Abstract
We define two novel species of the genus Staphylococcusthat are phenotypically similar to and have near identical 16S rRNA gene sequences to Staphylococcus aureus. However, compared to S. aureus and each other, the two species, Staphylococcus argenteus sp. nov. (type strain MSHR1132T = DSM 28299T = SSI 89.005T) and Staphylococcus schweitzeri sp. nov. (type strain FSA084T = DSM 28300T = SSI 89.004T), demonstrate: 1) at a whole-genome level considerable phylogenetic distance, lack of admixture, average nucleotide identity <95 %, and inferred DNA–DNA hybridization <70 %; 2) different profiles as determined by MALDI-TOF MS; 3) a non-pigmented phenotype for S. argenteus sp. nov.; 4) S. schweitzeri sp. nov. is not detected by standard nucA PCR; 5) distinct peptidoglycan types compared to S. aureus; 6) a separate ecological niche for S. schweitzeri sp. nov.; and 7) a distinct clinical disease profile for S. argenteus sp. nov. compared to S. aureus.
There are over 40 species of the genus Staphylococcus, of which the coagulase-positive Staphylococcus aureus is a major cause of human clinical disease. The population structure of S. aureus is well-understood and comprises clonal complexes (CCs) with <2 % nucleotide divergence. Recently two Staphylococcuslineages have been recovered from human clinical infections (Holt et al., 2011; Ng et al., 2009; Ruimy et al., 2010; Tong et al., 2010), from non-human primates (Schaumburg et al., 2012), and from bats in Africa (Akobi et al., 2012). These lineages have previously been identified as S. aureus according to phenotype, but on the basis of multi-locus sequence typing (MLST) and a single genome sequence, the lineages are allied to but significantly diverged from S. aureus. We describe here investigations including the use of whole-genome sequence analysis to justify classification as three separate species, S. aureus, and two novel species of the genus Staphylococcus.
Strain MSHR1132T was isolated from blood cultures of an Indigenous patient from Darwin, Australia (Holt et al., 2011); and strain FSA084T was isolated from the nares of a red-tailed monkey (Cercopithecus ascanius) from Gabon, Africa (Schaumburg et al., 2012). Both strains grew on tryptone soy agar (TSA) at 37 °C with large, round, smooth colonies similar to typical S. aureus. Colonies of FSA084T have a yellowish-pigmented appearance while those of MSHR1132T are non-pigmented, displaying a creamy white appearance. The difference in pigmentation between typical S. aureus and strain MSHR1132T is particularly evident after growing on chocolate agar (Oxoid) for 48 h at 37 °C (Holt et al., 2011). Strains MSHR1132T and FSA084T are both catalase-positive, coagulase-positive by tube coagulase test and colonies demonstrate β-haemolysis on blood agar. Gram staining tests revealed Gram-stain-positive cocci in clusters for both strains. MLST revealed MSHR1132T as ST1850 and FSA084T as ST2022. We also selected for further investigation five strains that clustered according to MLST with MSHR1132T (LBSA043, JABA32044, M260, M051, H115100079) and five that clustered with FSA084T (FSCB1B, FSCB5, FSA096, FSA090, FSA037). The MSHR1132T lineage strains were all recovered from human hosts from northern Australia (Brennan et al., 2013; McDonald et al., 2006), Fiji (Jenney et al., 2014) and the UK. The FSA084T lineage strains were recovered from non-human primates in Gabon and Côte d’Ivoire, Africa (Schaumburg et al., 2012). These additional strains demonstrated the same cell and colonial morphology as MSHR1132T and FSA084T respectively. All MSHR1132T lineage strains appeared non-pigmented.
