PCR analysis.
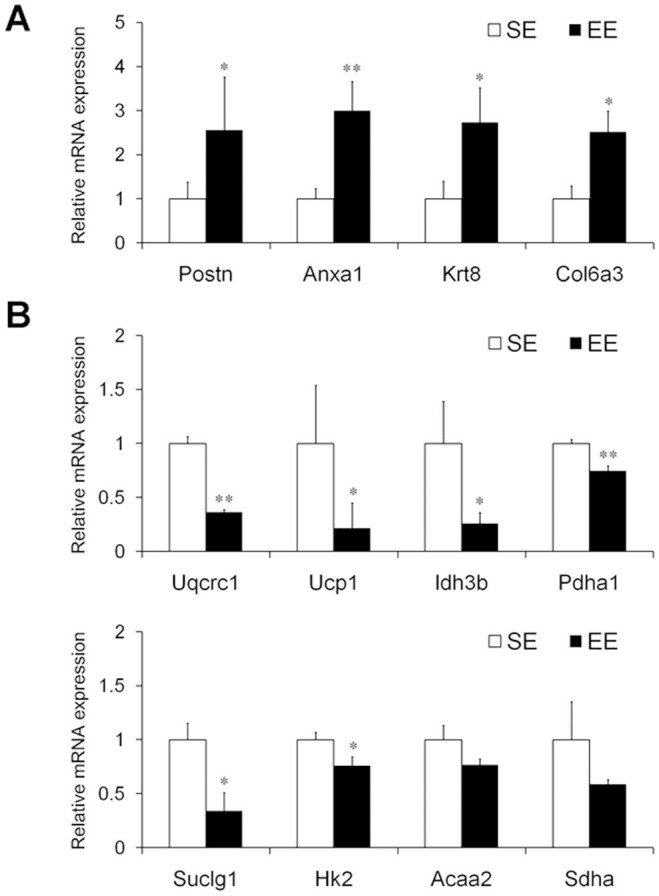
mRNA expression of 4 up-regulated genes (A; Postn, Anxa1, Krt8, and Col6a3) and 8 down-regulated genes (B; Uqcrc1, Ucp1, Idh3b, Pdha1, Suclg1, Hk2, Acaa2, and Sdha) in the Panc02 tumors of the standard environment (SE)- or the enriched environment (EE)-housed mice. All selected genes were confirmed to be changed in the same direction as that identified by the microarray analysis. Four out of the 4 genes were significantly up-regulated (A), and 6 out of the 8 genes were significantly down-regulated (B) following exposure to EE. Data are expressed as relative target gene expression levels compared with β-actin expression and presented as the mean ± SEM (n = 12 per group). * P < 0.05, ** P < 0.01 versus SE mice.
Functional annotation of differentially expressed genes in Panc02 tumors
We used a gene ontology (GO)-based enrichment analysis to classify the EE-induced gene expression changes in Panc02 tumors. The significantly enriched GO terms were ranked by their respective P values. Table 1 shows the top-ranked terms for the three different GO sub-ontologies: cellular component, biological process, and molecular function. The analysis for GO cellular component sub-ontology revealed that the EE-induced differentially expressed genes were mostly localized to the mitochondria. Interestingly, 68 of the 69 mitochondria-related genes were down-regulated by the EE, suggesting that the EE may have inhibited or at least downregulated mitochondrial metabolism in the tumors. The top 5 GO biological process terms and molecular function terms were also linked to mitochondrial function and included “generation of precursor metabolites”, “cellular respiration”, etc., providing further support of the mitochondria as a target for systemic regulation in EE.
Table 1. Gene ontology annotation of differentially expressed genes in Panc02 tumors induced by enriched environment.
| No. | Term | Count | P | PFDR |
|---|---|---|---|---|
| Top 5 GO cellular component terms | ||||
| 1 | Mitochondrion | 69 | 3.50E-40 | 4.24E-37 |
| 2 | Mitochondrial part | 46 | 4.44E-34 | 5.40E-31 |
| 3 | Mitochondrial envelope | 32 | 5.80E-22 | 7.06E-19 |
| 4 | Mitochondrial inner membrane | 29 | 6.68E-22 | 8.13E-19 |
| 5 | Mitochondrial membrane | 31 | 1.36E-21 | 1.65E-18 |
| Top 5 GO biological process terms | ||||
| 1 | Generation of precursor metabolites and energy | 39 | 1.83E-39 | 2.76E-36 |
| 2 | Oxidation reduction | 41 | 3.66E-26 | 5.50E-23 |
| 3 | Cellular respiration | 17 | 2.47E-21 | 3.72E-18 |
| 4 | Electron transport chain | 20 | 5.76E-21 | 8.66E-18 |
| 5 | Acetyl-CoA metabolic process | 14 | 1.99E-20 | 3.00E-17 |
| Top 5 GO molecular function terms | ||||
| 1 | Coenzyme binding | 19 | 1.39E-15 | 1.91E-12 |
| 2 | Cofactor binding | 21 | 3.44E-15 | 4.55E-12 |
| 3 | NAD or NADH binding | 9 | 2.92E-09 | 3.87E-06 |
| 4 | Iron-sulfur cluster binding | 9 | 8.49E-09 | 1.12E-05 |
| 5 | Metal cluster binding | 9 | 8.49E-09 | 1.12E-05 |
GO: gene ontology; PFDR: false discovery rate corrected P value.
