Abstract
The spore-forming gut bacterium Clostridium difficile is the leading cause of antibiotic-associated diarrhea in hospitalized patients. The major virulence factors are two large glucosylating cytotoxins. Hypervirulent strains (e.g. ribotype 027) with higher morbidity and mortality additionally produce the binary CDT toxin (Clostridium difficile transferase) that ADP-ribosylates actin and induces microtubule-based cell protrusions. Nanobodies are robust single domain antibodies derived from camelid heavy chain antibodies. Here we report the generation of functional nanobodies against the enzymatic CDTa and the heptameric receptor binding subunit CDTb. The nanobodies were obtained from a variable-domain repertoire library isolated from llamas immunized with recombinant CDTa or CDTb. Five CDTa-specific nanobodies blocked CDTa-mediated ADP-ribosylation of actin. Three CDTa-specific and two CDTb-specific nanobodies neutralized the cytotoxicity of CDTa+b. These nanobodies hold promise as new tools for research, diagnosis and therapy of C. difficile associated disease.
Clostridium difficile is a Gram-positive gut bacterium that provokes severe and relapsing diarrhea with lethality rates of up to 20%1,2. C. difficile is the major agent of antibiotic-associated colitis3. Disease severity is determined by the immune status of the patient and by three secretory enzyme toxins that inactivate host cell proteins by posttranslational modification with ADP-ribose (CDT) or glucose (ToxA, ToxB)4,5,6. ToxA and ToxB disrupt the cellular cytoskeleton by glucosylating Rho proteins and are considered the major virulence factors7,8. Hypervirulent strains, including ribotypes 027 and 078 responsible for outbreaks of severe C. difficile associated disease (CDAD), additionally produce C. difficile transferase (CDT)3,9,10,11,12. CDT is a binary toxin that ADP-ribosylates actin4,13. This induces formation of long microtubule-based protrusions in epithelial cells that facilitate adherence of bacteria and increase colonization14,15.
CDT is structurally related to iota toxin and C2 toxins of C. perfringens and C. botulinum4,16. These binary toxins are composed of a secreted actin-ADP-ribosylating enzymatic subunit (CDTa) and a separately secreted binding component (CDTb)17. Upon activation by proteolytic cleavage, CDTb binds to lipolysis-stimulated lipoprotein receptor (LSR), a type 1 membrane protein expressed by gut epithelial cells18. Akin to anthrax lethal toxin, CDTb is thought to form heptamers that interact with the enzymatic subunit to form a larger toxin complex18. Following endocytosis of the toxin complex, the low pH of the endosomal compartment induces a conformational shift of the binding component, which inserts into the membrane and forms a pore allowing translocation of the enzymatic component to the cell cytosol. Here CDTa ADP-ribosylates actin at Arg177, causing disruption of the actin cytoskeleton and formation of microtubule-based protrusions13,14. It has been proposed that CDTa increases pathogenicity of C. difficile by enhancing bacterial adherence to enteric epithelium and colonization of the distal gastrointestinal tract13,14. A landmark clinical trial recently showed clinical benefit of systemically applied humanized monoclonal antibodies directed against ToxA and ToxB, reducing the relapse rate from 25% to 7% of patients19. Comparable reagents for the binary toxin are still missing.
Camelids have evolved antibodies composed only of heavy chains20. The antigen binding paratope of these antibodies is composed of just one variable domain designated VHH21,22. VHHs possess an unusually long complementarity determining region 3 (CDR3) which can form finger-like extensions to penetrate into and block the active site of an enzyme that is usually inaccessible to conventional antibodies23,24. VHHs are easily cloned and expressed as recombinant single domain antibodies, which exhibit a high degree of chemical and thermal stability. Recombinant VHHs are also known as nanobodies because of their dimensions in the nanometer range (2 nm × 3 nm)21,22. These nanobodies are highly soluble, robust, and are easily converted into multivalent and/or multispecific formats25. As monomers or chains of linked monomers, nanobodies can neutralize their target antigens by binding only. In these formats, nanobodies lack Fc-mediated effector functions, such as complement activation or Fc-receptor mediated phagocytic clearance. The latter, however, can easily be supplied by genetic fusion of nanobodies to the Fc domains of conventional antibodies, thereby converting nanobodies into a format resembling the original heavy chain antibody21. Nanobodies have potential applications in various disease areas, including infectious and inflammatory diseases21,22. Hussack et al. recently reported the selection of nanobodies directed against the repetitive receptor binding domains (RBD) of ToxA and ToxB26. Some of the ToxA-specific nanobodies neutralized the cytotoxic activities of ToxA in cell culture models when added in 100–1.000 fold molar excess. However, none of the ToxB-specific nanobodies was capable of neutralizing ToxB-mediated cytotoxicity in cell culture models.
The goal of this study was to produce and characterize nanobodies directed against the enzymatic and binding components of CDT. We successfully selected several functional nanobodies against both of these subunits from phage display libraries generated from immunized llamas. Two of these nanobodies - one directed against CDTa (l+8), the other directed against CDTb (l-15.1g) - effectively blocked cytotoxicity of the binary toxin at equimolar concentrations. These nanobodies hold promise as new tools for research, diagnostic and therapy of C. difficile associated disease.
Results
Llama immunizations and selection of CDT-specific nanobodies
Three llamas were immunized with recombinant CDTa or CDTb (Fig. 1). ELISA analyses confirmed a robust heavy chain antibody response (Supplementary Fig. S1). VHH-Phage display libraries were generated by PCR-amplification of the VHH-repertoire from blood lymphocytes obtained 10–20 days after the last boost immunization and cloning into the pHEN2 vector (Supplementary Table S1). CDT-specific VHHs were selected by sequential panning of the phage libraries on immobilized CDTa or CDTb. After each round of selection, 24 clones were sequenced. The results revealed selection of clones derived from several distinct VHH families (Table 1), with CDR3 lengths ranging from 3 to 21 amino acid residues.
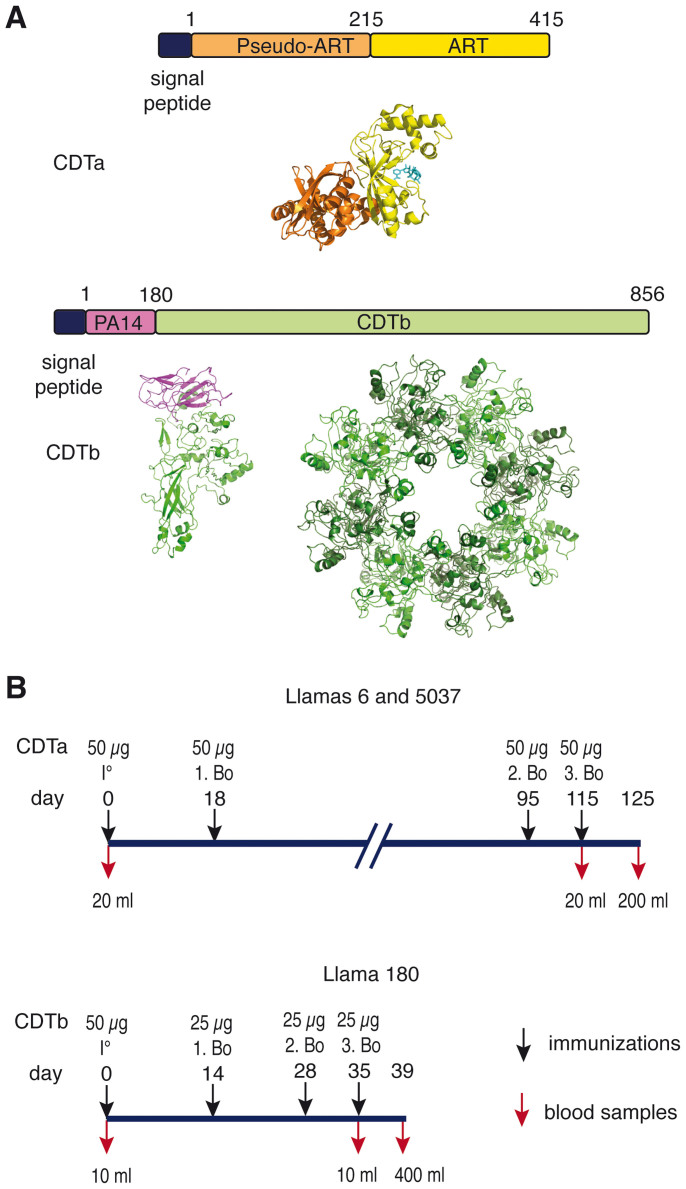
Figure 1. Immunization scheme of llamas with CDTa and CDTb.