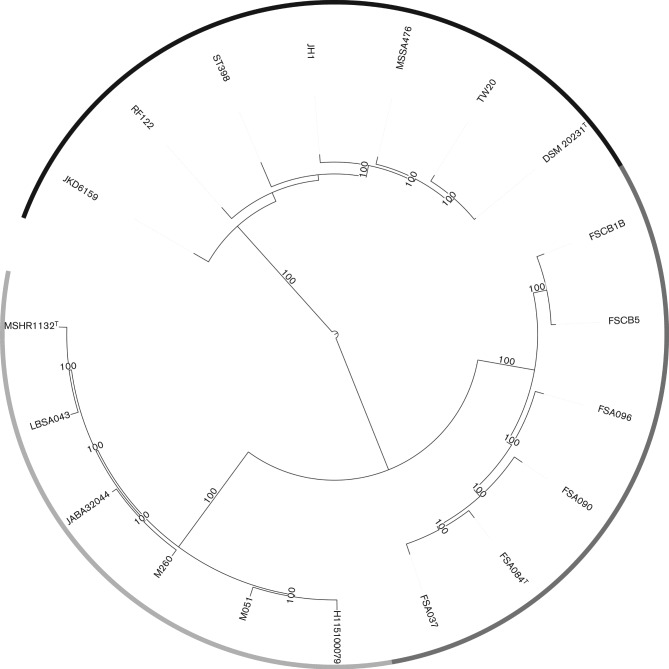
Whole-genome sequencing using the Illumina HiSeq platform of the 12 strains, followed by core-genome single nucleotide polymorphism (SNP)-based maximum-likelihood trees demonstrated that MSHR1132T lineage strains, FSA084T lineage strains, and reference S. aureusgenomes, form three distinct clusters with 100 % bootstrap support (Fig. 1). The details and GenBank accession numbers for these strains are provided in Table 1. Compared to S. aureus, the 16S rRNA gene sequence (1474 nt) is identical in strain MSHR1132T and differs at one position in strain FSA084T. However, pairwise average nucleotide identity (ANI), as calculated using JSpecies (Richter & Rosselló-Móra, 2009), across the genomes within and between these groups was consistent with separate species designations (Table 2). Previously it has been demonstrated that an ANI <95 % corresponds well to a DNA–DNA hybridization (DDH) value of <70 % (Goris et al., 2007). Similarly, an analysis using the Genome blast Distance Phylogeny (Meier-Kolthoff et al., 2013) to calculate genome-to-genome distances clearly demonstrated three separate groups with mean inferred DDH values of 34 % and 36 % between S. aureus and the MSHR1132T and FSA084T lineages, respectively, 46 % between MSHR1132T and FSA084T lineages, and >80 % within lineages (Table 3). An analysis of orthologous core genes shared by all three groups using Bayesian Analysis of Population Structure (BAPS) software (Cheng et al., 2013) demonstrated three BAPS clusters and an absence of admixture between the groups (Fig. 1). All MSHR1132T lineage strains lacked the carotenoid pigment operon.
Fig. 1.
Maximum-likelihood phylogenetic tree of whole-genome sequence alignments for MSHR1132T lineage strains (LBSA043, JABA32044, M260, M051, H115100079), FSA084T lineage strains (FSCB1B, FSCB5, FSA096, FSA090, FSA037), and reference S. aureus strains (JKD6159, RF122, ST398, JH1, MSSA476, TW20, DSM 20231T). The outer circle shading indicates the three Bayesian Analysis of Population Structure (BAPS) groups: S. aureus (black), MSHR1132T lineage (light grey) and FSA084T lineage (dark grey). Branches with bootstrap support of 100 % are indicated.
Table 1. Strains and GenBank accession numbers for whole-genome sequences.
| Strain | Place of origin | GenBank accession nos of whole-genome sequences (for short-read and annotated assemblies, respectively) |
| Staphylococcus argenteus sp. nov. | ||
| JABA32044V6S1 | Fiji | ERS140248 |
| CCEE01000001–CCEE01000018 | ||
| LBSA043 | Northern Australia | ERS140026 |
| CCEM01000001–CCEM01000011 | ||
| M051 | Northern Australia | ERS140254 |
| CCEN01000001–CCEN01000011 | ||
| M260 | Northern Australia | ERS140095 |
| CCEF01000001–CCEF01000019 | ||
| H115100079 | UK | ERS154949 |
| CCEP01000001–CCEP01000012 | ||
| MSHR1132T | Northern Australia | FR821777 |
| Staphylococcus schweitzeri sp. nov. | ||
| FSA037 | Gabon | ERS140147 |
| CCEH01000001–CCEH01000058 | ||
| FSA084T | Gabon | ERS140266 |
| CCEL01000001–CCEL01000035 | ||
| FSA090 | Gabon | ERS140239 |
| CCEO01000001–CCEO01000054 | ||
| FSA096 | Gabon | ERS140159 |
| CCEK01000001–CCEK01000047 | ||
| FSCB1B | Côte d’Ivoire | ERS140162 |
| CCEG01000001–CCEG01000026 | ||
| FSCB5 | Côte d’Ivoire | ERS140167 |
| CCEQ01000001–CCEQ01000038 | ||
| Staphylococcus aureus | ||
| DSM 20231T | AMYL01000000 | |
| JH1 | CP000736 | |
| RF122 | AJ938182 | |
| ST398 | AM990992 | |
| MSSA476 | BX571857 | |
| JKD6159 | CP002114 | |
| TW20 | FN433596 |
Table 2. Genome-wide average nucleotide identities (ANI) and inferred DNA–DNA hybridization (DDH) values for pairwise comparisons of strains from each of the three groups.