To determine the intratumoral pathways regulated by exposure to EEs, we adopted the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations of the differentially expressed genes. A total of 12 KEGG pathways were significantly overrepresented (false discovery rate [FDR]-corrected P < 0.05). As shown in Table 2, most of these overrepresented KEGG pathways (8/12) were linked to mitochondria-related functions. The top-ranked terms included two main mitochondrial functions: “citrate cycle” and “oxidative phosphorylation (OXPHOS)”. These two terms included a total of 36 differentially expressed genes, which were involved in almost all of the reactions involving the conversion of pyruvate into acetyl-CoA and the subsequent citrate cycle (Supplementary Fig. S1), as well as in all complexes of the electron transport chain. Notably, all of the 36 differentially expressed genes related to “citrate cycle” or “OXPHOS”, were down-regulated by EE (Supplementary Table S2). The decreased expression of the mitochondrial genes may have led to defective mitochondrial functioning in the tumor cells.
Table 2. KEGG pathways of differentially expressed genes in Panc02 tumors induced by enriched environment.
| No. | Term | Count | P | PFDR |
|---|---|---|---|---|
| 1 | Citrate cyclea | 17 | 4.30E-23 | 4.52E-20 |
| 2 | Parkinson's disease | 22 | 8.86E-18 | 9.32E-15 |
| 3 | Oxidative phosphorylationa | 21 | 1.03E-16 | 1.22E-13 |
| 4 | Alzheimer's disease | 23 | 4.62E-16 | 4.66E-13 |
| 5 | Huntington's disease | 23 | 5.20E-16 | 5.88E-13 |
| 6 | Pyruvate metabolisma | 9 | 4.38E-08 | 4.61E-05 |
| 7 | Propanoate metabolisma | 8 | 8.62E-08 | 9.07E-05 |
| 8 | Cardiac muscle contraction | 9 | 7.25E-06 | 0.007632 |
| 9 | Fatty acid metabolisma | 7 | 2.39E-05 | 0.025154 |
| 10 | Valine, leucine and isoleucine degradationa | 7 | 2.72E-05 | 0.028625 |
| 11 | Glycolysis/Gluconeogenesisa | 8 | 2.74E-05 | 0.028881 |
| 12 | Glyoxylate and dicarboxylate metabolisma | 5 | 4.59E-05 | 0.048242 |
aMitochondria-related pathways.
PFDR: false discovery rate corrected P value.
Discussion
The impacts of psycho-social factors on the development of pancreatic cancer have been attracting increasing research interest. Several recent studies have found that exposure to distress could significantly promote the progression of pancreatic cancer xenografts8,24,25. However, to date, no research has focused on the inhibitory effects of social interactions and psychological factors on pancreatic cancer. EE, which confers eustress or positive stress to rodents, has been reported in association with anti-tumor effects in mouse melanoma, colorectal cancer and breast cancer10,19. In the current study, we extended this eustress model to the study of pancreatic cancer and reported that EE exposure robustly inhibits the growth of mouse pancreatic cancer in both subcutaneous and orthotopic models. These findings suggest a potential therapeutic relevance of eustress in pancreatic cancer.