(A) Schematic diagram of the toxin subunits used for immunization. CDTa and CDTb are independently produced as secreted proteins with an N-terminal signal peptide that is cleaved during secretion. CDTa consists of two tandem ADP-ribosyltransferase (ART) domains. The N-terminal ART domain does not possess enzymatic activity and mediates binding to CDTb; the C-terminal ART domain mediates ADP-ribosylation of actin at Arg 177 and contains a deep NAD+-binding cleft. Proteolytic cleavage of its N-terminal activation domain (PA14) allows CDTb to form oligomers that bind CDTa and assists its delivery to the host cell cytosol. 3D models were generated with PyMol using coordinates of the pdb file 2wn7 for CDTa with NAD+ (cyan). CDTb was modeled onto the 3D structure of iota toxin (pdb code 2j42) using SwissModel and assembled into the multimer by alignment with anthrax toxin (pdb code 3hvd) using PyMol. (B) Three llamas (Lama glama) were immunized subcutaneously with purified recombinant CDTa or CDTb emulsified with Specol adjuvant, boosted and bled as indicated in the time scale.
Table 1. Characteristics of CDTa-specific and CDTb-specific families of nanobodies.
| specificity | llama | family | isolates | variants | CDR3 | affinity (Kd) | epitope | IC50 tox | IC50 ART | |
|---|---|---|---|---|---|---|---|---|---|---|
| CDTa | 6 | l+8 | 2 | 1 | ECGGYGAH | 1 nM | 1 | 2 nM | 2 nM | |
| 6 | l-14 | 2 | 1 | TYRPNTFTPAEYDY | 1.5 nM | 2 | > 1 µM | 4 nM | ||
| 6 | l-15 | 4 | 2 | GGFTEAYSGTYYPDS | 1.5 nM | 3 | > 1 µM | 4 nM | ||
| 6 | l+18 | 8 | 5 | GGSGYGCYAFTPAYGMDY | 1.5 nM | 2,3 | 50 nM | 4 nM | ||
| 5037 | s-21 | 18 | 4 | ADQRVGYIEYYSGSSGKEYDY | 3 nM | 2,3 | 50 nM | 4 nM | ||
| CDTb | 180 | l-3 | 13 | 9 | RGY | > 50 nM | 1 | > 1 µM | - | |
| 180 | l-15.1 | 13 | 10 | KVGANILTRSIGYKY | 0.5 nM | 1 | 4 nM | - | ||
| 180 | l-15.3 | 1 | 1 | TNKBYTDGRLEEYDY | 9 nM | 2 | > 1 µM | - | ||
| 180 | l-19 | 1 | 1 | DGRSNLRANYDSADYGMAY | 15 nM | 3 | > 1 µM | - | ||
Family names indicate the presence of a short (s) or long hinge (l), the absence (-) or presence (+) of a disulfide bond connecting CDR2 and CDR3, and the length of the CDR3 in numbers of amino acid residues. Isolates indicates the number of clones selected per family, variants indicates the number of clones carrying distinct but evidently related amino acid sequences. Variant amino acid positions in the CDR3 are indicated in red. Epitopes are numbered arbitrarily (nanobodies that block the binding of one another are considered to recognize the same or overlapping epitopes). “IC50 tox” indicates the nanobody concentration required to inhibit 50% rounding of HT29 cells upon incubation with 2 nM CDTa+b for 4 h at 37°C. “IC50 ART” indicates the nanobody concentration required to inhibit ADP-ribosylation of actin by 2 nM CDTa by 50%.
Production and reformatting of nanobodies
Recombinant nanobodies carrying a C-terminal chimeric His6x c-myc tag were expressed as periplasmic secretory proteins in E. coli HB2151 cells. SDS-PAGE analyses of periplasmic lysates obtained 3–4 h after induction of transcription with IPTG usually revealed prominent bands at 14–20 kDa corresponding to recombinant nanobodies (Fig. 2). Nanobodies were readily purified from E. coli periplasmic lysates by immobilized metal affinity chromatography (IMAC). ELISA analyses with crude periplasmic lysates on immobilized CDTa or CDTb using peroxidase-conjugated anti-c-myc antibodies confirmed the specificity of five CDTa-selected nanobody families for CDTa (Fig. 2) and of four CDTb-selected nanobody families for CDTb (Supplementary Fig. S2). Note that production levels of nanobodies in the E. coli periplasm varied considerably, even among members of the same nanobody family (Fig. 2). Moreover, nanobodies in periplasma lysates often yielded double bands in SDS-PAGE analyses (Fig. 2, arrows). Only the upper band could be detected with c-myc-specific antibodies in Western blot analyses and only the upper band could be purified by IMAC (not shown), indicating that the lower band lacked the C-terminal tags, most likely as a consequence of proteolytic cleavage. Further, nanobodies purified from E. coli periplasm still contained substantial levels of endotoxin (lipopolysaccharide), a potential concern for use of nanobodies in subsequent cellular assays.
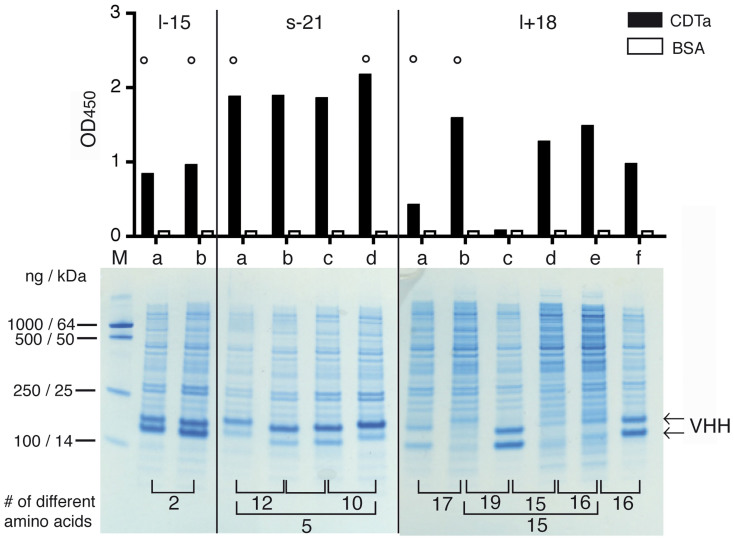
Figure 2. Production of monovalent nanobodies in the E. coli periplasm and verification of binding specificities.
E. coli HB2151 cells were transformed with individual VHH-encoding pHEN2 vectors and protein expression was induced with IPTG for 3 h. Periplasmic lysates were prepared by osmotic shock, clarified by centrifugation, and analyzed by SDS-PAGE and Coomassie staining (bottom). Binding specificity was analyzed by ELISA using CDTa or BSA as a control protein (top). Representative results are shown for three families of CDTa-specific nanobodies. Results for CDTb-specific families are shown in Supplementary Fig. S2. Family names indicate the presence of a long or short hinge, the absence or presence of an additional cysteine pair in CDRs 2 and 3, and the number of amino acid residues in the CDR3 (see Table I). Family members with distinct amino acid sequences were designated a, b, c, d etc. in order of decreasing number of isolates. Numbers on the bottom indicate the number of different amino acids between two family members. Nanobodies marked by a circle were selected for subcloning into a eucaryotic expression vector.
