Wurm et al. identified Fra-2/AP-1 as a key regulator of terminal epidermal differentiation. Loss of Fra-2 in suprabasal keratinocytes is sufficient to cause skin barrier defects due to reduced expression of differentiation genes. The induction of epidermal differentiation by Fra-2 is controlled by a dual mechanism involving Ezh2-dependent methylation and activation by ERK1/2-dependent phosphorylation.
Keywords: Fra-2/AP-1, Ezh2/PRC2, nonhistone substrate, epidermal differentiation complex, ERK1/2
Abstract
Altered epidermal differentiation characterizes numerous skin diseases affecting >25% of the human population. Here we identified Fra-2/AP-1 as a key regulator of terminal epidermal differentiation. Epithelial-restricted, ectopic expression of Fra-2 induced expression of epidermal differentiation genes located within the epidermal differentiation complex (EDC). Moreover, in a papilloma-prone background, a reduced tumor burden was observed due to precocious keratinocyte differentiation by Fra-2 expression. Importantly, loss of Fra-2 in suprabasal keratinocytes is sufficient to cause skin barrier defects due to reduced expression of differentiation genes. Mechanistically, Fra-2 binds and transcriptionally regulates EDC gene promoters, which are co-occupied by the transcriptional repressor Ezh2. Fra-2 remains transcriptionally inactive in nondifferentiated keratinocytes, where it was found monomethylated and dimethylated on Lys104 and interacted with Ezh2. Upon keratinocyte differentiation, Fra-2 is C-terminally phosphorylated on Ser320 and Thr322 by ERK1/2, leading to transcriptional activation. Thus, the induction of epidermal differentiation by Fra-2 is controlled by a dual mechanism involving Ezh2-dependent methylation and activation by ERK1/2-dependent phosphorylation.
The gene expression programs that regulate somatic tissue development are controlled by coordinated interactions of transcription factors, cofactors, and chromatin regulators (Young 2011). Deregulation of these gene expression programs can cause a broad range of diseases (Lee and Young 2013). In skin epidermis, the induction of keratinocyte differentiation to form a stratified epithelium is essential for the acquisition of the epidermal barrier function as well as tissue homeostasis (Blanpain et al. 2007). The frequent incidence of human disorders characterized by skin barrier defects—such as atopic dermatitis, psoriasis, ichthyosis vulgaris, and epidermal cancers—underlines the importance of coordinated transcriptional programs in keratinocytes (Rogers et al. 2006; Smith and Barker 2006; Blanpain et al. 2007; de Cid et al. 2009; Deady et al. 2014). Basal keratinocytes that leave the epidermal progenitor layer and differentiate first express early differentiation proteins, such as keratin 1 (K1) and K10, and, upon subsequent migration to outer epidermal layers, express terminal differentiation proteins, such as filaggrin (Flg), loricrin (Lor), trichohyalin-like 1 (Tchhl1), and the late cornified envelope proteins (LCEs) (Fuchs and Horsley 2008). The majority of these terminal differentiation proteins are encoded by genes located within the epidermal differentiation complex (EDC) on human chromosome 1q21, which corresponds to mouse chromosome 3 (Kypriotou et al. 2012). In basal keratinocytes, EDC genes are transcriptionally repressed by a chromatin remodeling complex referred to as the Polycomb repressor complex (PRC) (Simon and Kingston 2009; Beck and Blanpain 2012). PRC2—composed of Ezh2 (or Ezh1), Eed, Suz12, and Rbbp7/4—is recruited to chromatin, and Ezh2 mediates the trimethylation of histone H3 on Lys27 (H3K27me3). This histone mark presents a docking site for PRC1, which actively participates in gene silencing by monoubiquitination of H2A at Lys119 and subsequent chromatin compaction (Simon and Kingston 2009). It was previously reported that loss of Ezh2 in the epidermis induced precocious epidermal differentiation by up-regulation of EDC genes. Mechanistically, it was proposed that EDC gene expression was due to increased AP-1 transcription factor activity in the absence of the repressive H3K27me3 mark (Ezhkova et al. 2009). How AP-1 and PRC proteins fine-tune EDC gene expression remains to be determined.
AP-1 proteins consist of members of the Fos (c-Fos, FosB, Fra-1, and Fra-2) and Jun (c-Jun, JunB, and JunD) families and act as sequence-specific homodimeric and heterodimeric transcription factors, which are activated by mitogen-activated protein (MAP) kinases, including JNK, p38, and ERK (Eferl and Wagner 2003). Deregulated AP-1 expression is found in skin biopsies of patients with inflammatory/proliferative skin diseases, such as psoriasis, and epithelial cancers, such as squamous cell carcinomas (SCCs) (Zenz et al. 2005; Guinea-Viniegra et al. 2012, 2014; Briso et al. 2013; Eckert et al. 2013). The generation of genetically engineered mouse models (GEMMs) has provided insights into the important functions of Jun proteins as well as c-Fos in epidermal homeostasis, inflammation, and cancer (Zenz et al. 2003; Meixner et al. 2008; Guinea-Viniegra et al. 2009, 2012, 2014; Bakiri et al. 2011; Briso et al. 2013; Schonthaler et al. 2013). In contrast, the functions of the Fos-related proteins Fra-1 and Fra-2 in the epidermis are rather poorly understood.
Here we show that Fra-2 activity is necessary for epidermal barrier acquisition and EDC gene expression. Mechanistically, we demonstrate that Fra-2 regulates EDC gene expression by direct promoter binding upon loss of Ezh2-mediated methylation and activation through ERK1/2-dependent phosphorylation.
Results
Fra-2 induces terminal epidermal differentiation
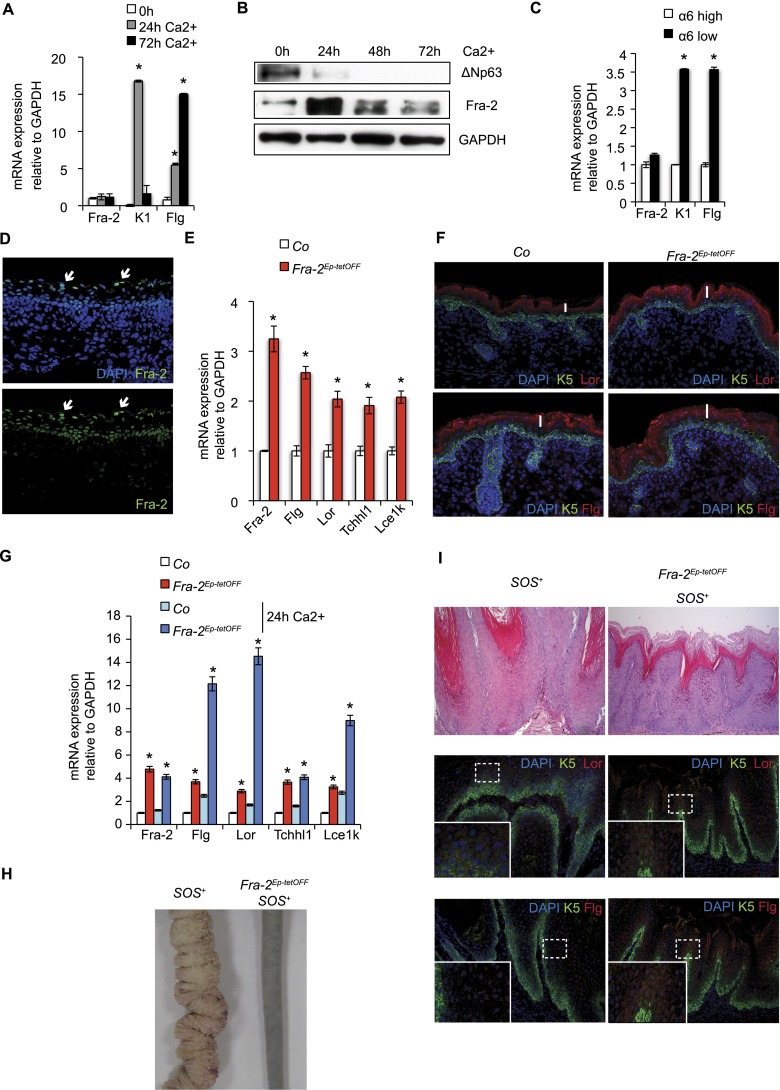
To define the role of Fra-2 in epithelial homeostasis, Fra-2 expression was evaluated in cultured mouse keratinocytes (mKCs) at basal conditions and upon calcium (Ca2+)-induced differentiation. mRNA levels of Fra-2 did not change during mKC differentiation (Fig. 1A). However, increased Fra-2 protein levels were detected upon Ca2+ treatment (Fig. 1B). Gene expression analyses of FACS-purified epidermal keratinocytes obtained from newborn pups indicated no difference of Fra-2 mRNA between basal mKCs (α6 integrin high) and differentiated mKCs (α6 integrin low) (Fig. 1C). In skin tissue sections, immunofluorescence (IF) staining revealed that Fra-2 protein was expressed in all epidermal layers. However, a gradient with the highest expression in terminally differentiated keratinocytes was observed (Fig. 1D, arrows).