An EE is a more complex housing condition compared with standard laboratory animal housing, typically including a combination of increased physical space, larger groups, objects used for activities or hiding, and running wheels. The components forming EE can be classified into two categories; inanimate and social stimulations21. Either inanimate26 or social stimulation27 has been reported to be profound enough to affect the CNS. In particular, social experience has been shown to promote adult neurogenesis28 and induce synaptic plasticity to promote functional recovery following stroke in rodents29. However, here we found that neither inanimate stimulation nor social stimulation alone was sufficient to achieve similar tumor suppressive effects as compared with EE, suggesting that the mechanisms involved in the regulation of peripheral cancer growth in EE may be more complicated than those associated with the regulation of some types of brain function. Similarly, although physical exercise has been reported to achieve beneficial effects in the brain, such as promoting hippocampal neurogenesis and improving spatial learning ability11,30, voluntary wheel running did not significantly reduce tumor weight in our study. This observation is consistent with a previous study showing that physical exercise alone does not account for the EE-induced melanoma resistance10. In that study, the authors also examined several biomarkers in the mouse serum and found that the runners possess distinct alterations in their serum biomarkers as compared with those of the EE mice. In the runners, leptin does not change, while adiponectin is significantly decreased, in contrast to the increase observed in the EE mice. They also found that running influenced gene expression in the arcuate nucleus and led to a different expression pattern as compared with EE, especially for BDNF, which is a responsive marker of EE in the brain. Our current study complements this prior study by demonstrating that novelty, social contact and exercise are all beneficial to mice, but each single factor is not sufficient to achieve anti-tumor effects.
The mechanisms underlying EE-induced anti-tumor effects remain elusive. In a previous study, Cao et al. proposed a specific brain-adipocyte BDNF/leptin axis model to explain the mode by which EE influences peripheral cancer growth10. According to this hypothesis, EE leads to the activation of sympathetic innervations of fat tissue, causing an increased adiponectin/leptin ratio in the plasma. Adiponectin and leptin are two major types of adipokines crucial for energy homeostasis. The mechanism by which the ratio of adiponectin/leptin reduces the cancer burden in EE mice remains to be investigated. To understand the molecular mechanisms underlying EE-induced tumor suppression, we compared the global gene expression patterns in pancreatic cancer xenografts of EE and SE mice. In addition to microarray analysis, which allowed for quantifications of mRNA levels, the iTRAQ proteomic approach was also applied to confirm the gene expression changes at the protein level. Only those genes showing expression changes at both levels were selected for further enrichment analyses. Thus, the results of our GO and KEGG pathway analyses are convincing. The EE-induced changes in gene expressions were enriched in the mitochondria, which are notably related to glucose metabolism. These genes encode the enzymes that play key roles in pyruvate decarboxylation, the citrate cycle and electron transport. Interestingly, all of these genes were down-regulated by EE, including key enzymes of the citrate cycle (citrate synthase, isocitrate dehydrogenase and oxoglutarate dehydrogenase) and in pyruvate decarboxylation (pyruvate dehydrogenase system) (Supplementary Fig. S1). The possibility may exist that the decreased expression of mitochondria-related genes in the tumors of EE mice is due to the slower progression of tumors. Nevertheless, previous studies have demonstrated that EE could affect mitochondrial function in the cerebral cortex31 and white adipose tissue32, supporting an intercellular effect of EE on mitochondria. Further experiments including the comparison of gene expression profile between large and small tumors within the control group will help to elucidate the impact of EE on mitochondria in tumor cells.
Cancer has long been recognized as a metabolic disease that is characterized by bioenergetic switching from the citrate cycle to glycolysis. Although Otto Warburg proposed that the oxidative glycolytic phenotype is the result of the impairment of mitochondrial OXPHOS, studies have found that mitochondrial function in many cancers is indeed intact and may even be up-regulated33,34. The mitochondrial citrate cycle serves to coordinate several key metabolic pathways35, including glutamine metabolism, which is critical for pancreatic cancer progression36. It also converts glucose and other metabolites to the precursors used in the synthesis of fatty acids, nucleic acids and amino acids. Because the unconstrained replicative potentials of cancer cells require large amounts of intermediates for macromolecular biosynthesis37, the over-expression or the amplification of bioenergetic genes is expected to occur in cancer cells. Indeed, many mitochondrial metabolism enzymes, including CS38, MDH239, UQCRSF140,41,42, UQCRH43, NDUFS644, ATP5B45,46, COX5B47,48, etc., have been reported to be overexpressed in cancers, such as pancreatic, endometrial, gastric, breast, ovarian, cervical, and liver cancers. These genes were all down-regulated by EE in our study. Thus, we propose that the global inhibition of mitochondrial metabolic genes might be a strategy for promoting cancer cell death in EE mice. Accordingly, chemical inhibitors targeting enzymes of the citrate cycle and OXPHOS have been reported to possess anticancer potentials33. For example, one recent paper documented that in pancreatic cancer, a subpopulation of cells responsible for tumor relapse relied on mitochondrial oxidative phosphorylation for survival. Treatment with oxidative phosphorylation inhibitors could effectively inhibit tumor recurrent in vivo49. Mitochondria-originated tumor inhibition may be caused by bioenergetic depletion or by apoptosis induced by mitochondrial dysfunction50,51. Further experiments, such as comparisons of mitochondrial contents, mitochondrial membrane potentials and reactive oxygen species levels in pancreatic cancers of EE and non-EE mice, will allow for functional validation of the findings of this study. Nevertheless, our data, together with the findings of previous reports, lead us to speculate that EE may target the mitochondrial metabolic pathway, at least, as one of the mechanisms, to control tumor growth.