In order to avoid these problems of inconsistent production levels, proteolytic degradation of C-terminal tags, and contamination by endotoxin, we subcloned the nanobody coding region into a eucaryotic expression vector (pCSE2.5) that had been optimized for secretory protein production in suspension cultures of HEK-6E cells in serum free medium27,28. SDS-PAGE analyses of crude HEK cell culture supernatants harvested 6 d after transfection, indeed, revealed consistently high yield production levels of nanobodies in both, monovalent and bivalent formats (Fig. 3). Qualitative comparisons of nanobody affinities were performed by ELISA using serial dilutions of nanobodies. The results revealed nanobody affinities for CDTa (Fig. 3) and CDTb (Supplementary Fig. S3) ranging from 0.5 nM to 15 nM, i.e. affinities comparable to those reported for nanobodies selected from immunized llamas for other target antigens21,24,29.
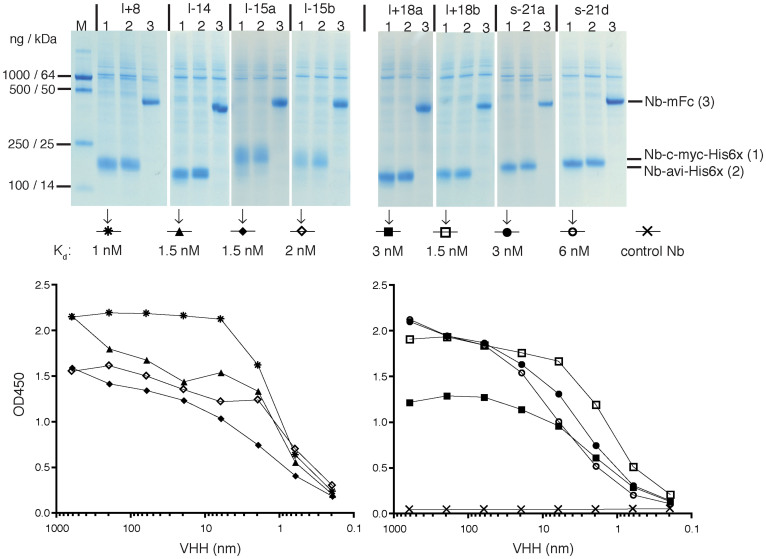
Figure 3. Production of reformatted nanobodies in transfected HEK-6E cells and comparative analyses of binding affinities.
HEK-6E cells were transfected with cDNA-expression constructs encoding nanobodies fused C-terminally to chimeric tags (c-myc-His6x or avi-His6x) or to the Fc domain of mouse IgG2c (mFc). Transfected cells were cultured in serum free medium for 6 d. Cell supernatants were clarified by centrifugation and analyzed by SDS-PAGE and Coomassie staining (top). Qualitative comparison of nanobody affinities was performed by ELISA using serial dilutions of monovalent nanobodies and wells coated with CDTa. After 20 minutes of incubation, wells were washed three times to remove unbound proteins and bound nanobodies were detected with a peroxidase-conjugated monoclonal antibody directed against the c-myc tag. A c-myc tagged nanobody directed against ARTC229 was used as negative control. Symbols used for the respective nanobodies and the calculated dissociation constants are indicated in the middle. Representative results are shown for five families of CDTa-specific nanobodies. Results for CDTb-specific families are shown in Supplementary Fig. S3.
Mapping of binding epitopes
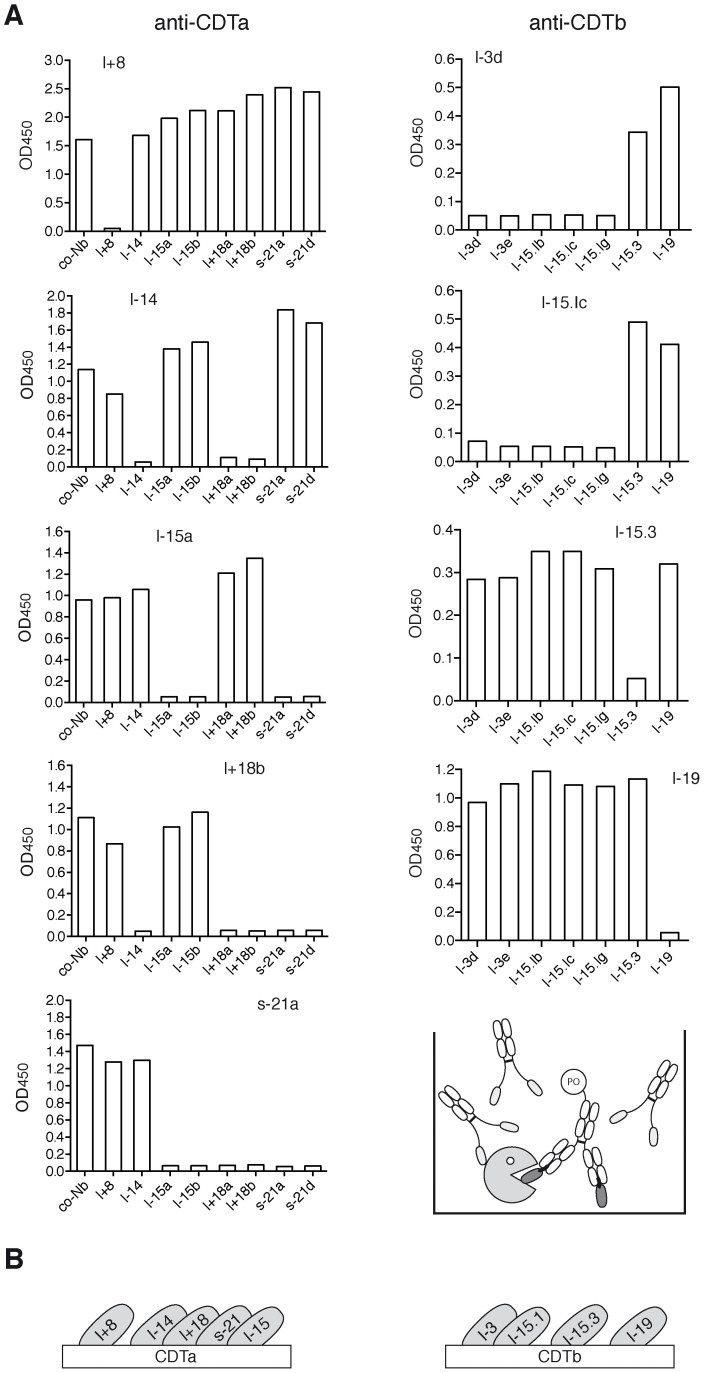
In order to assess whether the selected nanobodies recognize overlapping or distinct epitopes on CDTa or CDTb, binding analyses were performed in the presence of excess unlabeled nanobodies carrying an epitope tag distinct from that used for detection of the binding nanobody (Fig. 4). The results permit grouping of nanobodies according to their relative epitope specificities (Table 1). Nanobody l+8 bound CDTa independently of the other four nanobodies, while the latter showed partially overlapping epitopes: l+18 blocked binding of l-14 and s-21, but did not block binding of l-15; whereas s-21 blocked binding of l+18 and l-15, but not l-14. Nanobodies l-15.3 and l-19 bound CDTb independently of each other and of the other two nanobodies. Nanobody families l-3 and l-15.3 blocked one another. In case of CDTa, for which a 3D-structure is available (pdb code 2wn7), molecular modeling analyses yielded plausible binding modes for nanobodies l+8, l-14, and l+18 at the active site of CDTa (Supplementary Fig. S4).
Figure 4. Mapping of the epitope specificity of nanobodies by cross-blocking analyses.
For analysis of epitope specificities, selected nanobodies were subjected to competitive binding analyses (A). Antigens were immobilized on 96-well plates and pre-incubated with nanobody-Fc fusion proteins (2 µg/well) for 60 min at RT before addition of c-myc tagged nanobodies precomplexed with a peroxidase-conjugated anti-c-myc mAb (50 ng nanobody/well). The schematic diagram illustrates the experimental set-up, PO: peroxidase. (B) Schematic diagrams illustrating the relative localization of the binding epitopes of the analyzed nanobodies.