Figure 1.
Fra-2 induces terminal epidermal differentiation. (A) Fra-2 mRNA expression during Ca2+-induced in vitro mKC differentiation. K1 and Flg indicate induction of early and terminal epidermal differentiation markers. n = 3; (*) P < 0.05; bars represent mean ± standard deviation (SD). (B) Fra-2 protein expression during Ca2+-induced in vitro mKC differentiation. Loss of ΔNp63 indicates induction of differentiation. n = 4. (C) Fra-2 mRNA expression of basal (α6 integrin high) and differentiated (α6 integrin low) mKCs isolated from newborn pups. K1 and Flg indicate induction of early and terminal epidermal differentiation markers. n = 3; (*) P < 0.05; bars represent mean ± SD. (D) Fra-2 IF of wild-type skin at E17.5. n = 6; 40× magnification. (E) Differential EDC gene expression of Flg, Lor, Tchhl1, Lce1k, and Fra-2 expression in epidermal samples of mutants ectopically expressing Fra-2 (Fra-2Ep-tetOFF) compared with control littermates at E17.5. n = 6 per genotype; (*)P < 0.05; bars represent mean ± SD. (F) Basal cell marker expression (K5, green) and late differentiation marker expression (Lor, red, top panel; Flg, red, bottom panel) in Co and Fra-2Ep-tetOFF skin at E17.5. n = 6 per genotype; 40× magnification. White bars indicate the thickness of differentiated epidermal layers, which is quantified in Supplemental Figure S1C. (G) Differential EDC gene expression and Fra-2 expression in primary Fra-2Ep-tetOFF mKCs compared with control mKCs under basal conditions and upon Ca2+ treatment for 24 h in vitro. n = 3; (*) P < 0.05; bars represent mean ± SD. (H) Macroscopic appearance of SOS+ and Fra-2Ep-tetOFF SOS+ papillomas at 5 wk of age. n = 6 per genotype. (I) Analyses of SOS+ and Fra-2Ep-tetOFF SOS+ papillomas. H&E (top panel), basal cell marker expression (K5, green) and late differentiation marker expression (Lor, red, middle panel; Flg, red, bottom panel) in SOS+ and Fra2Ep-tetOFF SOS+ papillomas at 5 wk of age. 20× magnification. The insert depicts 40× magnification. Lor and Flg mean fluorescence intensity is quantified in Supplemental Figure S1D. n = 6 per genotype.
To analyze a potential functional role of Fra-2 in regulating epidermal differentiation, GEMMs with epithelial-specific ectopic expression as well as specific deletion of Fra-2 were generated. The expression of genes located with in the EDC—such as Flg, Lor, Tchhl1, and Lce1k—was analyzed in these model systems. While the proliferation of basal mKCs remained unaffected upon ectopic expression of Fra-2 in K5-expressing epithelia (Fra-2Ep-tetOFF) (Supplemental Fig. S1A,B), a significant increase of EDC gene transcripts was detected in epidermal samples of embryos at embryonic day 17.5 (E17.5) (Fig. 1E). The increase of EDC gene expression was accompanied by an increase of terminally differentiated epidermal layers in Fra-2Ep-tetOFF mutants (Fig. 1F; quantified in Supplemental Fig. S1C). These findings suggest that Fra-2 induces terminal epidermal differentiation.
Since moderate changes in EDC gene expression were detected upon ectopic Fra-2 expression, the consequences of increased Fra-2 expression on mKC homeostasis under stress conditions were next investigated. Primary mKCs isolated from Fra-2Ep-tetOFF mutants underwent precocious differentiation characterized by increased expression of terminal differentiation genes at basal conditions, which was more pronounced upon Ca2+ treatment (Fig. 1G).
To validate these findings in a more physiologically relevant GEMM, Fra-2 was ectopically expressed in papilloma-prone K5-SOS-F transgenic mice (Fra-2Ep-tetOFF SOS+). Skin papilloma growth was strongly suppressed, with significantly smaller lesions in Fra-2Ep-tetOFF SOS+ mice (Fig. 1H,I, H&E, top panel). Importantly, increased expression of EDC genes was detected by IF (Fig. 1I, middle and bottom panels; quantified in Supplemental Fig. S1D) and mRNA analyses (Supplemental Fig. S1E), while no change in proliferation was observed (Supplemental Fig. S1F,G). These results demonstrate that epidermal Fra-2 can induce EDC gene expression under stress conditions and functions as a suppressor of papilloma growth due to the induction of epidermal differentiation.
Fra-2/AP-1 is necessary for epidermal differentiation
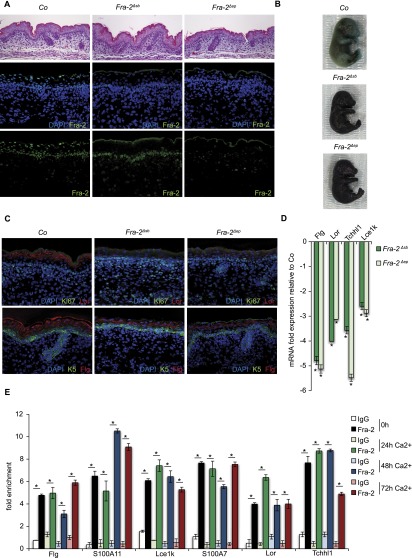
GEMMs with Fra-2 deletion in the differentiated compartment of the epidermis (suprabasal deletion with FoxN1-Cre; Fra-2Δsb) and with Fra-2 deletion in the entire epidermis (basal deletion with K5-Cre; Fra-2Δep) were generated (Fig. 2A). While macroscopically indistinguishable from control littermates at E17.5, Toluidine blue dye penetration assays revealed a persistent defect in skin barrier formation in both Fra-2Δsb and Fra-2Δep mutants (Fig. 2B). The observed skin barrier defect was likely a consequence of incomplete epidermal differentiation involving a significant reduction in the expression of EDC genes (Fig. 2C,D; Supplemental Fig. S2A). Thus, Fra-2 appears necessary for proper terminal epidermal differentiation. The proliferation status of basal mKCs remained unaffected in Fra-2Δsb and Fra-2Δep mutants (Supplemental Fig. S2B,C). Although Fra-2 is also expressed in basal cells, the phenotype of Fra-2Δsb mutants suggests that Fra-2 mainly functions in the differentiated compartment of the epidermis.
Figure 2.