The top 5 pathways (ranked by the P values) enriched in the KEGG analysis included three terms related to the major neurodegenerative diseases of Parkinson's disease, Alzheimer's disease and Huntington's disease. Mitochondrial dysfunction and oxidative stress have long been considered to be central mechanisms underlying neurodegeneration52. Thus, the genes related to OXPHOS are included in the neurodegenerative disease pathways in the KEGG database. The enrichments of these three neurodegenerative disease pathways in our study were mainly because of the differentially expressed genes related to OXPHOS. EE rearing was initially used to improve the cognitive, motor and sensory functions of animals. EE exposures may have compensated for the impaired performances observed in rodent models of neurodegenerative diseases17,53,54. Thus, it is possible that the mitochondria may be central to the mechanisms underlying the EE-induced beneficial effects observed both in neurodegenerative diseases and cancer. Given the important role of mitochondrial dysfunction in the pathogenesis of neurodegenerative disease, our current data also suggest that it may be interesting to assess the influences of EE on bioenergetic changes in the CNS of rodent models of neurodegenerative diseases in the future.
In summary, our study revealed that EE housing conditions significantly inhibited mouse pancreatic cancer progression. We have provided experimental evidence in favor of the application of positive stress or of benign environmental stimulation in future pancreatic cancer therapy. In addition, our findings suggest the involvement of the mitochondria in the anti-tumor effects of EE.
Methods
Experimental animals and environmental housing conditions
All animal experiments were performed in accordance with ethical guidelines under the protocols approved by the Medical Experimental Animal Care Commission at Shanghai Jiaotong University. Male 3-week-old C57BL/6 mice were obtained from Sino-British SIPPR/BK Lab Animal Ltd. (Shanghai, China) and maintained in a temperature- and humidity-controlled room with a 12 h light/dark cycle.
The grouping of the animals in the different housing conditions was as follows: the SE group was housed in groups of 4 mice per cage in standard laboratory cages (26 × 16 × 13 cm); the EE group was housed in groups of 12 mice per cage in large cages (61 × 43 × 21 cm) containing a running wheel, small huts, tunnels, wood toys, and nesting materials; the IC group was housed in groups of 4 mice per cage in EE cages supplemented with a variety of objects similar to the EE group; the SC group was housed in groups of 12 mice per cage in EE cages without any supplements; the VR group was housed with running wheels under SE conditions. The housing supplements for the EE and IC groups were moved to different locations within each cage twice per week and were replaced with new toys once per week. In all groups, the mice were allowed ad libitum access to food and water.
Subcutaneous and orthotopic implantations with mouse Panc02 pancreatic cancer cells
The murine pancreatic cancer cell line Panc02 (also known as Pan02) was obtained from the Division of Cancer Treatment and Diagnosis (DCTD) Tumor Repository, Frederick National Laboratory for Cancer Research (Frederick, MD, USA) and was maintained in DMEM medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5% CO2.
To investigate the effects of the housing conditions on pancreatic tumor growth, male 3-week-old C57BL/6 mice were randomized for exposures to the different housing conditions (n = 12 per group) for 3 weeks. Subcutaneous tumors were prepared by implanting Panc02 cells (6 × 105 per mouse) in the right flanks of the mice. The mice were returned to their respective cages after tumor implantation. From day 7 after tumor implantation, the tumor volumes were measured once a week with a vernier caliper and according to the formula V = π/6 × width2 × length. Thirty-five days after Panc02 cell injection, the mice were sacrificed. The tumors were excised, weighted, and processed for the following analyses.
For the orthotopic models of pancreatic cancer, the mice pancreases were implanted with Panc02 cells as described previously55 after 3 weeks of EE housing or SE housing. Briefly, the animals were anesthetized by the intraperitoneal administration of pelltrobarbitalum natricum (50 mg per kg body weight), and then, a ~7 mm incision in the left abdominal/flank region was made. Approximately 1 × 105 Panc02 cells suspended in 50 μl of a Matrigel (BD Biosciences, Bedford, MA, USA) mixture (1:1 volume) were injected into the head/neck region of the pancreas. The peritoneum and skin incisions were sequentially closed with 4-0 silk stitches. After recovery from the anesthesia, the mice were returned to their respective cages for an additional 3 weeks. At the end of the experiment, the mice were sacrificed and the tumor-bearing pancreases were excised from each mouse. The orthotopic tumors were then identified and weighed.