Values are mean with standard deviation. ANI was calculated using JSpecies (Goris et al., 2007) and inferred DDH with Genome blast Distance Phylogeny (Meier-Kolthoff et al., 2013).
| Group | Pairwise comparison with: | |||
| 1 | 2 | 3 | ||
| 1. S. argenteus sp. nov. | ANI | 98.8 (0.14) | 92.0 (0.08) | 87.4 (0.20) |
| DDH | 89.1 (1.37) | 46.4 (0.16) | 33.5 (0.37) | |
| 2. S. schweitzeri sp. nov. | ANI | 98.0 (0.44) | 88.6 (0.14) | |
| DDH | 85.6 (4.32) | 36.3 (0.23) | ||
| 3. S. aureus | ANI | 97.4 (0.49) | ||
| DDH | 84.8 (8.00) | |||
Table 3. Key biochemical tests used for identification of staphylococcal species.
Species: 1, S. aureus [number of strains (n) = 18]; 2, S. schweitzeri sp. nov. (n = 6); 3, S. argenteus sp. nov. (n = 6). Results were obtained in triplicate for each of MSHR1132T lineage (S. argenteus sp. nov.) strains, FSA084T lineage (S. schweitzeri sp. nov.) strains, and 18 ATCC strains of S. aureus using a Vitek2 GP Card (bioMérieux); see text for details of strains. Values are the proportion (%) of tests that were either positive or negative for each group of strains.
| Biochemical test | 1 | 2 | 3 | |||
| d-Xylose | – | (100) | – | (100) | – | (100) |
| Arginine dihydrolase 1 | + | (100) | + | (100) | + | (100) |
| β-Galactosidase | – | (96) | – | (100) | – | (94) |
| Phosphatase | + | (100) | + | (100) | + | (94) |
| β-Glucuronidase | – | (100) | – | (100) | – | (100) |
| l-Pyrrolidonyl arylamidase | + | (100) | + | (94) | + | (94) |
| Urease | – | (100) | – | (100) | – | (56) |
| Polymixin B resistance | + | (94) | + | (100) | + | (100) |
| Lactose | – | (97) | – | (100) | – | (100) |
| N-Acetyl-d-glucosamine | + | (98) | – | (67) | – | (72) |
| Maltose | + | (100) | + | (100) | + | (100) |
| Novobiocin resistance | – | (98) | – | (67) | – | (100) |
| Growth in 6.5 % NaCl | + | (100) | + | (100) | + | (100) |
| d-Mannitol | + | (100) | + | (100) | + | (100) |
| d-Mannose | + | (100) | + | (100) | + | (94) |
| Raffinose | – | (100) | – | (100) | – | (100) |
| Sucrose | + | (100) | + | (100) | + | (100) |
| Trehalose | + | (83) | + | (100) | + | (100) |
| d-Ribose | – | (96) | + | (72) | – | (83) |
| d-Galactose | + | (82) | – | (56) | + | (100) |
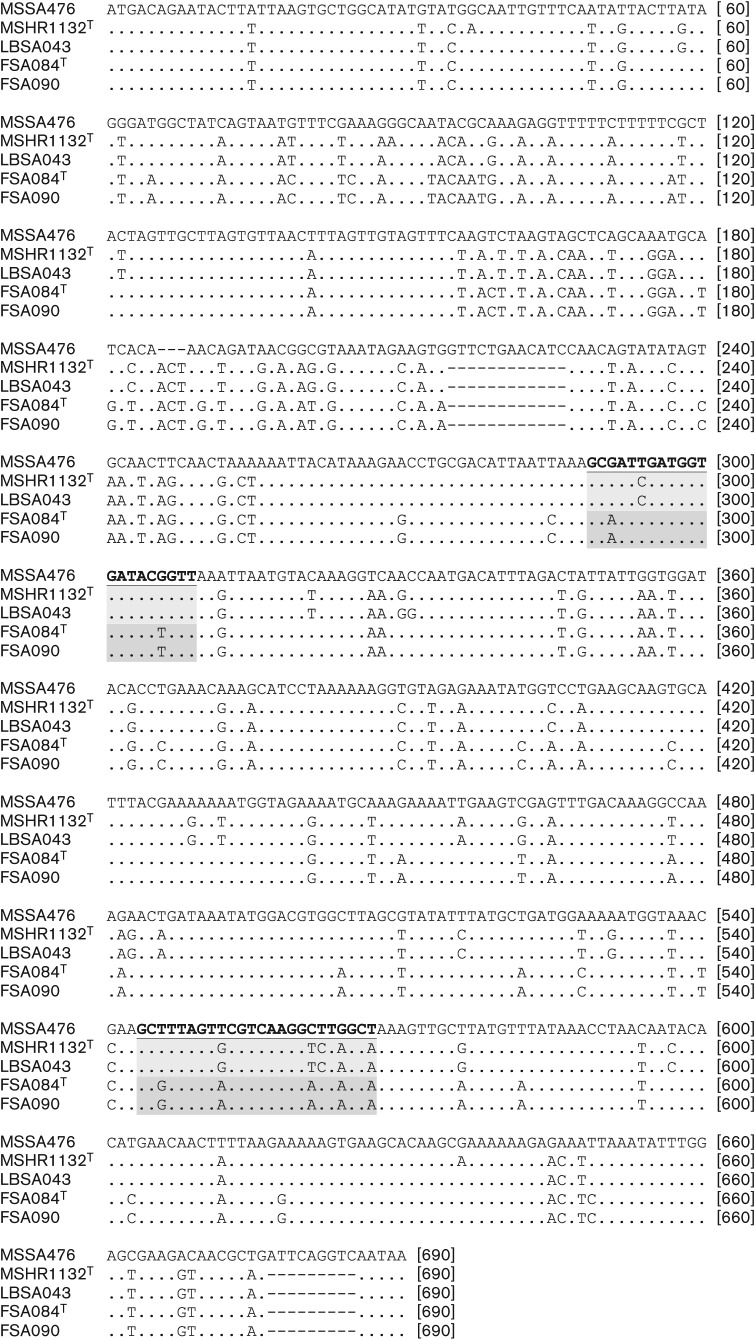