Fra-2/AP-1 is necessary for epidermal differentiation regulating EDC genes. (A) H&E and Fra-2 IF of Co (Fra-2f/f), Fra-2Δsb (Fra-2f/f; FoxN1 Cre+/T), and Fra-2Δep (Fra-2f/f; K5 Cre+/T) embryos at E17.5. n = 6 per genotype; 40× magnification. (B) Toluidine blue dye penetration assay in Co, Fra-2Δsb, and Fra-2Δep embryos at E17.5. n = 6 per genotype. (C) Proliferation/basal cell marker expression (Ki67, green, top panel; K5, green, bottom panel) and late differentiation marker expression (Lor, red, top panel; Flg, red, bottom panel) in Co, Fra-2Δsb, and Fra-2Δep embryos at E17.5. n = 6 per genotype; 40× magnification. (D) Differential EDC gene expression in Fra-2Δsb and Fra-2Δep embryos compared with control littermates at E17.5. n = 4 per genotype; (*) P < 0.05; bars represent mean ± SD. (E) ChIP analyses of Fra-2 at EDC promoters during wild-type mKC differentiation (0–72 h Ca2+). n = 5; (*) P < 0.05; bars represent mean ± SD.
To test whether Fra-2 directly induces EDC genes by promoter binding, chromatin immunoprecipitation (ChIP) experiments in basal mKCs and during differentiation were performed. mRNA from these same samples was collected to confirm induction of epidermal differentiation and correlate binding with gene expression (Supplemental Fig. S2D). Fra-2 associated with EDC promoters at conserved AP-1 consensus sites (Fig. 2E). Surprisingly, Fra-2 was bound at EDC gene promoters under not only differentiation conditions but also basal conditions when EDC transcripts are not expressed (Supplemental Fig. S2D). Collectively, these findings indicate that even though expressed and bound at target gene promoters, Fra-2 remains inactive in basal cells, suggesting that transcriptional repressors might coregulate EDC genes together with AP-1 transcription factors.
Fra-2 and Ezh2 coregulate epidermal differentiation genes and physically interact
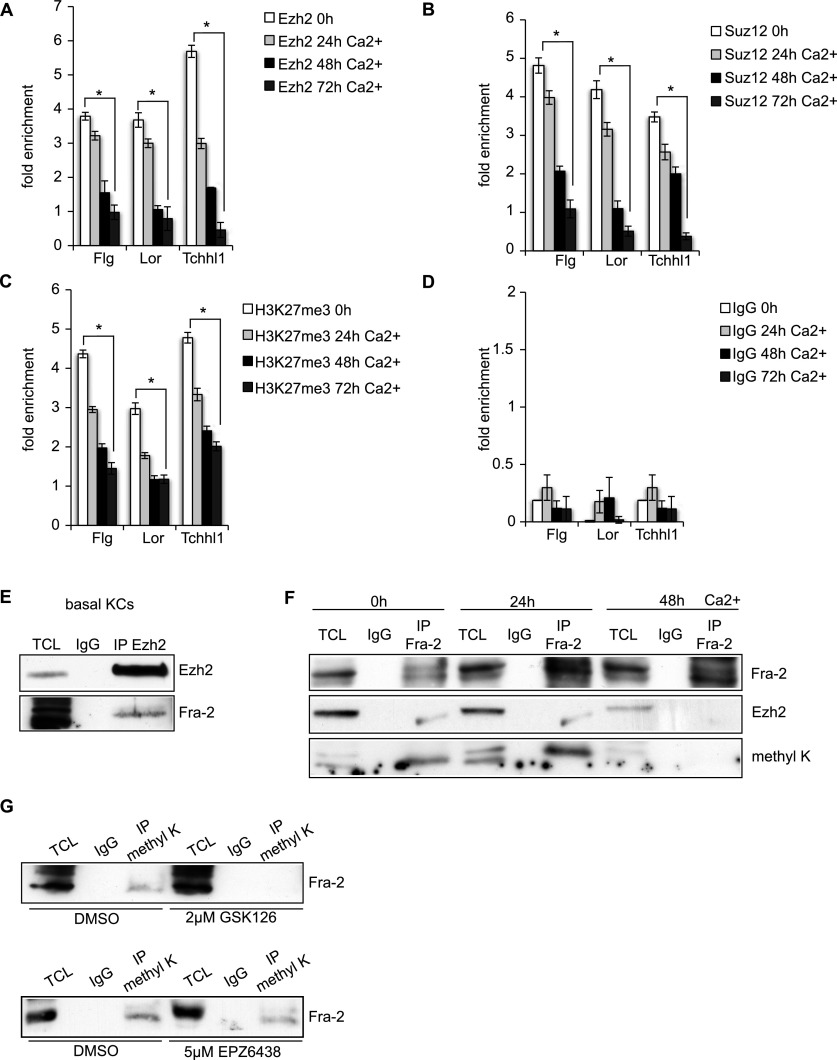
To study the mechanism by which Fra-2 induces EDC gene activation, the presence of transcriptional repressors at EDC gene promoters in basal cells was investigated. Since PRC2 represses EDC genes (Ezhkova et al. 2009; Mejetta et al. 2011), ChIP studies were performed using antibodies against the PRC2 subunits Ezh2 and Suz12 and the repressive histone mark H3K27me3. The transcription start sites (TSSs) of Fra-2-bound EDC genes were enriched for Ezh2 and Suz12 and positive for H3K27me3 in basal cells. Importantly, the associations of PRC2 subunits as well as H3K27me3 levels were reduced upon Ca2+-induced differentiation (Fig. 3A–D). These findings are consistent with the fact that Ezh2 and Suz12 protein levels were drastically reduced during mKC differentiation (Supplemental Fig. S3A). In addition to the reduction of the repressive H3K27me3 modification, histone modifications associated with transcriptional activation, such as H3K4me3 and H3K27me3S28p, were increased upon mKC differentiation (Supplemental Fig. S3B,C). These results show that Fra-2/AP-1 and Ezh2/PRC2 co-occupy EDC promoters in basal keratinocytes and that the removal of PRC2 subunits correlates with Fra-2-mediated transcriptional initiation.
Figure 3.
Fra-2 and Ezh2 coregulate epidermal differentiation genes and physically interact. (A–D) ChIP analyses of Ezh2 (A), Suz12 (B), H3K27me3 (C), and negative control IgG (D) during mKC differentiation (0–72 h Ca2+). n = 3; (*) P < 0.05; bars represent mean ± SD. (E) Co-IP of Ezh2 and Western blot analysis of Ezh2 and Fra-2 in basal mKCs. n = 5. (TCL) Total cell lysate. (F) Co-IP of Fra-2 during mKC differentiation (0–48 h Ca2+) and Western blot analysis of Fra-2, Ezh2, and methylated lysine (methyl K). n = 5. (G) Immunoprecipitation of methyl K and Western blot of Fra-2 upon Ezh2 inhibition with GSK126 and EPZ6438 in basal mKCs. n = 3.
Besides modifying histones, Ezh2 methylates nonhistone substrates, such as some transcription factors, thereby regulating their transcriptional activity (He et al. 2012a; Lee et al. 2012; Xu et al. 2012; Kim et al. 2013). Therefore, we investigated a potential histone methylation-independent role of Ezh2 on Fra-2 during epidermal differentiation employing coimmunoprecipitations (co-IPs). Ezh2 was found to coprecipitate with Fra-2 in protein lysates isolated from basal keratinocytes (Fig. 3E). Co-IP experiments using an anti-Fra-2 antibody revealed that Fra-2 coprecipitated with Ezh2 in basal mKCs and that the interaction was reduced upon mKC differentiation, consistent with the reduction of Ezh2 protein levels (Fig. 3F). Since Ezh2 methylates specific lysine residues of histone proteins and Ezh2 interacts with Fra-2, the methylation status of Fra-2 during mKC differentiation was investigated. Using immunoprecipitated Fra-2 and an antibody against pan-methyl lysine, methylated Fra-2 was detected (Fig. 3F). In agreement with PRC2 reduction upon differentiation, Fra-2 methylation was absent in differentiated mKCs (Fig. 3F). To elucidate whether chromatin-bound Fra-2 was methylated and interacted with Ezh2, Fra-2 ChIP experiments followed by both DNA and protein recovery were performed. Quantitative PCR along with Western blot analyses indicated that at least a fraction of Fra-2 bound at EDC promoters was found methylated and interacted with Ezh2 (Supplemental Fig. S3D,E).