Five of five CDTa-specific nanobodies inhibit the enzymatic activity of CDTa
In order to assess whether the CDTa-selected nanobodies can interfere with the enzymatic activity of CDTa, we analyzed CDTa-catalyzed ADP-ribosylation of actin in HEK cell lysates in the presence of excess nanobodies (Fig. 5). The results show that all five CDTa-specific nanobody families completely blocked ADP-ribosylation of actin catalyzed by CDTa. Titration analyses indicate that the most potent nanobodies l+8, l-14 and l+18e blocked ADP-ribosylation at equimolar ratios with CDTa.
Figure 5. CDTa-specific nanobodies inhibit the enzymatic activity of CDTa.
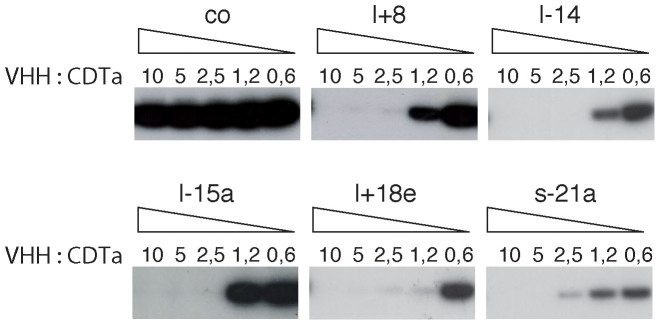
HEK cells were lysed with Triton-X-100 and cell lysates were clarified by centrifugation. Before addition to the lysates, CDTa was preincubated for 20 min with serial dilutions of CDTa-specific or control (co) nanobodies at the indicated molar ratios of VHH:CDTa. 32P-NAD+ was added as a substrate for CDTa-catalyzed ADP-ribosylation of actin. Reactions were stopped after 15 min by addition of SDS-PAGE sample buffer. Proteins were size fractionated by SDS-PAGE and covalently incorporated radioactivity was detected by exposing the dried gel to an X-ray film.
CDTa-specific nanobody l+8 and CDTb-specific nanobody l-15.1g effectively inhibit CDT-induced cytotoxicity
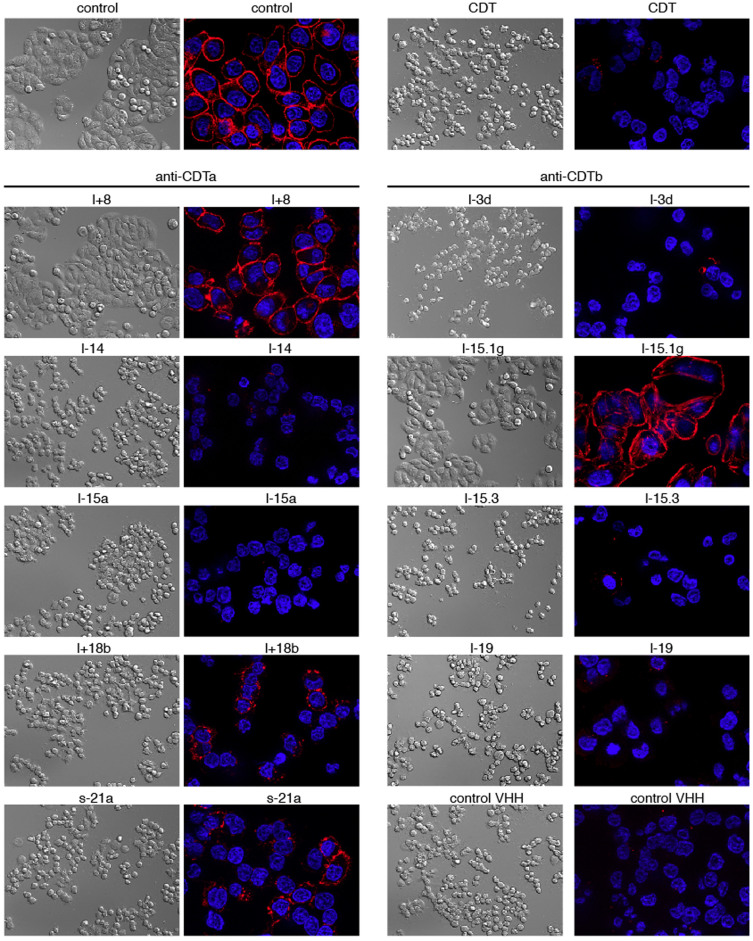
Treatment of cells expressing LSR, the CDT receptor, with a mixture of recombinant CDTa and CDTb results in profound cytotoxicity as evidenced by rounding and detachment of cells18 (Supplementary Fig. S5). In order to assess whether any of the selected nanobodies can interfere with CDT-mediated cytotoxicity, we assessed the integrity of HT29 cells by microscopy following incubation with CDTa + CDTb for 4 h at 37°C in the absence or presence of nanobodies in 16 fold molar excess (Fig. 6). Cells were fixed and counterstained with fluorescently labeled Phalloidin which binds to filamentous actin, but not to monomeric actin. The results show that the CDTa-specific nanobody l+8 and CDTb-specific nanobody l-15.1g effectively protected HT29 cells from the cytotoxic effects of CDTa + CDTb, nanobodies l-15.1b, l+18b and s-21a showed partial protection, and the other nanobodies did not show any detectable effect on CDTa+b mediated disruption of filamentous actin (Fig. 6). Titration analyses revealed effective neutralization by l+8 and by l-15.1g at equimolar concentrations with CDTa+b, while neutralization with l+18b and l-15.1b required higher concentrations of nanobodies (Supplementary Fig. S6). The results of radio-ADP-ribosylation analyses confirm that nanobodies l+8, l+18b and l-15.1g effectively prevent and that nanobody l-15.1b partially prevents ADP-ribosylation of actin in response to treatment of HT29 cells with CDTa+b (Supplementary Fig. S7).
Figure 6. CDTa-specific nanobody l+8 and CDTb-specific nanobody l-15.1g inhibit CDTa+b induced cytotoxicity.
Before addition to cultures of adherent HT29 cells, CDTa and CDTb were preincubated with a 16 fold excess of CDTa- or CDTb-specific nanobodies. Four hours after addition of toxins, cells were fixed and cellular morphology was assessed by differential interference microscopy. The cellular cytoskeleton was stained with the F-actin-staining dye phalloidin-rhodamine, cell nuclei were stained with the DNA staining dye Hoechst 33342. Apotome-images were captured with a Zeiss Axiovert 200M microscope. Control cells (top) were incubated for four hours in the absence or presence of CDTa+b.
CDTa-specific nanobody l+8 inhibits CDT-induced disruption of transepithelial resistance
The transepithelial electrical resistance (TEER) is a sensitive measure of the integrity of a monolayer of epithelial cells. Disruption of the cell cytoskeleton can be detected by a loss of TEER. In order to assess whether the CDTa-specific nanobody l+8 could also inhibit CDTa+b induced breakdown of TEER, we measured TEER in established monolayers of MDCK C7 cells incubated with CDTa + CDTb in the presence of l+8 or a control nanobody. The results show that l+8 effectively inhibits CDTa-induced loss of TEER (Supplementary Fig. S8).
Discussion
The goal of this study was to produce functional nanobodies directed against the enzymatic and binding components of CDT. We successfully selected several functional nanobodies against both of these subunits from phage display libraries generated from immunized llamas.
It has been reported that nanobodies derived from immunized llamas display a propensity to bind crevices on protein surfaces, e.g. the active site crevice of enzymes23,24. In most cases reported to date where llamas have been immunized with an enzyme, a substantial fraction of the selected nanobodies functionally block the enzymatic activity of their target21. Our results with the CDTa-selected nanobodies are in accord with these previous reports.