PCR amplification of the nucA gene that is used as a standard confirmatory marker for S. aureus is positive in strain MSHR1132T but negative in strain FSA084T. An examination of the nucA gene and in particular the primer sites for nucA (Brakstad et al., 1992) reveal one and two mismatches for the forward primer, and five and five mismatches for the reverse primer, for MSHR1132T and FSA084T, respectively (Fig. 2). The presence of mismatches at the 3′ end of primers for strain FSA084T most likely contributes to the lack of amplification of product for strain FSA084T. There were two in-frame deletions of 9 and 12 bp and one in-frame insertion of 3 bp in both MSHR1132T and FSA084T nucA sequences compared to S. aureus.
Fig. 2.
Sequence alignment of representative nucA gene sequences. The standard primer sites (Brakstad et al., 1992) are indicated in bold underline (S. aureus), light grey shading (S. argenteus sp. nov.) or dark grey shading (S. schweitzeri sp. nov.). MSSA476 represents a reference S. aureussequence, MSHR1132T and LBSA043 represent S. argenteus sp. nov., and FSA084T and FSA090 represent S. schweitzeri sp. nov.
Biochemical profiling was performed with the Vitek2 GP card platform (bioMérieux) according to the manufacturer’s instructions. We tested in triplicate each of the 12 strains together with 18 strains of S. aureus from the ATCC collection (ATCC 12600T, 13565, 13709, 14458, 19095, 19636, 23235, 25904, 25923, 27664, 29213, 29247, 33591, 33592, 43300, 49230, 49775, 51811) (Table 3). The biochemical test profiles for both MSHR1132T lineage strains and FSA084T lineage strains, are consistent with S. aureus, with mean probabilities of >95 % of identity as S. aureus. Although no test definitively discriminated between the three groups, the following may be helpful in identifying these lineages. The FSA084T lineage strains were positive for d-ribose in 72 % of tests compared to 4 % and 17 % for S. aureus and MSHR1132T lineage strains, respectively. The MSHR1132T lineage strains were positive for urease in 56 % of tests compared to 0 % for both S. aureus and FSA084T lineage strains. S. aureus was positive for N-acetyl-d-glucosamine in 98 % of tests compared to 33 % and 28 % for FSA084T lineage strains and MSHR1132T lineage strains, respectively.