To interrogate a role of Ezh2 in Fra-2 methylation, Ezh2 was pharmacologically targeted using two small molecules, GSK126 and EPZ6438, inhibitors of the methyltransferase domain of Ezh2. The methylation status of Fra-2 protein in basal keratinocytes was assessed by immunoprecipitation for methylated lysine and Western blot for Fra-2. In both cases, Fra-2 methylation was decreased upon Ezh2 inhibition (Fig. 3G).
These data demonstrate that Fra-2 and Ezh2 interact and that Fra-2 is methylated on lysine residues in basal cells, correlating with transcriptional inactivity at the EDC locus. Interestingly, the pharmacological inhibition of Ezh2 was not sufficient to induce terminal mKC differentiation. However, GSK126 inhibitor-treated cells differentiated prematurely upon Ca2+ treatment (Supplemental Fig. S3F). These findings indicate that an additional stimulus is required to induce Fra-2 activity.
Fra-2 lysine methylation alters Fra-2 transcriptional activity in basal mKCs
To investigate the molecular nature of Fra-2 lysine methylation, endogenous Fra-2 was immunoprecipitated from basal mKC protein extracts followed by mass spectrometry (MS) analyses. In two independent analyses (see the Materials and Methods), MS analyses identified monomethylation and dimethylation of Lys104 (K104), as shown in Figure 4, A–D.
Figure 4.
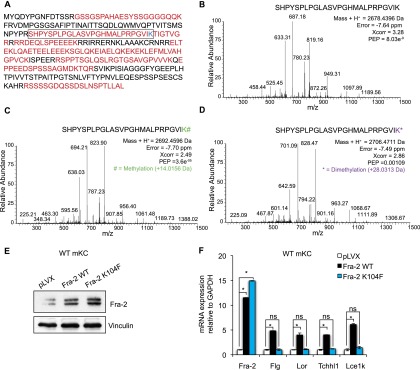
Fra-2 transcriptional activity is altered by Lys104 methylation. (A) Sequence coverage of Fra-2 identified by liquid chromatography-tandem MS (LC-MS/MS) analysis, in which red residues represent the identified peptides. (B) MS/MS spectrum of unmodified Fra-2 89–104 peptide (framed in A). Mass error, identification score, and posterior error probability (PEP) are shown for the Fra-2 89-104 peptide. (C) MS/MS spectrum of the Fra-2 89–104 peptide in which monomethylation was unambiguously assigned to K104 residue. Mass error, identification score, and PEP are shown for the Fra-2 89–104 peptide. (D) MS/MS spectrum of the Fra-2 89–104 peptide in which dimethylation was unambiguously assigned to the K104 residue. Mass error, identification score, and PEP are shown for the Fra-2 89–104 peptide. (E) Fra-2 protein expression of primary mKCs infected with lentiviruses expressing Fra-2 wild type (Fra-2 WT) or Fra-2 K104F compared with empty vector-infected cells (pLVX). n = 3. (F) Differential Fra-2 and EDC gene expression of primary mKCs expressing Fra-2 wild type and Fra-2 K104F compared with empty vector-infected cells. n = 3.
Site-directed mutagenesis was next performed to substitute K104 with phenylalanine (K104F), mimicking lysine methylation (Huq et al. 2007). Primary wild-type mKCs were infected with lentiviruses expressing full-length, Flag-tagged, Fra-2 (Fra-2 wild type), Fra-2 K104F, or empty vector (pLVX). Efficient Fra-2 expression was verified by Western blot and quantitative RT–PCR (qRT–PCR) with primers specific for total and ectopically expressed Fra-2 (Fig. 4E,F; Supplemental Fig. S4A). While lentivirus-mediated expression of Fra-2 wild type induced EDC gene expression in primary wild-type mKCs, expression of Fra-2 K104F did not (Fig. 4F). These experiments were also performed in primary mKCs carrying Fra-2 loxed alleles infected with Adeno-Cre (AdCre) viruses for in vitro Cre-mediated Fra-2 inactivation and Adeno-GFP (AdGFP) control viruses. Cre-mediated recombination was assessed by measuring nonfunctional Fra-2Δ-GFP fusion gene expression, which is produced upon excision of exon 3 of Fra-2 and fusion of exon 2 with a GFP reporter (Supplemental Fig. S4B; Eferl et al. 2007). Efficient Fra-2 wild-type and Fra-2 K104F expression was assessed by exogenous Fra-2-Flag and total Fra-2 expression in these cells (Supplemental Fig. S4C,D). While lentiviral expression of Fra-2 wild type restored the differentiation defect of primary Fra-2 knockout keratinocytes, the expression of the Fra-2 K104F mutant did not (Supplemental Fig. S4D). These results indicate that lysine methylation of K104 decreases the transcriptional activity of Fra-2 on EDC genes in primary mKCs.
Fra-2 is phosphorylated and stabilized by ERK1/2 upon mKC differentiation
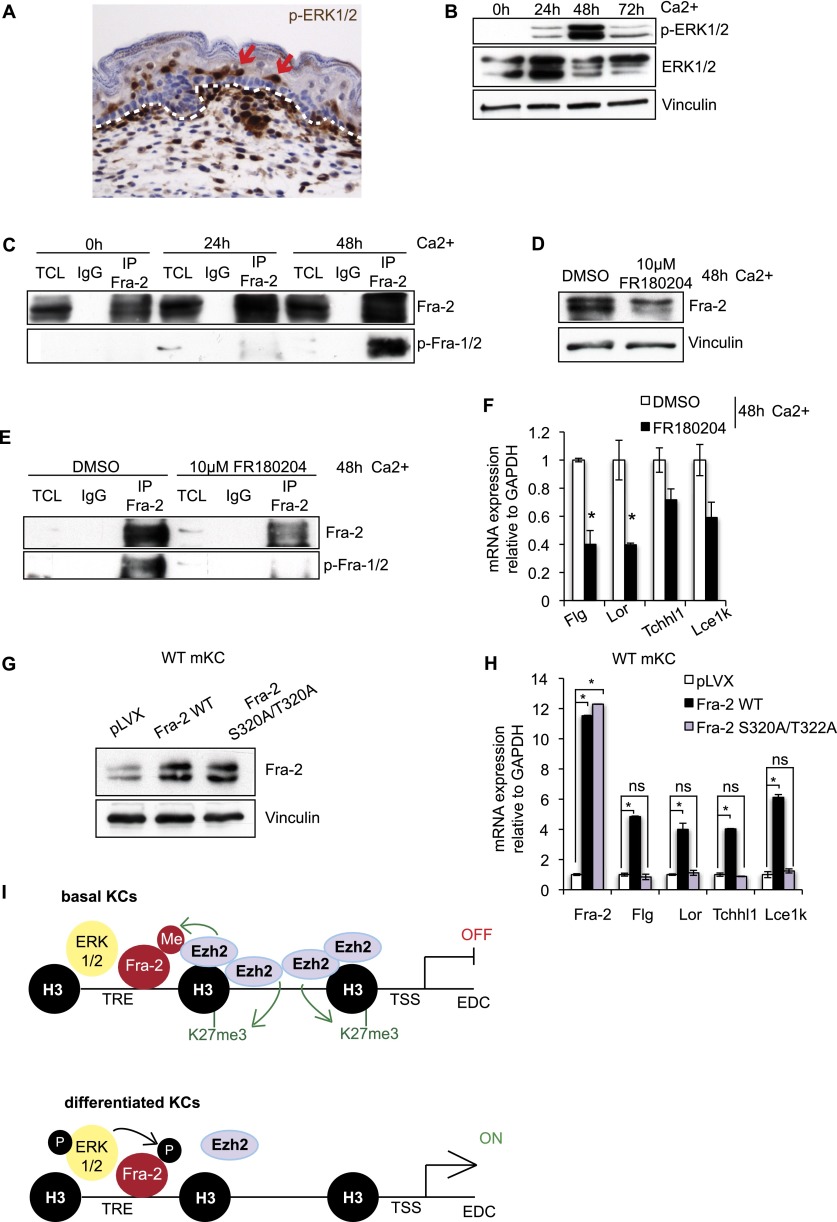
Since Fra-2 mRNA remained unaltered during mKC differentiation, although an increase of total Fra-2 protein was detected, we hypothesized that Fra-2 was stabilized at the protein level by post-translational modifications. ERK1/2 kinases were shown to stabilize Fra-2 protein by C-terminal Ser320 (S320) and Thr322 (T322) phosphorylation (Alli et al. 2013). We therefore investigated ERK1/2 activation during mKC differentiation. ERK1/2 phosphorylation was detected in suprabasal keratinocytes of developing embryos at E17.5 (Fig. 5A) and upon Ca2+-induced differentiation in vitro (Fig. 5B). To investigate the C-terminal Fra-2 serine phosphorylation, a phospho-Fra-1 antibody reacting with a conserved peptide (Ser267 on Fra-1 and Ser320 on Fra-2) was used on immunoprecipitated Fra-2 at different time points of mKC differentiation. C-terminal Fra-2 phosphorylation was increased upon Ca2+-induced differentiation, correlating with increased ERK1/2 phosphorylation (Fig. 5B,C).