Co-crystallization studies have revealed that the exceptionally long CDR3 of nanobodies can form finger-like protrusions that extend into and block the active site24,30. The crystal structure of CDTa, indeed, contains a deep NAD+-binding pocket31. It is, thus, conceivable that some of the CDTa-specific nanobodies reported here extend their CDR3 (containing 8, 14, 15, 18 and 21 amino acid residues, Table 1) into the deep NAD+-binding cleft of CDTa. Although co-crystal structures of actin in complex with CDTa are not yet available, the 3D structure of actin in complex with the enzymatic component of the related iota toxin of C. perfringens revealed an extended contact area between actin and iota toxin adjacent to the NAD+-binding crevice32. It is, thus, also possible that some of the selected nanobodies sterically block ADP-ribosylation of actin without directly blocking the NAD+-binding site. Molecular modeling analyses for the binding epitopes of nanobodies l+8, l-14, and l+18, indeed yielded plausible binding sites consistent with the experimental data (Supplementary Fig. S4). The predicted binding sites indicate that these three nanobodies interfere with both, binding of NAD+ and actin. These models can be evaluated by site-directed mutagenesis of CDTa, and ultimately, by co-crystallization of the nanobodies and CDTa.
Blocking the binding of a toxin to its host cell receptor is generally considered the most suitable strategy to neutralize toxins with antibodies or nanobodies17,26. Hussack et al. recently generated nanobodies directed against the receptor binding domain of C. difficile ToxA and ToxB, each of which are composed of multiple repetitive carbohydrate binding domains at their C-termini26. When added in 3000 fold molar excess, six of seven selected nanobodies inhibited the cytotoxicity of ToxA whereas none of seven selected nanobodies could block the cytotoxicity of ToxB. Neutralization of ToxA at lower nanobody concentrations was enhanced significantly when nanobodies recognizing independent epitopes of ToxA where added in combination rather than individually. In our study, we observed effective neutralization of CDTa+b for two of our nanobodies (l+8 and l-15.1g) even at equimolar concentrations (Supplementary Fig. S6). The more effective neutralization of our nanobodies may reflect structural differences of the binary toxin and the large cytotoxins33. While the receptor binding domain of the large holotoxins contains multiple repetitive lectin-like carbohydrate-binding modules, CDTb, in contrast, does not contain any repetitive binding modules and presumably binds its receptor LSR via a unique site18. CDTb is thought to form heptamers that are stabilized by binding to LSR34. The CDTb-specific nanobody thus could neutralize the cytotoxicity either by steric interference with the assembly of CDTb into heptamers or by interfering with binding of CDTb to CDTa or to LSR. In either case, high affinity binding of a nanobody might be effective even at equimolar concentrations.
Surprisingly, three of five nanobodies selected for binding to CDTa also neutralized cytotoxicity of the binary toxin (Fig. 6). All of these nanobodies also effectively blocked ADP-ribosylation of actin catalyzed by monomeric CDTa (Fig. 5). Interaction of CDTa with CDTb is thought to be mediated via the N-terminal pseudo-ART domain, while the C-terminal ART domain catalyzes ADP-ribosylation of actin31. Since all five nanobodies block ADP-ribosylation of actin, it is unlikely that these nanobodies sterically interfere with binding of CDTa to CDTb. Rather, it is more likely that these nanobodies bind to the catalytic domain and then interfere with translocation of CDTa to the cytosol.
Although the precise translocation mechanism is not yet known, it is thought to require unfolding of the enzymatic domain, translocation of the unfolded polypeptide chain through a narrow channel and subsequent refolding of the translocated chain into an active enzyme in the cytosol35,36. Nanobodies contain a canonical disulfide bond connecting the two ß-chains of the immunoglobulin domain21. Similar to some other llama derived nanobodies, CDTa-specific nanobody l+8 contains an additional pair of cysteine residues that likely form a disulfide bridge connecting the CDR1 and CDR3. Co-translocation of the nanobody to the cytosol, presumably would require unfolding and reduction of these disulfide bonds. Forced expression of nanobodies in the cytosol has been achieved in numerous cases21,22. Indeed, fusion of nanobodies directed against cytosolic or nuclear target proteins to fluorescent proteins indicate that nanobodies fold correctly in the reducing environment of the cytosol and bind their target with high specificity and affinity37,38.
In this context it is important to note that nanobodies directed against the receptor binding domain of a toxin, in general would be ineffective once the cytotoxic enzymatic domain has entered the cell and dissociated from the receptor binding domain. In contrast, nanobodies directed against the enzymatic domain could still neutralize cytotoxicity after entry, provided that the nanobodies either are expressed by the cell itself or are efficiently translocated to the cytosol as effectively as the enzymatic component39.
Expression by the host cell could be achieved by delivery of nanobody-encoding cDNA either by transfection or infection with recombinant viruses40,41. Translocation to the cytosol could be achieved by fusion to translocation domains, e.g. the pseudo-ART domain of CDTa42 or by cationic transfection polymers43.
The role of the binary CDT toxin for pathogenicity of C. difficile strains is still a controversial issue12,34,44. Many of the clinically most aggressive strains of C. difficile express the binary toxin in addition to one or both of the large cytotoxins ToxA and ToxB45. Moreover, recent studies suggest that CDT may increase pathogenicity by enhancing adherence of bacteria to epithelial cells that form CDTa-induced protrusions14,15. Another study showed a high correlation between detections of the CDTa gene and a high recurrence rate of C. difficile colitis46. However, good diagnostic tools to assess the presence of CDT in stool samples are still lacking. The nanobodies described here provide a promising basis for the development of more sensitive diagnostic tools, e.g. by use of immobilized nanobodies to enrich toxins from stool samples or culture supernatants. These nanobodies may also provide useful experimental tools to assess the relative role of CDTa vs. the two large glucosylating cytotoxins in experimental models of CDAD.
The standard therapy for C. difficile infection is treatment with metronidazole, vancomycin, or fidaxomicin. 90% of all treated patients respond to this therapy, but of these patients 15–30% suffer from relapsing infection3,6,47. Therefore, there is a need for the development of alternative therapeutic options to treat C. difficile infections48. Recent studies have provided promising results for two different treatment strategies: systemic treatment with toxin-neutralizing antibodies and fecal microbiome transplantation19,49,50. The nanobodies reported here may provide a complementary approach.
A recent clinical study revealed the therapeutic potential of passive immunotherapy with neutralizing antibodies19: 101 patients received a single infusion of a ToxA-neutralizing monoclonal antibody in combination with a ToxB-neutralizing monoclonal antibody. This reduced the recurrence rate to 7% in the antibody group vs. 25% in the placebo group19. Similarly, the systemic administration of the neutralizing CDT-specific nanobodies described here may have therapeutic benefit. In comparison to conventional monoclonal antibodies, nanobodies provide several advantages, including a lower cost of production and more effective tissue penetration21,22. As monomers or dimers, however, nanobodies have a short half life and are rapidly eliminated from circulation by renal filtration. Two strategies have been applied successfully to increase the in vivo half life of nanobodies: 1) genetic fusion to an albumin-specific nanobody and 2) genetic fusion to the Fc domain of a conventional IgG antibody21,25. Both strategies utilize the recycling function of the neonatal Fc receptor which binds independently to albumin and to IgG antibodies, protecting both from intracellular degradation51. Fusion to the Fc domain also reconstitutes the phagocytic clearance of the Ag-Ab complexes via Fcγ-receptors52. Thus, the modular nature of nanobodies allows tailoring of nanobodies to specific needs, e.g. by fusion to other nanobodies or to the Fc-domains of conventional antibodies.