We attempted to discriminate 12 MSHR1132T lineage strains (an additional six strains to those already described), 12 FSA084T lineage strains (an additional six strains to those already described), and 22 consecutive standard clinical strains of S. aureusby using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Microflex LT MALDI-TOF instrument; Bruker Daltonik) (Table 4). We prepared samples using liquid phase formic acid extraction, according to the manufacturer’s recommendations, and compared the spectral profiles gained to the existing Bruker standard clinical database of profiles using MALDI Biotyper 2.1 software (Bruker Daltonik) with default settings. Strains of S. aureuswere confidently identified. The MSHR1132T lineage strains and FSA084T lineage strains profiles were most similar to the S. aureusprofile, but identity scores were much lower than for the strains of S. aureus (P<0.0001 for both compared to S. aureus) and fell below the manufacturer’s recommended threshold for a species level identification. We generated new reference profiles with three MSHR1132T lineage strains and three FSA084T lineage strains and repeated the analysis of all 46 strains. All strains were then confidently identified into their different groups. These findings are consistent with the three groups being separate species based on cell proteomic analysis.
Table 4. Comparison of MALDI-TOF MS identity scores using the standard clinical database and an amended database with reference profiles from S. argenteus sp. nov. and S. schweitzeri sp. nov. groups.
Identity score values are graded as highly probable species identification (score value 2.300–3.000), secure genus and probable species identification (2.000–2.299) and probable genus identification (1.700–1.999). Values are mean (standard deviation) of identity scores.
| Standard database | Amended database | |||
| Best hit | Identity score | Best hit | Identity score | |
| S. aureus (n = 22) | S. aureus | 2.295 (0.067) | S. aureus | 2.295 (0.067) |
| S. argenteus sp. nov. (n = 12) | S. aureus | 2.071 (0.102) | S. argenteus sp. nov. | 2.700 (0.066) |
| S. schweitzeri sp. nov. (n = 12) | S. aureus | 1.847 (0.095) | S. schweitzeri sp. nov. | 2.676 (0.072) |
Analyses of fatty acids, respiratory quinones and peptidoglycans were carried out by the Identification Service of the Leibniz-Institut DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen), Braunschweig, Germany. S. aureus ATCC 29213, MSHR1132T and FSA084T were cultured and tested for fatty acid composition under identical conditions. The fatty acid profiles of strains MSHR1132T and FSA084T were similar and dominated by anteiso-C15 : 0 and anteiso-C17 : 0 and corresponded in their composition to S. aureus ATCC 29213 (Table 5). Strains MSHR1132T and FSA084T contained the menaquinones MK-7, MK-8 and MK-9 at ratios of 11 : 70 : 11 plus a non-identified peak (MSHR1132T) and 7 : 80 : 13 (FSA084T). Menaquinones with seven to eight side-chains are characteristic for the genus Staphylococcus (Gotz et al., 2006). Both MSHR1132T and FSA084T showed the same peptidoglycan type (A3α type, A11.8 type, l-Lys–l-Ala–(Gly)4-5) (Schleifer & Kandler, 1972; Schumann, 2011) (http://www.dsmz.de/?id=449). This peptidoglycan type is reported from Staphylococcus vitulinus (Švec et al., 2004; Zakrzewska-Czerwińska et al., 1995), Staphylococcus lentus (Schleifer et al., 1983) and Staphylococcus sciuri (Kloos et al., 1976), and is distinct from that of S. aureus (A3α type, A11.2 type, l-Lys–(Gly)4-5) (Gotz et al., 2006).
Table 5. Cellular fatty acid contents of Staphylococcus aureus, Staphylococcus argenteus sp. nov. and Staphylococcus schweitzeri sp. nov.