Figure 5.
Fra-2 is phosphorylated and stabilized by ERK1/2 upon mKC differentiation. (A) p-ERK1/2 immunohistochemistry in the skin of wild-type embryos at E17.5. Arrows point toward suprabasal expression, and the dashed white line indicates basement membrane. n = 3. (B) ERK1/2 activation shown by p-ERK1/2 Western blot during in vitro mKC differentiation (0–72 h Ca2+). n = 3. (C) Immunoprecipitation of Fra-2 during mKC differentiation (0–48 h Ca2+) and Western blot analyses of Fra-2 and p-Fra-1/2. n = 3. (D) Western blot analysis of Fra-2 upon Ca2+ treatment (48 h) and ERK1/2 inhibition with FR180204. n = 3. (E) Immunoprecipitation of Fra-2 upon Ca2+ treatment (48 h) and ERK1/2 inhibition with FR180204 and Western blot of Fra-2 and p-Fra-1/2. n = 3. (F) mRNA expression analyses of EDC genes upon Ca2+ treatment (48 h) and ERK1/2 inhibition with FR180204. n = 3; (*) P < 0.05; bars represent mean ± SD. (G) Fra-2 protein expression of primary mKCs infected with lentiviruses expressing Fra-2 wild type (Fra-2 WT) or mutant Fra-2 deficient for C-terminal phospho-acceptor sites (Fra-2 S320A/T322A) compared with empty vector-infected cells. n = 3. (H) Differential Fra-2 and EDC gene expression of primary mKCs expressing Fra-2 wild type or Fra-2 S320A/T322A compared with empty vector-infected cells. n = 3. (I) Schematic model of EDC gene regulation by Fra-2, Ezh2, and ERK1/2: In basal cells, methylated Fra-2 (Fra-2-Me) bound at TPA-responsive elements (TREs) of EDC gene promoters, and interacting with Ezh2 prevents EDC gene expression. Upon mKC differentiation, Fra-2 becomes phosphorylated (Fra-2-P) by ERK1/2 and loses its methylation, and Ezh2 protein levels and H3K27 methylation at TSSs of EDC promoters are reduced, which together induce EDC gene expression.
To evaluate the role of ERK1/2 on Fra-2 phosphorylation, a small-molecule inhibitor of ERK1/2 (FR180204) was used. ERK1/2 inhibitor treatment led to reduced Fra-2 protein levels, reduced Fra-2 phosphorylation, and reduced EDC gene expression upon Ca2+ treatment (Fig. 5D–F). These data indicate that Fra-2 is stabilized by C-terminal phosphorylation during mKC differentiation and that ERK1/2 activation correlates with Fra-2 phosphorylation.
We next functionally validated the role of C-terminal Fra-2 phosphorylation on EDC gene expression and mutated S320 and T322 to alanine (S320A/T322A), which cannot be phosphorylated. Primary mKCs were infected with lentiviruses expressing Fra-2 wild type, Fra-2 S320A/T322A, or empty vector. Efficient Fra-2 expression was verified by Western blot and qRT–PCR with primers specific for total and ectopically expressed Fra-2 (Fig. 5G,H; Supplemental Fig. S5A). While lentivirus-mediated expression of Fra-2 wild type induced EDC gene expression, expression of Fra-2 S320A/T322A did not (Fig. 5H). These findings were independently confirmed in Fra-2 knockout mKCs. Importantly, lentivirus-mediated expression of Fra-2 S320A/T322A mutants did not restore the differentiation defect of primary Fra-2 knockout mKCs, while expression of Fra-2 wild type did (Supplemental Fig. S5B–D). These findings indicate that the ability of Fra-2 to induce EDC gene expression is reduced upon mutation of C-terminal phospho-acceptor sites.
In conclusion, these results show an inverse kinetic of lysine methylation—loss of interaction with Ezh2—and ERK1/2-mediated serine phosphorylation of Fra-2. We hypothesize that Fra-2 phosphorylation occurs independently of Fra-2 methylation, since the inhibition of Ezh2 and the loss of Fra-2 methylation did not result in an increase in phospho-Fra-2 levels or ERK activation (Supplemental Fig. S5E,F).
At EDC promoters, methylated but not phosphorylated Fra-2 correlates with EDC gene inactivation, while loss of Fra-2 methylation, ERK1/2 activation, and Fra-2 phosphorylation correlates with active Fra-2-dependent gene expression (Fig. 5I).
Discussion
Here we report that Fra-2 is an Ezh2 substrate in basal cells and acts as a key regulator of terminal epidermal differentiation. Mechanistically, Fra-2 binds conserved AP-1 consensus sites in EDC promoters and is able to induce transcriptional activation when phosphorylated by ERK1/2. We found that, unlike other master regulators of terminal epidermal differentiation such as Klf4, which is expressed suprabasally (Segre et al. 1999), Fra-2 is expressed and bound at EDC promoters in basal cells, where it is unphosphorylated, is methylated on K104, and remains transcriptionally inactive. Using genetic loss-of-function approaches in mice, we demonstrate that loss of Fra-2 in suprabasal cells is sufficient to cause skin barrier defects. These data support the observation that even though it is expressed in all epidermal keratinocytes, Fra-2 mainly functions in the differentiated layers of the epidermis. Furthermore, we show that ectopic Fra-2 expression is sufficient to induce terminal epidermal differentiation, leading to suppression of papilloma growth by inducing precocious keratinocyte differentiation.