Behavioral test: elevated plus maze (EPM)
The EPM test for assessing anxiety-related behavior in mice was conducted as described previously56 with modifications for use in our laboratory. The EPM apparatus consisted of two open arms (30 × 5 cm), two closed arms (30 × 5 cm) with high wells (20 cm), and a center platform by which all four arms were connected. The maze was elevated at 80 cm above the floor. The EPM test was performed during the light photoperiod of the mice. Each mouse was placed on the center platform facing an open arm and was allowed to explore freely. Behaviors were video recorded for 10-min sessions, and the behavioral parameters for each mouse were then collected using the SuperMaze software (version 2.0; Shanghai Xinruan Information Tech Co., Shanghai, China). The time spent on the open arms and the percentages of the distances travelled in the open arms were used as indices of anxiety-like behavior. Between each test session, the maze was cleaned with 70% ethanol.
EPM test was performed twice before and once after Panc02 cells implantation for each mouse (Figure 1B). To avoid the influence of prior EPM tests on mice behavior, we followed a test-retest paradigm described previously57. Briefly, mice were retested for EPM with an interval of at least 2–3 weeks, and the retests were performed in a different room.
Dissection of the hypothalamus and hippocampus
Brains were quickly harvested from each animal immediately after anesthesia overdose, and the hypothalamus and the hippocampi were then dissected out on an ice-cooled plate. With the brain resting on its dorsal surface on the plate, the hypothalamus was dissected out according to a previous description58. Subsequently, the hippocampus was exposed according the technique described by Haqihara et al.59, and then isolated carefully. Immediately after dissection, the isolated brain sections were transferred into individual microtubes and snap-frozen in liquid nitrogen. All samples were stored at −80°C until further processing.
Immunohistochemistry
Panc02 tumors were fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at 5 μm. For PCNA immunohistochemical staining, the sections were incubated with anti-PCNA antibody (eBioscience, San Diego, CA, USA) at 4°C overnight. Primary antibody staining was visualized using the ImPress Universal kit (Vector Laboratories, Burlingame, CA, USA) with NovaRed (Vector Laboratories) as a substrate. The sections were then counterstained with hematoxylin. To quantify immunohistochemical staining, we counted the number of positive cell per 20× microscope field with 10 random fields per animal. The data were expressed as the percentage changes in the positive cell number over SE mice.
Gene expression microarray
Total RNA from the homogenized tumors was extracted using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. The RNA samples from 6 randomly selected SE mice or EE mice were pooled and then sent to Shanghai OE Biotech Co., Ltd. (Shanghai, China) for cDNA synthesis, cRNA amplification and labeling, and subsequent hybridization to the Agilent Whole Mouse Genome 4×44k Microarray (Agilent Technologies, Santa Clara, CA, USA). The microarray slides were scanned with an Agilent scanner, and the output images were digitalized using the Feature Extraction software (Agilent Technologies). The raw data were then normalized using quantile normalization according to the procedures recommended by Agilent Technologies with the GeneSpring software (version 12.0, Agilent Technologies). The probes that were flagged as "Detected" or "Compromised" were retained. The genes were considered to be differentially expressed if they possessed fold changes of > 2.0 or of < 0.5 in the comparison of EE versus SE. The microarray data have been deposited in the National Center for Biotechnology Gene Expression Omnibus database (accession GSE58306, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE58306).
Real-time quantitative PCR
The total RNA was extracted as described above, and subjected to reverse transcription using the FastQuant RT Kit (with gDNase) (Tiangen Biotech, Beijing, China). Quantitative real-time PCR was performed with the FastStart Universal SYBR Green Master (Roche Diagnostics, Mannheim, Germany) using the StepOne Plus Real-time PCR System (Applied Biosystems, Foster City, CA, USA). The relative expression levels of the target genes were calculated after normalization against a reference gene (β-actin). The primer sequences used for the quantitative PCR are provided in Supplementary Table S3.