Strain: 1, S. aureus ATCC 29213; 2, S. argenteus sp. nov. MSHR1132T; 3, S. schweitzeri sp. nov. FSA084T. Values are percentages of total fatty acids.
| Fatty acid | 1 | 2 | 3 |
| C14 : 0 | 0.3 | 0.2 | 0.2 |
| C16 : 0 | 2.1 | 2.1 | 1.9 |
| C17 : 0 | 0.4 | 0.4 | 0.3 |
| C18 : 0 | 6.3 | 5.8 | 6.0 |
| C18 : 1ω9c | 2.0 | 2.4 | 2.1 |
| C18 : 1ω7c | 0.2 | 0.0 | 0.2 |
| C18 : 2ω6,9c/anteiso-C18 : 0* | 0.8 | 0.9 | 0.7 |
| C19 : 0 | 0.5 | 0.4 | 0.3 |
| C20 : 0 | 2.6 | 1.6 | 1.7 |
| C20 : 1ω9c | 0.4 | 0.9 | 0.6 |
| C20 : 2ω6,9c | 0.2 | 0.0 | 0.3 |
| iso-C13 : 0 | 0.1 | 0.0 | 0.0 |
| iso-C14 : 0 | 0.4 | 0.5 | 0.6 |
| iso-C15 : 0 | 5.4 | 7.2 | 5.8 |
| iso-C16 : 0 | 0.8 | 1.1 | 1.3 |
| iso-C17 : 0 | 3.1 | 4.6 | 4.0 |
| iso-C18 : 0 | 0.3 | 0.4 | 0.5 |
| iso-C19 : 0 | 0.9 | 1.0 | 1.0 |
| anteiso-C13 : 0 | 0.1 | 0.0 | 0.1 |
| anteiso-C15 : 0 | 50.2 | 48.5 | 47.1 |
| anteiso-C17 : 0 | 20.4 | 19.6 | 22.3 |
| anteiso-C19 : 0 | 2.9 | 2.4 | 3.2 |
| Total | 100 | 100 | 100 |
Differentiation between these two fatty acids was not possible.
In Conclusion, although MSHR1132T lineage strains and FSA084T lineage strains share identical or near identical 16S rRNA gene sequences, have similar fatty acid and menaquinone compositions to S. aureus, and are phylogenetically the closest known relatives of S. aureus, there are strong justifications for assigning these lineages to two novel species of the genus Staphylococcus, for which the names Staphylococcus argenteus sp. nov. (type strain MSHR1132T) and Staphylococcus schweitzeri sp. nov. (type strain FSA084T) are proposed. These justifications are: 1) phylogenetic distance, lack of admixture, ANI <95 %, and inferred DDH <70 %; 2) different profiles as determined by MALDI-TOF MS; 3) non-pigmented phenotype of S. argenteus sp. nov.; 4) S. schweitzeri sp. nov. cannot be detected by standard nucA PCR; 5) distinct peptidoglycan types compared to S. aureus; 6) a separate ecological niche for S. schweitzeri sp. nov., which has only once been recovered from human hosts to date (Schaumburg et al., 2012); 7) distinct clinical disease profile for S. argenteus sp. nov. compared to S. aureus (Tong et al., 2013).
Description of Staphylococccus argenteus sp. nov.
Staphylococcus argenteus (ar.gen′te.us. L. masc. adj. argenteus silver, silvery).
Colonies are large, 2 mm in diameter, round, convex, smooth, creamy white and demonstrate β-haemolysis on blood agar. The difference in pigmentation between typical S. aureus and S. argenteus is particularly evident after growing on chocolate agar for 48 h at 37 °C. Cells are Gram-stain-positive, coccoid, 1 µm in diameter, and form clusters. Facultatively anaerobic. Cells are catalase-positive and coagulase-positive by tube coagulase test. Biochemically positive for alkaline phosphatase, arginine dihydrolase, l-pyrrolidonyl arylamidase, galactose, maltose, mannitol, mannose, methyl β-d-glucopyranoside, sucrose, and trehalose; and negative for urease, α-glucosidase, phosphatidylinositol phospholipase C, β-galactosidase, alanine-phenylalanine-proline arylamidase, l-aspartic acid arylamidase, α-mannosidase, β-glucuronidase, l-leucine arylamidase, proline arylamidase, α-galactosidase, alanine arylamidase, tyrosine arylamidase, amygdalin, xylose, α-cyclodextrin, sorbitol, ribose, lactose, N-acetylglucosamine, pullulan, raffinose, salicin (Vitek2 GP Card). The peptidoglycan is of the type A3α, A11.8, l-Lys–l-Ala–(Gly)4-5. The menaquinones MK-7, MK-8 and MK-9 are at ratios of 11 : 70 : 11 and the predominant fatty acids are anteiso-C15 : 0 and anteiso-C17 : 0.