We speculate that Fra-2 is kept transcriptionally inactive in basal cells due to methylation by Ezh2. Upon Ca2+-induced differentiation, the concomitant loss of interaction with Ezh2, Fra-2 methylation, and ERK1/2-mediated phosphorylation correlates with EDC gene expression and keratinocyte differentiation. Using Ca2+ stimulation to induce keratinocyte differentiation, we demonstrate that basal keratinocytes differentiate prematurely upon inhibition of Ezh2. These findings are in agreement with published results of in vivo gene targeting of Ezh2 in the epidermis. Ezh2-null keratinocytes differentiate prematurely and express higher levels of EDC genes due to increased AP-1 transcription factor activity (Ezhkova et al. 2009). It has been shown previously that Ezh2 can modify the activity of some transcription factors through methylation (He et al. 2012a; Lee et al. 2012; Xu et al. 2012; Kim et al. 2013). Employing MS-based approaches, we identified monomethylation and dimethylation on K104 of Fra-2 in basal keratinocytes. Additionally, we demonstrated by lentiviral expression of a Fra-2 mutant mimicking lysine methylation that this modification decreases the transcriptional activity of Fra-2 on EDC genes. Interestingly, K104 of Fra-2 has been found acetylated in an independent study (Park et al. 2013), suggesting a potential role of this residue in regulating the transcriptional activity of this protein. We hypothesize that Fra-2-K104 methylation influences the interaction of Fra-2 with transcriptional coactivators, as has been described for methylated GATA4 (He et al. 2012a).
During the past decade, several independent studies established that Ezh2 is overexpressed in solid tumors, and high expression is associated with aggressive cancers and progression (Hock 2012). In human cancers, it remains unclear whether H3K27 methylation is the only way by which Ezh2 controls tumorigenesis. Since Ezh2 has other substrates beyond histone H3, it was proposed that Ezh2 might facilitate tumorigenesis by regulating the activity of nonhistone substrates such as transcription factors (He et al. 2012b; Kim et al. 2013). Interestingly, skin papillomas of poorly differentiated K5-SOS-F mice show high levels of Ezh2 and express Fra-2 (data not shown). Importantly, ectopic expression of Fra-2 in this background resulted in reduced papilloma growth due to precocious differentiation. These findings suggest that the ratio of expression of Ezh2 to its substrates is crucial in regulating epidermal differentiation and tumorigenesis. Further research will be necessary to assess the expression of Ezh2 and Fra-2 in human skin diseases characterized by epidermal differentiation defects in order to evaluate the possible clinical benefit of blocking Ezh2 as a therapeutic strategy.
Additionally, we identified an important function of the MAP kinases ERK1/2 in terminal mKC differentiation by phosphorylating and stabilizing Fra-2. ERK1/2 are activated upon Ca2+-induced differentiation via PKCα, and the inhibition of ERK1/2 results in reduced expression of terminal keratinocyte differentiation genes (Schmidt et al. 2000; Seo et al. 2004). We provide further mechanistic insights demonstrating that Fra-2 is phosphorylated by ERK1/2 and acts as an ERK1/2-dependent inducer of terminal differentiation by regulating EDC gene expression. Additionally, in vivo analyses of ERK1/2 phosphorylation in developing embryos showed a clear nuclear pattern in suprabasal cells, indicating essential functions of activated ERK1/2 in epidermal barrier acquisition. Previously published studies revealed that ERK2 phosphorylates Fra-2 on five serine and threonine residues (Alli et al. 2013). Targeted mutagenesis of these residues demonstrated that the C-terminal phosphorylation of Fra-2 on Ser320 and Thr322 results in increased protein stability (Alli et al. 2013). Using an antibody against phospho-Ser320, which is conserved among Fra-1, Fra-2, and c-Fos, we found that C-terminal Fra-2 phosphorylation is increased upon mKC differentiation, correlating with ERK1/2 activation and Fra-2 transcriptional activity at EDC target genes. In addition, we functionally validated the role of C-terminal Fra-2 phosphorylation in primary mKCs by expression of phospho-deficient Fra-2 mutants, which exhibited reduced transcriptional activity.
In summary, we propose a novel mechanism for how Fra-2/AP-1 is regulated through lysine methylation by Ezh2 and C-terminal serine phosphorylation by ERK1/2 (Fig. 5I). It will be interesting to explore in future studies whether this methyl–phospho switch is a general mechanism of transcription factor regulation and study the possible implications in skin diseases and beyond.
Materials and methods
Mice
All mouse experiments were performed in accordance with local and institutional regulations. Fra-2Δsb mice were generated by crossing Fra-2f/f mice (Eferl et al. 2007) with mice expressing Cre recombinase under the control of the FoxN1 promoter (Soza-Ried et al. 2008). Fra-2Δep mice were generated by crossing Fra-2f/f mice with mice expressing Cre recombinase from the K5 promoter (Zenz et al. 2005). Control (Cre-negative mice), Fra-2Δsb mutants (Fra-2f/f; FoxN1 Cre+/T), and Fra-2Δep mutants (Fra-2f/f; K5 Cre+/T) were identified by genotyping PCR on tail biopsies.
Fra-2Ep-tetOFF mice were generated by crossing mice carrying a Flag-tagged Fra-2 cDNA knock-in controlled by a tetracycline operator downstream from the col1a1 gene (col Fra-2+/KI) (Bozec et al. 2013) to mice expressing tTA from the K5 promoter (Diamond et al. 2000). Control (col Fra-2+/KI; K5tTA+/+) and Fra-2Ep-tetOFF mutants (col Fra-2+/KI; K5tTA+/T) were identified by genotyping PCR on tail biopsies. Mice were generated and maintained in the absence of doxycycline so that transgene expression was turned on during embryonic development.
Fra-2Ep-tetOFF SOS+ mice were generated by crossing Fra-2Ep-tetOFF mice to K5-SOS-F mice (Sibilia et al. 2000) (EGFRwa/wa and K5 SOS+/T). SOS+ (col Fra-2+/KI, K5tTA+/+, EGFRwa/+, and SOS+/T) and Fra-2Ep-tetOFF SOS+ (col Fra-2+/KI, K5tTA+/T, EGFRwa/+, and SOS+/T) mice were generated and maintained in the absence of doxycycline so that transgene expression was turned on during embryonic development.
Histology, immunohistochemistry, and IF analyses
Back skins from embryos at E17.5 and tails skins from adult mice were fixed overnight at 4°C with buffered 4% PFA and embedded in paraffin. Skin sections were cut at 3 μm. H&E and immunohistochemistry were performed as previously described (Zenz et al. 2005). p-ERK1/2 (Thr202/Tyr204) (1:1000; no. 4370, Cell Signaling) and Ki67 (1:200; Dako) primary antibodies were used for immunohistochemistry. Sections were analyzed by light microscopy (DM2500, Leica).
Back skins from embryos at E17.5 and tail skins from adult mice were frozen, embedded in OCT compound, and cut at 10 μm. Slides were blocked in PBS, 0.1% Triton X, 1% BSA, and 1% donkey serum; washed in PBS and 0.1% Triton X; mounted with Mowiol + DAPI (VectaShield); and analyzed by confocal microscopy. Primary antibodies used for IF were Fra-2 (1:50; CNIO Monoclonal Antibody Unit), K5 (1:1000; Covance), Flg (1:1000; Covance), Lor (1:1000; Covance), Ki67 (1:200; Dako), and Ezh2 (1:1000; no. 5246, Cell Signaling). Alexa fluor secondary antibodies conjugated to 488-nm or 555-nm absorbing fluorophores (Invitrogen) were used.
Mutagenesis and cloning
For the generation of Fra-2 methyl-mutations and phospho-mutations, site-directed mutagenesis was carried out on Flag-tagged mouse Fra-2 using the QuikChange Lightening site-directed mutagenesis kit (Stratagene, Agilent Technologies) following the manufacturer’s protocol. After sequencing, Flag-tagged Fra-2 wild type, Fra-2 K104F, and Fra-2 S320A/T322A were subsequently cloned into pLVX for lentiviral expression.
Cell culture
For long-term culture, mKCs were isolated from the back skin of newborn mice using trypsin (Gibco) and cultured in medium containing 0.05 mM Ca2+ and 15% FBS as previously described (Nowak and Fuchs 2009). To induce differentiation in culture, calcium was raised from 0.05 mM to 1.5 mM for the indicated time periods. Ezh2 methyltransferase activity was inhibited using GSK126 (GlaxoSmithKline) and EPZ6438 (Epizyme) inhibitors at 5 μM for 6–48 h. Erk1/2 kinase activity was inhibited using FR180204 (Tocris Bioscience) at 5 μM for up to 48 h.