Proteomic analysis
The global protein expression profile was determined using the iTRAQ quantitative proteomic approach. Protein preparation, iTRAQ labeling, strong cation exchange (SCX), nanoLC-MS/MS analysis, and protein identification and quantitation were performed following the procedures detailed in a previous study60. Briefly, the tumor protein lysates from 6 randomly selected SE or EE mice were pooled, and then labeled with the iTRAQ labeling reagents 114 and 116 (iTRAQ Reagent 4-Plex Kit, Applied Biosystems), respectively. The iTRAQ-labeled samples were fractionated by SCX chromatography, and subsequently analyzed using the NanoLC system (NanoLC-2D Ultra, Eksigent, Dublin, CA, USA). Protein identifications and quantitations were performed with the ProteinPilot software (version 4.1; AB SCIEX, Foster City, CA, USA) and the mouse UniProtKB/Swiss-Prot proteome database. The FDR for the peptide identifications was estimated using a decoy database search strategy. We used a strict unused protein score cutoff of > 1.3 (confidence > 95%) for the protein identifications. Only those proteins identified from at least two peptides were included for the quantitative assay. The proteins were considered to be differentially expressed if their iTRAQ ratios were > 1.5 or < 0.67 in the comparison of EE versus SE.
Bioinformatics analysis
The results of the microarray and iTRAQ assays were exported into Microsoft Excel for manual data interpretation. The web-accessible Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to find the intersecting of up- or down-regulated genes that were identified by both the microarray and the iTRAQ assay. The commonly up-regulated genes identified by both assays were then combined with the commonly down-regulated genes. The fold change data of the combined set of differentially expressed genes were log2 transformed before being imported into the TIGR MeV software (available at http://www.tm4.org/mev.html) for hierarchical clustering analysis. The GO and KEGG pathway enrichment analyses of the differentially expressed genes were performed using the web-accessible functional annotation tool from the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.7 (http://david.abcc.ncifcrf.gov). The GO terms and KEGG pathways with FDR-corrected P values of < 0.05 were considered to be enriched.
Statistical Analysis
The results were presented as the mean ± SEM unless indicated otherwise. Statistical analyses were performed using the SPSS software version 19.0 (SPSS Inc., Chicago, IL, USA). The significance of difference between groups was assessed by the Student's t test for single comparisons or by the analysis of variance (ANOVA) with the Student-Newman-Keuls tests for multiple comparisons. A value of P < 0.05 (two-tailed) was considered statistically significant.
Author Contributions
H.T. conceived the research and took overall supervision in the study. G.L., Y.G., Y.F., Y.W., H.L., Y.S. and X.C. performed experiments. G.L. and Y.G. performed data analysis. Y.G. and H.T. wrote the manuscript. M.Y., X.Y., W.P. and J.G. contributed to the discussion of results and to the review of the manuscript. All authors approved the final manuscript.
Supplementary Material
Supplementary information
Supplementary Table S1
Acknowledgments
This work was supported by grants from the Chinese State Key Laboratory of Oncogenes and Related Genes (91-11-02 to H.T.; 91-13-06 to Y.G.) and the Young Scientist Fund of National Natural Science Foundation of China (81201823 to Y.G.).
References
- Almhanna K. & Philip P. A. Defining new paradigms for the treatment of pancreatic cancer. Curr Treat Options Oncol 12, 111–125 (2011). [DOI] [PubMed] [Google Scholar]
- Guillaumond F., Iovanna J. L. & Vasseur S. Pancreatic tumor cell metabolism: Focus on glycolysis and its connected metabolic pathways. Arch Biochem Biophys 545C, 69–73 (2014). [DOI] [PubMed] [Google Scholar]
- Chida Y., Hamer M., Wardle J. & Steptoe A. Do stress-related psychosocial factors contribute to cancer incidence and survival? Nat Clin Pract Oncol 5, 466–475 (2008). [DOI] [PubMed] [Google Scholar]
- Lin Y. et al. Striking life events associated with primary breast cancer susceptibility in women: a meta-analysis study. J Exp Clin Cancer Res 32, 53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary K. E. et al. Sex differences in associations between psychosocial factors and aberrant crypt foci among patients at risk for colon cancer. Gend Med 8, 165–171 (2011). [DOI] [PubMed] [Google Scholar]
- Milsum J. H. A model of the eustress system for health/illness. Behav Sci 30, 179–186 (1985). [DOI] [PubMed] [Google Scholar]
- Hamer M., Chida Y. & Molloy G. J. Psychological distress and cancer mortality. J Psychosom Res 66, 255–258 (2009). [DOI] [PubMed] [Google Scholar]