The type strain MSHR1132T ( = DSM 28299T = SSI 89.005T) was isolated from the blood culture of a 55-year-old Indigenous Australian female in 2006 in Darwin, Northern Territory, Australia. The type strain has also been deposited in the Robert Koch Institute (Germany) and the National Collection of type Cultures, Public Health England (UK).
Description of Staphylococcus schweitzeri sp. nov.
Staphylococcus schweitzeri (schwei′tzer.i. N.L. gen. n. schweitzeri of Schweitzer, named after Albert Schweitzer, founder of a hospital in Lambaréné, Gabon, and Nobel Peace Prize Laureate in 1952).
Colonies are round, 1.7 mm in diameter, convex, smooth, yellow and demonstrate β-haemolysis on blood agar. Cells are Gram-stain-positive, coccoid, 1 µm in diameter, and form clusters. Facultatively anaerobic. Cells are catalase-positive and coagulase-positive by tube coagulase test. Biochemically positive for alkaline phosphatase, arginine dihydrolase, l-pyroglutamic acid arylamidase, maltose, mannitol, mannose, methyl β-d-glucopyranoside, sucrose, trehalose, ribose and N-acetylglucosamine, but negative for phosphatidylinositol phospholipase C, α-glucosidase, β-galactosidase, urease, alanine-phenylalanine-proline arylamidase, l-aspartic acid arylamidase, α-mannosidase, β-glucuronidase, l-leucine arylamidase, proline arylamidase, α-galactosidase, alanine arylamidase, tyrosine arylamidase, amygdalin, xylose, α-cyclodextrin, sorbitol, galactose, lactose, pullulan, raffinose and salicin (Vitek2 GP Card; bioMérieux). The peptidoglycan is of the type A3α, A11.8, l-Lys–l-Ala–(Gly)4-5. The menaquinones MK-7, MK-8 and MK-9 are at ratios of 7 : 80 : 13 and the predominant fatty acids are anteiso-C15 : 0 and anteiso-C17 : 0.
The type strain FSA084T ( = DSM 28300T = SSI 89.004T) was isolated from the nares of a non-human primate (Cercopithecus ascanius) from Gabon, Africa within 12 h after the death of the animal in 2010. The type strain has also been deposited in the Robert Koch Institute (Germany) and the National Collection of type Cultures, Public Health England (UK).
Acknowledgements
The work described here was funded by the Wellcome Trust through core funding for the Sanger Institute Pathogen Variation Group and by the Deutsche Forschungsgemeinschaft (DFG, EI 247/8-1). S. Y. C. T. is an Australian National Health and Medical Research Council Career Development Fellow (1065736).
Abbreviations:
- ANI
average nucleotide identity
- DDH
DNA–DNA hybridization
- MALDI-TOF
matrix-assisted laser desorption ionization–time of flight
- MLST
multi-locus sequence typing
References
- Akobi B., Aboderin O., Sasaki T., Shittu A. (2012). Characterization of Staphylococcus aureus isolates from faecal samples of the Straw-Coloured Fruit Bat (Eidolon helvum) in Obafemi Awolowo University (OAU), Nigeria. BMC Microbiol 12, 279. 10.1186/1471-2180-12-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakstad O. G., Aasbakk K., Maeland J. A. (1992). Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol 30, 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan L., Lilliebridge R. A., Cheng A. C., Giffard P. M., Currie B. J., Tong S. Y. (2013). Community-associated meticillin-resistant Staphylococcus aureus carriage in hospitalized patients in tropical northern Australia. J Hosp Infect 83, 205–211. 10.1016/j.jhin.2012.10.014 [DOI] [PubMed] [Google Scholar]
- Cheng L., Connor T. R., Sirén J., Aanensen D. M., Corander J. (2013). Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30, 1224–1228. 10.1093/molbev/mst028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57, 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Gotz F., Bannerman T., Schleifer K.-H. (2006). The genera Staphylococcus and Macrococcus. In The Prokaryotes, 3rd edn, Vol. 4 Bacteria: Firmicutes, Cyanobacteria, pp. 5–75. Edited by Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. New York, NY: Springer; 10.1007/0-387-30744-3_1 [DOI] [Google Scholar]