Isolation and short-term culture of primary mKCs from the tail skin of wild-type mice, Fra-2f/f mice, Fra-2Ep-tetOFF mice, and littermate controls were performed as described (Blanpain et al. 2004). Keratinocyte medium was changed to K-SFM (Gibco) 12 h after plating. Fra-2Ep-tetOFF keratinocytes were cultured in the absence of doxycycline. Calcium concentration was raised to 1.5 mM for differentiation experiments.
Lentiviruses were produced in 293T cells by HEPES-buffered saline-CaCl2 phosphate-mediated transient cotransfection of pLVX plasmids and pLP1, pLP2, and pLP/VSVG packaging plasmids. Primary wild-type cells were infected with lentiviruses on days 1 and 2 after plating. Primary Fra-2f/f cells were infected with AdGFP/AdCre viruses (University Iowa) on days 1 and 4 and with lentiviruses on days 2 and 3 after plating. Cells were collected for mRNA and protein analyses 5 d after the last lentiviral infection.
Protein isolation
Protein isolation from epidermal tissue or cultured cells for Western blot and immunoprecipitation was performed in RIPA buffer (50 mM Tris HCl at pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% NP-40) containing protease and phosphatase inhibitors (Roche).
Western blot
Western blot analysis was performed according to standard procedures using the following antibodies: Fra-2 (1:100; CNIO Monoclonal Antibody Unit), Ezh2 (1:1000; no. AC22 Cell Signaling), p-Fra-1 (Ser265) (1:1000; no. 3880, Cell Signaling), pan-methyl lysine (1:200; ab174719, Abcam), p-Ser (1:500; P5747, Sigma), p-ERK1/2 (Thr202/Tyr204) (1:1000; no. 4370), ERK1/2 (1:1000; no. 4695, Cell Signaling), ΔNp63 (1:500; sc-8609, Santa Cruz Biotechnology), Suz12 (1:1000; ab12073, Abcam), Flg (1:1000; Covance), Lor (1:1000; Covance), and vinculin (1:1000; V9131, Sigma).
Co-IP
Two micrograms to 5 μg of Fra-2 (CNIO Monoclonal Antibody Unit), Ezh2 (5246, Cell Signaling), and pan-methyl lysine (ab174719, Abcam) antibodies were incubated with 20 μL of Dynabeads protein G (Invitrogen) for 3 h to overnight. One milligram of precleared cell extract per immunoprecipitation was used as input material and incubated with antibody-conjugated beads overnight at 4°C under rotation. Immunoprecipitations were extensively washed with RIPA buffer and eluted with standard Laemmli buffer for Fra-2, methyl lysine, and p-Fra-1 Western blot analysis or with XT sample buffer without β-mercaptoethanol for Ezh2 Western blot analysis.
Immuno-complex elution and digestion for MS
Captured proteins were released from the antibody-coupled beads by incubation at 45°C with 200 μL of 8 M urea in 100 mM Tris-HCl (pH 8.0). Protein samples were processed using the filter-aided sample preparation (FASP) method (Wisniewski et al. 2009b) and digested overnight at 25°C using endoproteinase Lys-C (Wako Pure Chemical Industries) at a 1:50 enzyme to protein ratio followed by digestion with trypsin (Promega) at a 1:100 enzyme to protein ratio for 6 h at 37°C. Peptides were subjected to tip-based strong anion exchange (SAX) fractionation using the described protocol (Wisniewski et al. 2009a). In addition, a second immunoprecipitation experiment was carried out in which the protein eluate was separated by SDS-PAGE. For in-gel digestion, bands were excised, and proteins were reduced with 15 mM TCEP in 50 mM ammonium bicarbonate for 45 min at 60°C and alkylated with 50 mM 2-chloroacetamide in 50 mM ammonium bicarbonate for 30 min at 25°C in the dark. Proteins were subsequently digested overnight at 37°C with trypsin at a 1:200 enzyme to protein ratio.
Liquid chromatography-tandem MS (LC-MS/MS) analysis
Desalted peptides were separated by reversed-phase chromatography using a nanoLC ultra system (Eksigent) directly coupled with a LTQ-Orbitrap Velos instrument (Thermo Fisher Scientific) via a nanoelectrospray source (ProxeonBiosystem). Peptides were loaded onto the column (Reprosil-Pur C18-AQ, 3 μm, 200 × 0.075 mm, Dr. Maisch GmbH) with a previous trapping column step (0.10 × 20 mm, Reprosil-Pur C18-AQ, 3 μm, 120 Å) during 10 min with a flow rate of 2.5 μL of loading buffer (0.1% FA) per minute. Elution from the column was made with a 120-min linear gradient or a 45-min linear gradient (in-solution and in-gel, respectively; buffer A: 4% ACN, 0.1%FA; buffer B: 100% ACN, 0.1% FA) at 300 nL/min. The peptides were directly electrosprayed into the mass spectrometer using a PicoTip emitter (360-μm OD/20-μm ID, 10-μm tip ID, new objective) and a 1.4-kV spray voltage with a heated capillary temperature of 325°C and S-Lens of 60%. Mass spectra were acquired in a data-dependent manner, with an automatic switch between the MS and MS/MS scans using a top 15 or top five method (in-solution and in-gel, respectively) with a threshold signal of 800 counts. MS spectra were acquired with a resolution of 60,000 (FWHM) at 400 m/z in the Orbitrap, scanning a mass range between 350 and 1500 m/z. Peptide fragmentation was performed using collision-induced dissociation (CID), and fragment ions were detected in the linear ion trap. The normalized collision energy was set to 35%, the Q-value was set to to 0.25, and the activation time was set to 10 msec. The maximum ion injection times for the survey scan and the MS/MS scans were 500 msec and 100 msec, respectively, and the ion target values were set to 1E6 and 5000, respectively, for each scan mode.
MS data analysis
Raw files were analyzed by Proteome Discoverer (version 1.4.1.14) against a mouse database (Swiss-Prot canonical, 17,002 sequences, Jan22_2014 release, including common contaminant proteins). Oxidation of methionines and monomethylation, dimethylation, and trimethylation of lysines were set as variable modifications, whereas carbamidomethylation of cysteines was considered as fixed modification in the SequestHT search engine. The minimal peptide length was set to six amino acids; a maximum of two missed cleavages were allowed. Peptides were filtered at 1% false discovery rate (FDR) by using Percolator.
FACS
Basal and differentiated mKCs were isolated from the back skins of newborn mice as described above. Back skins from six mice were pooled. Cell suspensions were stained for 30 min at 4°C using a phycoerythrin-conjugated rat anti-CD49f (integrin α6 chain) antibody (no. 555736, BD) as previously described (Nowak and Fuchs 2009). Cells were sorted on a FACSAria Ilu using CellQuest Pro software (BD).
RT-qPCR
RNA was isolated from epidermal tissue and cultured cells using Trizol reagent (Invitrogen). cDNA synthesis was performed using 2 μg of total RNA using Ready-To-Go You-Prime IT first strand beads (Amersham Pharmacia Biotech) and random primers (Invitrogen). RNA was isolated from FACS-purified cells using RNeasy mini kit (Qiagen). cDNA synthesis was performed using 100 ng of total RNA using SuperScript VILO cDNA synthesis kit. PCR products were quantified using GoTag PCR master mix (Promega), Ep-Realplex thermal cycler (Eppendorf), and the ΔΔCT method. Primer sequences are available on request.