- Schuller H. M., Al-Wadei H. A., Ullah M. F. & Plummer H. K. 3rd Regulation of pancreatic cancer by neuropsychological stress responses: a novel target for intervention. Carcinogenesis 33, 191–196 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantharajah J. & Hannan A. J. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci 7, 697–709 (2006). [DOI] [PubMed] [Google Scholar]
- Cao L. et al. Environmental and genetic activation of a brain-adipocyte BDNF/leptin axis causes cancer remission and inhibition. Cell 142, 52–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H., Kempermann G. & Gage F. H. Neural consequences of environmental enrichment. Nat Rev Neurosci 1, 191–198 (2000). [DOI] [PubMed] [Google Scholar]
- Benaroya-Milshtein N. et al. Environmental enrichment in mice decreases anxiety, attenuates stress responses and enhances natural killer cell activity. Eur J Neurosci 20, 1341–1347 (2004). [DOI] [PubMed] [Google Scholar]
- Baldini S. et al. Enriched early life experiences reduce adult anxiety-like behavior in rats: a role for insulin-like growth factor 1. J Neurosci 33, 11715–11723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampon C. et al. Enrichment induces structural changes and recovery from nonspatial memory deficits in CA1 NMDAR1-knockout mice. Nat Neurosci 3, 238–244 (2000). [DOI] [PubMed] [Google Scholar]
- Kempermann G., Kuhn H. G. & Gage F. H. More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495 (1997). [DOI] [PubMed] [Google Scholar]
- Sale A., Berardi N. & Maffei L. Enrich the environment to empower the brain. Trends Neurosci 32, 233–239 (2009). [DOI] [PubMed] [Google Scholar]
- Laviola G., Hannan A. J., Macri S., Solinas M. & Jaber M. Effects of enriched environment on animal models of neurodegenerative diseases and psychiatric disorders. Neurobiol Dis 31, 159–168 (2008). [DOI] [PubMed] [Google Scholar]
- Kovesdi E. et al. The effect of enriched environment on the outcome of traumatic brain injury; a behavioral, proteomics, and histological study. Front Neurosci 5, 42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachat-Kappes R. et al. Effects of enriched environment on COX-2, leptin and eicosanoids in a mouse model of breast cancer. PLoS One 7, e51525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L. & During M. J. What is the brain-cancer connection? Annu Rev Neurosci 35, 331–345 (2012). [DOI] [PubMed] [Google Scholar]
- Rosenzweig M. R., Bennett E. L., Hebert M. & Morimoto H. Social grouping cannot account for cerebral effects of enriched environments. Brain Res 153, 563–576 (1978). [DOI] [PubMed] [Google Scholar]
- Will B., Galani R., Kelche C. & Rosenzweig M. R. Recovery from brain injury in animals: relative efficacy of environmental enrichment, physical exercise or formal training (1990-2002). Prog Neurobiol 72, 167–182 (2004). [DOI] [PubMed] [Google Scholar]
- Mikula M. et al. Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct Integr Genomics 11, 215–224 (2010). [DOI] [PubMed] [Google Scholar]
- Al-Wadei H. A., Al-Wadei M. H., Ullah M. F. & Schuller H. M. Celecoxib and GABA cooperatively prevent the progression of pancreatic cancer in vitro and in xenograft models of stress-free and stress-exposed mice. PLoS One 7, e43376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T. et al. beta2-AR-HIF-1alpha: a novel regulatory axis for stress-induced pancreatic tumor growth and angiogenesis. Curr Mol Med 13, 1023–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C., Ginzberg L. L., O'Keefe J. & Burgess N. Grid cell firing patterns signal environmental novelty by expansion. Proc Natl Acad Sci U S A 109, 17687–17692 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karelina K., Norman G. J., Zhang N. & DeVries A. C. Social contact influences histological and behavioral outcomes following cerebral ischemia. Exp Neurol 220, 276–282 (2009). [DOI] [PubMed] [Google Scholar]
- Fowler C. D., Liu Y., Ouimet C. & Wang Z. The effects of social environment on adult neurogenesis in the female prairie vole. J Neurobiol 51, 115–128 (2002). [DOI] [PubMed] [Google Scholar]
- Silasi G., Hamilton D. A. & Kolb B. Social instability blocks functional restitution following motor cortex stroke in rats. Behav Brain Res 188, 219–226 (2008). [DOI] [PubMed] [Google Scholar]
- van Praag H., Christie B. R., Sejnowski T. J. & Gage F. H. Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc Natl Acad Sci U S A 96, 13427–13431 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lores-Arnaiz S., Lores Arnaiz M. R., Czerniczyniec A., Cuello M. & Bustamante J. Mitochondrial function and nitric oxide production in hippocampus and cerebral cortex of rats exposed to enriched environment. Brain Res 1319, 44–53 (2010). [DOI] [PubMed] [Google Scholar]