For therapeutic purposes, nanobodies can, in principle, be administered systemically and locally. In recent clinical trials, llama-derived nanobodies have been administered systemically by intravenous or subcutaneous injections to more than 700 humans, without any adverse side effects53. Nanobodies could also be administered topically to the gastrointestinal tract, e.g. by oral or rectal administration. Application via the oral route may require encapsulation to avoid excessive damage to nanobodies by low gastric pH and pancreatic proteases. Various encapsulation strategies have been established for the oral administration of therapeutic proteins43,54. Moreover, using site directed mutagenesis it has been possible to select nanobodies resistant to low pH and pancreatic proteases55. Expression of nanobodies as cell surface or secretory proteins by lactobaccili is another feasible option56,57. Finally, rectal administration, e.g. as a supplement to fecal microbiome transplantation, may also be possible50. To this end, immobilization of nanobodies on beads may aid absorption and rectal elimination of soluble toxins from the gastrointestinal tract.
In conclusion, the nanobodies described here provide a promising basis for the development of more efficient diagnostic and therapeutics tools for C. difficile mediated diseases.
Methods
Protein production and llama immunizations
The enzymatic (CDTa) and binding components (CDTb) of CDT (Fig. 1) were produced separately in E. coli or B. megaterium and were purified as previously described14,15. The binding component was activated by treatment with trypsin58. Three llamas (Lama glama) were immunized subcutaneously with purified recombinant proteins emulsified with Specol adjuvant29,39,59. The humoral immune response was monitored in serially diluted serum by ELISA on microtiter plates (Nunc MaxiSorp, Thermo Fisher Scientific, Waltham, MA) coated with recombinant CDTa or CDTb (100 ng/well). Bound antibodies were detected using mouse monoclonal antibodies directed against llama IgG2 (clone 198D) and IgG3 (clone 8E1) kindly provided by Dr. Judith Appelton, Cornell University, NY60 and peroxidase-conjugated goat anti-mouse IgG (GE Healthcare, Chalfont St Giles, UK). Animals were bled 10–14 d after the 3rd or 4th boost.
Construction of phage display libraries and selection of CDT-specific VHHs
Mononuclear cells were isolated from 120 ml of blood by Ficoll-PaqueTM (GE Healthcare, Chalfont St Giles, UK) gradient centrifugation. RNA purified from these cells by TRIZOL reagent (Invitrogen, Carlsbad, CA) was subjected to cDNA synthesis with random hexamer primers. The VHH coding region was amplified by PCR with degenerate VHH-specific primers SHFmu: tcgcggccca gccggccatg gcgcaggtsm arctgcagga gtcwgg, SHRmu: atggtgatga tgattgtgcg gccgcgctgg ggtcttcgct gtggtgcg, LHFmu: tcgcggccca gccggccatg gccgatgtgc agctgcaggm gtcwggrgga gg, LHRmu: atggtgatga tgatgtgcgg ccgctggttg tggttttggt gtcttggg. PCR products were purified from agarose gels, digested sequentially with SfiI and NotI (NEB, Ipswich, MA) and cloned into the pHEN2 phagemid vector downstream of the PelB-leader peptide and upstream of the chimeric His6x-Myc epitope tag39,61. Transformation into XL1-Blue E. coli (Stratagene, La Jolla, CA) yielded libraries with sizes of 4.0 × 105–107 clones. Phage particles were precipitated with polyethylene glycol from culture supernatants of transformants infected with a 10-fold excess of M13K07 helper phage (GE Healthcare, Chalfont St Giles, UK). Panning of specific phage was performed using either toxins immobilized on microtiter plates (Nunc MaxiSorp, Thermo Fisher Scientific, Waltham, MA) or in solution with biotinylated toxins and subsequent capture on streptavidin-coated magnetic beads (Life Technologies, Carlsbad, CA). Phage particles (1.6 × 1014) incubated with toxins for 60 min with agitation at room temperature in PBS, 10% Carnation non fat dry milk powder (Nestlé, Glendale, CA). Following extensive washing, bound phages were eluted with 50 mM diethylamine and neutralized with 1 M Tris-HCl pH 8. Eluted phages were titrated and subjected to two more rounds of panning, following the same procedure. Phage titers were determined at all steps by infection of TG1 E. coli cells (Stratagene, La Jolla, CA). Plasmid DNA was isolated from single colonies and subjected to sequence analyses using pHEN2-specific forward and reverse primers.
Production of recombinant proteins and reformatting of nanobodies
Monomeric nanobodies were expressed in HB2151 E. coli cells (GE Healthcare, Chalfont St Giles, UK). Protein expression was induced with IPTG (Roche, Rotkreuz, Switzerland) when bacterial cultures had reached an OD600 of 0.5 and cells were harvested after further cultivation for 3–4 h at 37°C. Periplasmic lysates were generated by osmotic shock and removal of bacterial debris by high speed centrifugation. The coding region of selected nanobodies was subcloned using NcoI and NotI into the pCSE2.5 vector27 (kindly provided by Thomas Schirrmann, Braunschweig) so as to fuse C-terminal tags, including His6x-c-myc, avi-His6x, or the CH2 and CH3 domains of mouse IgG2c. Monomeric nanobodies and nanobody-Fc fusion proteins were expressed in transiently transfected HEK-6E cells. Six days post transfection, supernatants were harvested and cleared by centrifugation. Nanobodies were purified by immobilized metal affinity chromatography using Ni-NTA agarose (Sigma, St Louis, MO), nanobody-Fc fusion proteins were purified by affinity chromatography on protein G-sepharose (GE-Healthcare, Chalfont St Giles, UK).
ELISA
CDTa or CDTb (100 ng/100 µl PBS/well) were adsorbed to 96-well Nunc MaxiSorp plates (Thermo Fisher Scientific, Waltham, MA) at 4°C over night. Wells were washed twice with PBS and blocked for 2 hours with PBS containing 5% nonfat powdered milk at room temperature. Wells were incubated for 30 min with nanobody-containing periplasma lysates (diluted 1:10 in PBS). Following washing with PBS/0.05% Tween 20, bound nanobodies were detected with peroxidase-conjugated anti-c-Myc mAb 9E10 (Sigma, St Louis, MO).
Alternatively, wells were incubated with HEK-cell supernatants containing VHH-Fc fusion proteins and bound antibodies were detected with peroxidase conjugated anti-mouse IgG (GE Healthcare, Chalfont St Giles, UK). In some experiments monovalent nanobodies (10 ng) were dimerized by preincubation for 30 min with peroxidase-conjugated anti-c-Myc mAb 9E10 (200 ng). Bound peroxidase was visualized using 3,3'′,5,5'′-Tetramethylbenzidine (TMB) (Sigma, St Louis, MO) as substrate. For affinity analyses, wells were incubated for 60 min with serial dilutions of monovalent nanobodies in PBS containing 10 µg/ml BSA. Wells were washed three times with PBS/0.05% Tween 20. Bound antibodies were detected with peroxidase-conjugated anti-c-Myc mAb 9E10 and TMB as substrate. For epitope analyses, wells were preincubated with excess monovalent nanobodies or nanobody-Fc fusion proteins (500 ng/100 µl PBS, 1% BSA) for 30 min at RT before addition of preconjugated VHH-anti-c-myc and further incubation for 20 min at RT. The absorbance at 450 nm was measured using a Victor3 ELISA-reader (Perkin-Elmer, Waltham, MA).
ADP-ribosylation assays
To monitor ADP-ribosylation of actin, HEK cells were lysed for 30 min at 4°C in PBS containing 0.5% Triton-X-100 (107 cells/ml) and cell lysates were clarified by high speed centrifugation (15 min 13.000 g). CDTa (0.2 µM) was preincubated with serial dilutions of nanobodies (2 µM–0.12 µM) in PBS containing 10 µg/ml BSA for 20 min at RT. HEK cell lysates (50 µl) were incubated with toxin/nanobody (50 µl) in the presence 32P-NAD+ for 10 minutes at 37°C. The reaction was stopped by addition of SDS-PAGE sample buffer (NuPAGE®, Invitrogen, Carlsbad, CA). Proteins were size-fractionated on precast NuPAGE® gels by SDS-PAGE and stained with Coomassie Blue (Invitrogen, Carlsbad, CA). Radiolabeled proteins were detected by autoradiography for 15 h at −80°C using X-ray film (GE Healthcare, Chalfont St Giles, UK).