- Holt D. C., Holden M. T., Tong S. Y., Castillo-Ramirez S., Clarke L., Quail M. A., Currie B. J., Parkhill J., Bentley S. D., et al. (2011). A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3, 881–895. 10.1093/gbe/evr078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenney A., Holt D., Ritika R., Southwell P., Pravin S., Buadromo E., Carapetis J., Tong S., Steer A. (2014). The clinical and molecular epidemiology of Staphylococcus aureus infections in Fiji. BMC Infect Dis 14, 160. 10.1186/1471-2334-14-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloos W. E., Schleifer K. H., Smith R. F. (1976). Characterization of Staphylococcus sciuri sp. nov. and its subspecies. Int J Syst Bacteriol 26, 22–37 10.1099/00207713-26-1-22 [DOI] [Google Scholar]
- McDonald M., Dougall A., Holt D., Huygens F., Oppedisano F., Giffard P. M., Inman-Bamber J., Stephens A. J., Towers R., et al. (2006). Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol 44, 3720–3727. 10.1128/JCM.00836-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J. W., Holt D. C., Lilliebridge R. A., Stephens A. J., Huygens F., Tong S. Y., Currie B. J., Giffard P. M. (2009). Phylogenetically distinct Staphylococcus aureus lineage prevalent among indigenous communities in northern Australia. J Clin Microbiol 47, 2295–2300. 10.1128/JCM.00122-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruimy R., Angebault C., Djossou F., Dupont C., Epelboin L., Jarraud S., Lefevre L. A., Bes M., Lixandru B. E., et al. (2010). Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis 202, 924–934. 10.1086/655901 [DOI] [PubMed] [Google Scholar]
- Schaumburg F., Alabi A. S., Köck R., Mellmann A., Kremsner P. G., Boesch C., Becker K., Leendertz F. H., Peters G. (2012). Highly divergent Staphylococcus aureus isolates from African non-human primates. Environ Microbiol Rep 4, 141–146. 10.1111/j.1758-2229.2011.00316.x [DOI] [PubMed] [Google Scholar]
- Schleifer K. H., Kandler O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36, 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. H., Geyer U., Kilpper-Bälz R., Devriese L. A. (1983). Elevation of Staphylococcus sciuri subsp. lentus (Kloos et al.) to species status: Staphylococcus lentus (Kloos et al.) comb. nov. Syst Appl Microbiol 4, 382–387. 10.1016/S0723-2020(83)80022-8 [DOI] [PubMed] [Google Scholar]
- Schumann P. (2011). Peptidoglycan Structure. Methods Microbiol 38, 101–129. 10.1016/S0723-2020(83)80022-8 [DOI] [Google Scholar]
- Švec P., Vancanneyt M., Sedláček I., Engelbeen K., Štětina V., Swings J., Petráš P. (2004). Reclassification of Staphylococcus pulvereri Zakrzewska-Czerwińska et al. 1995 as a later synonym of Staphylococcus vitulinus Webster et al. 1994. Int J Syst Evol Microbiol 54, 2213–2215. 10.1099/ijs.0.63080-0 [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Lilliebridge R. A., Bishop E. J., Cheng A. C., Holt D. C., McDonald M. I., Giffard P. M., Currie B. J., Boutlis C. S. (2010). Clinical correlates of Panton-Valentine leukocidin (PVL), PVL isoforms, and clonal complex in the Staphylococcus aureus population of Northern Australia. J Infect Dis 202, 760–769. 10.1086/655396 [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Sharma-Kuinkel B. K., Thaden J. T., Whitney A. R., Yang S. J., Mishra N. N., Rude T., Lilliebridge R. A., Selim M. A., et al. (2013). Virulence of endemic nonpigmented northern Australian Staphylococcus aureus clone (clonal complex 75, S. argenteus) is not augmented by staphyloxanthin. J Infect Dis 208, 520–527. 10.1093/infdis/jit173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Czerwińska J., Gaszewska-Mastalarz A., Lis B., Gamian A., Mordarski M. (1995). Staphylococcus pulvereri sp. nov., isolated from human and animal specimens. Int J Syst Bacteriol 45, 169–172. 10.1099/00207713-45-1-169 [DOI] [PubMed] [Google Scholar]