- Cao L. et al. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metab 14, 324–338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathania D., Millard M. & Neamati N. Opportunities in discovery and delivery of anticancer drugs targeting mitochondria and cancer cell metabolism. Adv Drug Deliv Rev 61, 1250–1275 (2009). [DOI] [PubMed] [Google Scholar]
- Ward P. S. & Thompson C. B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala S. P., Ko Y. H. & Pedersen P. L. The pivotal roles of mitochondria in cancer: Warburg and beyond and encouraging prospects for effective therapies. Biochim Biophys Acta 1797, 1225–1230 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. P. & Sabatini D. M. Cancer cell metabolism: Warburg and beyond. Cell 134, 703–707 (2008). [DOI] [PubMed] [Google Scholar]
- Schlichtholz B. et al. Enhanced citrate synthase activity in human pancreatic cancer. Pancreas 30, 99–104 (2005). [DOI] [PubMed] [Google Scholar]
- Liu Q. et al. Malate dehydrogenase 2 confers docetaxel resistance via regulations of JNK signaling and oxidative metabolism. Prostate 73, 1028–1037 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun K. H., Kim S. Y., Yoon J. H., Song J. H. & Park W. S. Amplification of the UQCRFS1 Gene in Gastric Cancers. J Gastric Cancer 12, 73–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R. et al. Functional characterization of the 19q12 amplicon in grade III breast cancers. Breast Cancer Res 14, R53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S. J. et al. CA125 and UQCRFS1 FISH studies of ovarian carcinoma. Gynecol Oncol 90, 29–36 (2003). [DOI] [PubMed] [Google Scholar]
- Owens K. M., Kulawiec M., Desouki M. M., Vanniarajan A. & Singh K. K. Impaired OXPHOS complex III in breast cancer. PLoS One 6, e23846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto L. et al. Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha. Mol Cancer 7, 58 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. et al. Global profiling of proteolytically modified proteins in human metastatic hepatocellular carcinoma cell lines reveals CAPN2 centered network. Proteomics 12, 1917–1927 (2012). [DOI] [PubMed] [Google Scholar]
- Geyik E. et al. Investigation of the association between ATP2B4 and ATP5B genes with colorectal cancer. Gene 540, 178–182 (2014). [DOI] [PubMed] [Google Scholar]
- Krupar R. et al. Immunologic and metabolic characteristics of HPV-negative and HPV-positive head and neck squamous cell carcinomas are strikingly different. Virchows Arch 465, 299–312 (2014). [DOI] [PubMed] [Google Scholar]
- Dang C. et al. Identification of dysregulated genes in cutaneous squamous cell carcinoma. Oncol Rep 16, 513–519 (2006). [PubMed] [Google Scholar]
- Viale A. et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun 5, 3295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. et al. Isthmin targets cell-surface GRP78 and triggers apoptosis via induction of mitochondrial dysfunction. Cell Death Differ 21, 797–810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moura M. B., dos Santos L. S. & Van Houten B. Mitochondrial dysfunction in neurodegenerative diseases and cancer. Environ Mol Mutagen 51, 391–405 (2010). [DOI] [PubMed] [Google Scholar]
- Rodriguez J. J., Terzieva S., Olabarria M., Lanza R. G. & Verkhratsky A. Enriched environment and physical activity reverse astrogliodegeneration in the hippocampus of AD transgenic mice. Cell Death Dis 4, e678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg N. R., Haack A. K. & Meshul C. K. Enriched environment promotes similar neuronal and behavioral recovery in a young and aged mouse model of Parkinson's disease. Neuroscience 172, 443–452 (2011). [DOI] [PubMed] [Google Scholar]
- Kim M. P. et al. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat Protoc 4, 1670–1680 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan A. et al. Transgenic brain-derived neurotrophic factor expression causes both anxiogenic and antidepressant effects. Proc Natl Acad Sci U S A 103, 13208–13213 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamec R. & Shallow T. Effects of baseline anxiety on response to kindling of the right medial amygdala. Physiol Behav 70, 67–80 (2000). [DOI] [PubMed] [Google Scholar]
- Quennell J. H. et al. Leptin deficiency and diet-induced obesity reduce hypothalamic kisspeptin expression in mice. Endocrinology 152, 1541–1550 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara H., Toyama K., Yamasaki N. & Miyakawa T. Dissection of hippocampal dentate gyrus from adult mouse. J Vis Exp (33), 1543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. C. et al. Quantitative proteomic analysis identifies CPNE3 as a novel metastasis-promoting gene in NSCLC. J Proteome Res 12, 3423–3433 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Table S1