Cytotoxicity assays
Cytotoxicity assays were carried out by microscopic analyses of the cellular morphology of subconfluent HT29 human colon carcinoma cells grown on glass-bottom 96 well microtiter plates (ibidi, Martinsried, Germany). CDTa (2.4 ng/100 µl) and protease activated CDTb (5 ng/100 µl) were preincubated for 30 min at 37°C with serial dilutions of nanobodies before addition to HT29 cells. Cells were incubated for 4 h at 37°C, gently washed, and fixed in 4% paraformaldehyde. Fixed cells were counterstained with rhodamine-phalloidin and Hoechst 33342 (Molecular Probes, Life Technologies, Carlsbad, CA), and analyzed by microscopy with a Zeiss Axiovert 200M microscope equipped with digital interference contrast and an apotome (Zeiss, Oberkochen, Germany).
Additional cytotoxicity assays were carried out by monitoring the transepithelial resistance (TEER) of confluent monolayers of MDCK C7 cells62 grown on 0.4 µM filter-plates with a Volt Ohm meter (Merck Millipore, Billerica, MA, USA). Before addition to the culture medium, CDTa (10 nM) and CDTb (20 nM) were preincubated with excess nanobodies (200 nM) for 30 min at 37°C. TEER values were determined every hour (n = 3) and background values (medium alone) were substracted.
Molecular modeling
Molecular modeling of the binding epitopes of three VHHs to CDTa (pdb code 2wn7) were performed using BioLuminate (version 1.7, Schrodinger, New York, NY)63. Homology models were built with the antibody prediction module in BioLuminate using framework templates obtained from the curated antibody database. CDR loops were grafted from the database based on a combination of structure clustering, sequence similarity and stem residue geometry matching. BioLuminate automatically preserved the extra disulfide bond in nanobodies l+8 and l+18 that connects CDR2 and CDR3 if the template also had one in the aligned position. In other cases, the formation of the disulfide bond was forced manually before de novo loop prediction. Four models were built for l+8, six models for l-14, and five models for l+18. All of these models were used for docking to CDTa using Piper64. Prior to docking, NAD+ was removed from CDTa using the Protein Preparation Wizard65. An attractive potential (scaling the vdW attractive term by 1.1) was applied to the NAD+-binding site, which helped to restrain the sampling to this site. 70000 rotations were sampled and the top 30 poses were returned from each docking job. The following criteria were used to prune the docking poses: (a) docking poses with low surface complementarity (<0.3) were discarded; (b) VHHs binding with non-CDR regions were discarded; (c) l-14 and l+18 bind partially overlapping epitopes; (d) l+8 does not block or is not blocked by l-14 or l+18; (e) all VHHs block binding of NAD+ and/or actin. The most plausible docking poses that passed the above criteria were: the #16 pose from the 2nd homology model of l+8, the #4 pose from the 1st homology model of l-14, and the #1 pose from the 2nd homology model of l+18 (Supplementary Figure S4). The templates used for building these models were: (1) l+8: framework – 4JVP, CDR1 - 4KDT, CDR2 – 4JVP, CDR3 – 1MAM; (2) l-14: framework – 3STB, CDR1 – 3VG9, CDR2 – 2XA3, CDR3 – 2AJ3; (3) l+18: framework – 3STB, CDR1 – 1NFD, CDR2 – 4NBZ, CDR3 – de novo.
Author Contributions
M.U., A.M.E. and L.S. contributed equally. M.U., A.M.E., L.S., M.S., S.M., C.S., V.A., V.Z. and K.Z. performed experiments and analyzed results. M.S., J.B., H.R., F.H., H.-W.M., F.G., K.A. and F.K.-N. designed research and analyzed data. M.U., A.M.E. and F.K.-N. wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grants No 310/4 and No310/5 from the Deutsche Forschungsgemeinschaft and by grant 01DN12005 from the Bundesministerium für Bildung und Forschung to FKN and FG, by grants AK6/20 and AK6/23 to AK, by a stipend from the Studienstiftung des Deutschen Volkes to MU, and by stipends from the Graduiertenkolleg Entzündung und Regeneration to LS and MS. We thank Fabienne Seyfried, antibody core unit of the UMC-Hamburg, for excellent technical assistance. We thank Dr. Antonio Virgilio Failla, microscopy core facility of the UMC-Hamburg for assistance with the microscopy. We thank Guillermo Vila Melo and Gonzalo Perez Zabala for help with llama immunizations. We thank Drs. Bernhard Fleischer and Eva Tolosa, Hamburg for critical reading of the manuscript. The pCSE2.5 expression plasmid was kindly provided by Thomas Schirrmann, Braunschweig; monoclonal antibodies against llama IgG2 and IgG3 were kindly provided by Judith Appelton, Ithaca, HT-29 cells were kindly provided by Holger Kalthoff, Kiel.
Footnotes
F.H. and F.K.-N. receive a share of antibody sales via MediGate GmbH, a wholly owned subsidiary of the University Medical Center Hamburg-Eppendorf. F.G. is co-founder of Inmunova, a wholly owned subsidiary of the Instituto Leloir, which provides molecular assembly vaccines and llama immunizations.
References
- Rupnik M., Wilcox M. H. & Gerding D. N. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol 7, 526–536 (2009). [DOI] [PubMed] [Google Scholar]
- Huttunen R., Vuento R., Syrjanen J., Tissari P. & Aittoniemi J. Case fatality associated with a hypervirulent strain in patients with culture-positive Clostridium difficile infection: a retrospective population-based study. Int J Infect Dis 16, e532–535 (2012). [DOI] [PubMed] [Google Scholar]
- Kelly C. P. & LaMont J. T. Clostridium difficile--more difficult than ever. N Engl J Med 359, 1932–1940 (2008). [DOI] [PubMed] [Google Scholar]
- Perelle S., Gibert M., Bourlioux P., Corthier G. & Popoff M. R. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun 65, 1402–1407 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch C. & Aktories K. Microbial toxins and the glycosylation of rho family GTPases. Curr Opin Struct Biol 10, 528–535 (2000). [DOI] [PubMed] [Google Scholar]
- Viswanathan V. K., Mallozzi M. J. & Vedantam G. Clostridium difficile infection: An overview of the disease and its pathogenesis, epidemiology and interventions. Gut Microbes 1, 234–242 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne S. A. et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 (2010). [DOI] [PubMed] [Google Scholar]
- Lyras D. et al. Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker D. et al. Relatedness of human and animal Clostridium difficile PCR ribotype 078 isolates determined on the basis of multilocus variable-number tandem-repeat analysis and tetracycline resistance. J Clin Microbiol 48, 3744–3749 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyne L. Clostridium difficile--beyond antibiotics. N Engl J Med 362, 264–265 (2010). [DOI] [PubMed] [Google Scholar]
- McDonald L. C. et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353, 2433–2441 (2005). [DOI] [PubMed] [Google Scholar]
- Gerding D. N., Johnson S., Rupnik M. & Aktories K. Clostridium difficile binary toxin CDT: Mechanism, epidemiology, and potential clinical importance. Gut Microbes 5, 15–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulke I. et al. Characterization of the enzymatic component of the ADP-ribosyltransferase toxin CDTa from Clostridium difficile. Infect Immun 69, 6004–6011 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan C. et al. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog 5, e1000626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwan C. et al. Cholesterol- and sphingolipid-rich microdomains are essential for microtubule-based membrane protrusions induced by Clostridium difficile transferase (CDT). J Biol Chem 286, 29356–29365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories K. et al. Botulinum C2 toxin ADP-ribosylates actin. Nature 322, 390–392 (1986). [DOI] [PubMed] [Google Scholar]
- Barth H., Aktories K., Popoff M. R. & Stiles B. G. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol Mol Biol Rev 68, 373–402 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodorou P. et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT). Proc Natl Acad Sci U S A 108, 16422–16427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy I. et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N Engl J Med 362, 197–205 (2010). [DOI] [PubMed] [Google Scholar]
- Hamers-Casterman C. et al. Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993). [DOI] [PubMed] [Google Scholar]
- Wesolowski J. et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol 198, 157–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 82, 775–797 (2013). [DOI] [PubMed] [Google Scholar]
- Lauwereys M. et al. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J 17, 3512–3520 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Genst E. et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 103, 4586–4591 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijink B. M. et al. Improved tumor targeting of anti-epidermal growth factor receptor Nanobodies through albumin binding: taking advantage of modular Nanobody technology. Mol Cancer Ther 7, 2288–2297 (2008). [DOI] [PubMed] [Google Scholar]
- Hussack G. et al. Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J Biol Chem 286, 8961–8976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirrmann T. & Büssow K. Transient Production of scFv-Fc Fusion Proteins in Mammalian Cells. in Antibody Engineering Vol. 2. (eds. Kontermann R., & Dübel S., eds.) 387–398 (Springer-Verlag, Berlin Heidelberg; 2010). [Google Scholar]
- Zhang J., MacKenzie R. & Durocher Y. Production of chimeric heavy-chain antibodies. Methods Mol Biol 525, 323–336 (2009). [DOI] [PubMed] [Google Scholar]
- Koch-Nolte F. et al. Single domain antibodies from llama effectively and specifically block T cell ecto-ADP-ribosyltransferase ART2.2 in vivo. Faseb J 21, 3490–3498 (2007). [DOI] [PubMed] [Google Scholar]
- Desmyter A. et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol 3, 803–811 (1996). [DOI] [PubMed] [Google Scholar]
- Sundriyal A., Roberts A. K., Shone C. C. & Acharya K. R. Structural basis for substrate recognition in the enzymatic component of ADP-ribosyltransferase toxin CDTa from Clostridium difficile. J Biol Chem 284, 28713–28719 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge H. et al. Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens iota-toxin. Proc Natl Acad Sci U S A 105, 7399–7404 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A. H., Roberts A. K., Shone C. C. & Acharya K. R. Super toxins from a super bug: structure and function of Clostridium difficile toxins. Biochem J 436, 517–526 (2011). [DOI] [PubMed] [Google Scholar]
- Stiles B. G., Wigelsworth D. J., Popoff M. R. & Barth H. Clostridial binary toxins: iota and C2 family portraits. Front Cell Infect Microbiol 1, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A. & Collier R. J. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem 76, 243–265 (2007). [DOI] [PubMed] [Google Scholar]
- Feld G. K. et al. Structural basis for the unfolding of anthrax lethal factor by protective antigen oligomers. Nat Struct Mol Biol 17, 1383–1390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. C. et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell 154, 928–939 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhofer A. et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol 17, 133–138 (2010). [DOI] [PubMed] [Google Scholar]
- Alzogaray V. et al. Single-domain llama antibodies as specific intracellular inhibitors of SpvB, the actin ADP-ribosylating toxin of Salmonella typhimurium. Faseb J 25, 526–534 (2011). [DOI] [PubMed] [Google Scholar]
- Klein F. et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492, 118–122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutykhina I. L. et al. Passive immunization with a recombinant adenovirus expressing an HA (H5)-specific single-domain antibody protects mice from lethal influenza infection. Antiviral Res 97, 318–328 (2013). [DOI] [PubMed] [Google Scholar]
- Barth H. & Stiles B. G. Binary actin-ADP-ribosylating toxins and their use as molecular Trojan horses for drug delivery into eukaryotic cells. Curr Med Chem 15, 459–469 (2008). [DOI] [PubMed] [Google Scholar]
- Theiss A. L. et al. Nanoparticle-based therapeutic delivery of prohibitin to the colonic epithelial cells ameliorates acute murine colitis. Inflamm Bowel Dis 17, 1163–1176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman R. J. et al. Clostridium difficile binary toxin (CDT) and diarrhea. Anaerobe 17, 161–165 (2011). [DOI] [PubMed] [Google Scholar]
- Cartman S. T., Heap J. T., Kuehne S. A., Cockayne A. & Minton N. P. The emergence of ‘hypervirulence' in Clostridium difficile. Int J Med Microbiol 300, 387–395 (2010). [DOI] [PubMed] [Google Scholar]
- Stewart D. B., Berg A. & Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg 17, 118–124 (2013). [DOI] [PubMed] [Google Scholar]
- Marsh J. W. & Curry S. R. Therapeutic Approaches for Clostridium difficile Infections. Curr Protoc Microbiol 30, 9A 3 1–9 (2013). [DOI] [PubMed] [Google Scholar]
- Cocanour C. S. Best strategies in recurrent or persistent Clostridium difficile infection. Surg Infect (Larchmt) 12, 235–239 (2011). [DOI] [PubMed] [Google Scholar]
- Khoruts A. & Sadowsky M. J. Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol 4, 4–7 (2011). [DOI] [PubMed] [Google Scholar]
- Brandt L. J. et al. Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am J Gastroenterol 107, 1079–1087 (2012). [DOI] [PubMed] [Google Scholar]
- Andersen J. T. & Sandlie I. The versatile MHC class I-related FcRn protects IgG and albumin from degradation: implications for development of new diagnostics and therapeutics. Drug Metab Pharmacokinet 24, 318–332 (2009). [DOI] [PubMed] [Google Scholar]
- Chen K. et al. Endocytosis of soluble immune complexes leads to their clearance by FcgammaRIIIB but induces neutrophil extracellular traps via FcgammaRIIA in vivo. Blood 120, 4421–4431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockstaele F., Holz J. B. & Revets H. The development of nanobodies for therapeutic applications. Curr Opin Investig Drugs 10, 1212–1224 (2009). [PubMed] [Google Scholar]
- Laroui H. et al. Drug-loaded nanoparticles targeted to the colon with polysaccharide hydrogel reduce colitis in a mouse model. Gastroenterology 138, 843–853 (2010). [DOI] [PubMed] [Google Scholar]
- Hussack G., Hirama T., Ding W., Mackenzie R. & Tanha J. Engineered single-domain antibodies with high protease resistance and thermal stability. PLoS One 6, e28218 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K. K., Marcotte H., Alvarez B., Boyaka P. N. & Hammarstrom L. In situ gastrointestinal protection against anthrax edema toxin by single-chain antibody fragment producing lactobacilli. BMC Biotechnol 11, 126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbroucke K. et al. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal Immunol 3, 49–56 (2010). [DOI] [PubMed] [Google Scholar]
- Barth H. et al. Cellular uptake of Clostridium botulinum C2 toxin requires oligomerization and acidification. J Biol Chem 275, 18704–18711 (2000). [DOI] [PubMed] [Google Scholar]
- Boersma W. J., Bogaerts W. J., Bianchi A. T. & Claassen E. Adjuvant properties of stable water-in-oil emulsions: evaluation of the experience with Specol. Res Immunol 143, 503–512 (1992). [DOI] [PubMed] [Google Scholar]
- Daley L. P., Gagliardo L. F., Duffy M. S., Smith M. C. & Appleton J. A. Application of monoclonal antibodies in functional and comparative investigations of heavy-chain immunoglobulins in new world camelids. Clin Diagn Lab Immunol 12, 380–386 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarebski L. M., Urrutia M. & Goldbaum F. A. Llama single domain antibodies as a tool for molecular mimicry. J Mol Biol 349, 814–824 (2005). [DOI] [PubMed] [Google Scholar]
- Gekle M., Wunsch S., Oberleithner H. & Silbernagl S. Characterization of two MDCK-cell subtypes as a model system to study principal cell and intercalated cell properties. Pflugers Arch 428, 157–162 (1994). [DOI] [PubMed] [Google Scholar]
- Zhu K. et al. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins 82, 1646–1655 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Brenke R., Comeau S. R. & Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 (2006). [DOI] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R. & Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information