ChIP
ChIP was performed using cultured mKCs fixed in 1% formaldehyde. Nuclear lysates were obtained by lysis of cell membrane in a hypotonic buffer (50 mM, HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton-X-100) and clearance of detergents (10 mM Tris-HCL at pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Nuclei were resuspended in ChIP sonication buffer (100 mM NaCl, 10 mM TrisHCl at pH 8, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine). Sonication was performed in a Covaris ultrasound device. Whole nuclear extract was quantified with a BCA protein assay, and 0.5–1 mg of protein was used for preclearing with 10 μL of protein G Dynabeads. Immunoprecipitation was performed with 2–5 μg of antibodies coupled to protein G Dynabeads against Fra-2 (CNIO Monoclonal Antibody Unit), Ezh2 (no. 5246, Cell Signaling), Suz12 (ab12073, Abcam), H3K27me3 (ab6002, Abcam), H3K4me3 (ab8580, Abcam), and H3K27me3S28p (no. 491015, Life Technologies) overnight at 4°C. Rat or rabbit IgG was used as a negative control (Millipore). After washing once with low-salt buffer (1% Triton X-100, 0.1% SDS, 2 mM EDTA at pH 8.0, 150 mM NaCl, 20 mM Tris-HCl at pH 8.0), once with high-salt buffer (1% Triton X-100, 0.1% SDS, 2 mM EDTA at pH 8.0, 500 mM NaCl, 20 mM Tris-HCl at pH 8.0), once with LiCl buffer (1% NP-40, 1% Na-deoxycholate, 1 mM EDTA, 250 mM LiCl, 20 mM Tris-HCl at pH 8.0), and once with TE buffer (10 mM Tris at pH 8.0, 1 mM EDTA), protein/DNA complexes were eluted from beads, and DNA was decross-linked overnight at 65°C in elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl at pH 8). Eluate was treated with RNase A (Sigma) for 2 h at 37°C and with proteinase K (VWR) for 2 h at 55°C. DNA was recovered with phenol/chloroform extraction and amplified using GoTag PCR master mix (Promega) and primers specific for AP-1-binding sites and TSSs of EDC promoters using an Ep-Realplex light cycler (Eppendorf). Primer sequences are available on request.
Dye penetration assay
E17.5 embryos were collected, rinsed once in PBS and once in 100% methanol, stained in 0.1% Toluidine blue for 30 min at room temperature, and washed in PBS before image capture.
Statistical analysis
Results are presented as mean ± standard deviation (SD). P-values were calculated using Student’s t-test. An asterisk indicates P < 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Falk Weih for providing FoxN1 Cre transgenic mice, and Dr. David Santamaría for providing lentiviral expression plasmids. We are very grateful to the members of the Wagner and Ezhkova laboratory for constructive input and criticism. S.W. is funded by a FPU predoctoral fellowship from the Spanish Ministry of Education and a BIF travel fellowship. E.F.W is funded by the Banco Bilbao Vizcaya Argentaria (BBVA) Foundation and a European Research Council Advanced Grant (ERC FCK/2008/37). J.Z. is supported by the Shandong Provincial Natural Science Foundation, China. J.G.-V. is supported by the Spanish Ministry of Education (SAF2012_39670). J.M. is supported by Ramon y Cajal Programme (MINECO, RYC-2012-10651). The CNIO Proteomics Unit belongs to ProteoRed, PRB2-ISCIII, supported by grant PT13/0001. E.E. is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (R01 AR063724).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.249748.114.
References
- Alli NS, Yang EC, Miyake T, Aziz A, Collins-Hooper H, Patel K, McDermott JC. 2013. Signal-dependent fra-2 regulation in skeletal muscle reserve and satellite cells. Cell Death Dis 4: e692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakiri L, Reschke MO, Gefroh HA, Idarraga MH, Polzer K, Zenz R, Schett G, Wagner EF. 2011. Functions of Fos phosphorylation in bone homeostasis, cytokine response and tumourigenesis. Oncogene 30: 1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Blanpain C. 2012. Mechanisms regulating epidermal stem cells. EMBO J 31: 2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118: 635–648. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Horsley V, Fuchs E. 2007. Epithelial stem cells: turning over new leaves. Cell 128: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozec A, Bakiri L, Jimenez M, Rosen ED, Catala-Lehnen P, Schinke T, Schett G, Amling M, Wagner EF. 2013. Osteoblast-specific expression of Fra-2/AP-1 controls adiponectin and osteocalcin expression and affects metabolism. J Cell Sci 126: 5432–5440. [DOI] [PubMed] [Google Scholar]
- Briso EM, Guinea-Viniegra J, Bakiri L, Rogon Z, Petzelbauer P, Eils R, Wolf R, Rincon M, Angel P, Wagner EF. 2013. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev 27: 1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deady S, Sharp L, Comber H. 2014. Increasing skin cancer incidence in young, affluent, urban populations: a challenge for prevention. Br J Dermatol 171: 324–331. [DOI] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL, Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R, Helms C, et al. . 2009. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet 41: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond I, Owolabi T, Marco M, Lam C, Glick A. 2000. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol 115: 788–794. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Young CA, Jans R, Crish JF, Xu W, Rorke EA. 2013. AP1 transcription factors in epidermal differentiation and skin cancer. J Skin Cancer 2013: 537028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. 2003. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3: 859–868. [DOI] [PubMed] [Google Scholar]
- Eferl R, Zenz R, Theussl HC, Wagner EF. 2007. Simultaneous generation of fra-2 conditional and fra-2 knock-out mice. Genesis 45: 447–451. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. 2009. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136: 1122–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Horsley V. 2008. More than one way to skin. Genes Dev 22: 976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea-Viniegra J, Zenz R, Scheuch H, Hnisz D, Holcmann M, Bakiri L, Schonthaler HB, Sibilia M, Wagner EF. 2009. TNFα shedding and epidermal inflammation are controlled by Jun proteins. Genes Dev 23: 2663–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea-Viniegra J, Zenz R, Scheuch H, Jimenez M, Bakiri L, Petzelbauer P, Wagner EF. 2012. Differentiation-induced skin cancer suppression by FOS, p53, and TACE/ADAM17. J Clin Invest 122: 2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea-Viniegra J, Jimenez M, Schonthaler HB, Navarro R, Delgado Y, Concha-Garzon MJ, Tschachler E, Obad S, Dauden E, Wagner EF. 2014. Targeting miR-21 to treat psoriasis. Sci Transl Med 6: 225re1. [DOI] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. 2012a. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 26: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Korboukh I, Jin J, Huang J. 2012b. Targeting protein lysine methylation and demethylation in cancers. Acta Biochim Biophys Sin (Shanghai) 44: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H. 2012. A complex Polycomb issue: the two faces of EZH2 in cancer. Genes Dev 26: 751–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq MD, Tsai NP, Khan SA, Wei LN. 2007. Lysine trimethylation of retinoic acid receptor-α: a novel means to regulate receptor function. Mol Cell Proteomics 6: 677–688. [DOI] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al. . 2013. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23: 839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypriotou M, Huber M, Hohl D. 2012. The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp Dermatol 21: 643–649. [DOI] [PubMed] [Google Scholar]
- Lee TI, Young RA. 2013. Transcriptional regulation and its misregulation in disease. Cell 152: 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Lee JS, Kim H, Kim K, Park H, Kim JY, Lee SH, Kim IS, Kim J, Lee M, et al. . 2012. EZH2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol Cell 48: 572–586. [DOI] [PubMed] [Google Scholar]
- Meixner A, Zenz R, Schonthaler HB, Kenner L, Scheuch H, Penninger JM, Wagner EF. 2008. Epidermal JunB represses G-CSF transcription and affects haematopoiesis and bone formation. Nat Cell Biol 10: 1003–1011. [DOI] [PubMed] [Google Scholar]
- Mejetta S, Morey L, Pascual G, Kuebler B, Mysliwiec MR, Lee Y, Shiekhattar R, Di Croce L, Benitah SA. 2011. Jarid2 regulates mouse epidermal stem cell activation and differentiation. EMBO J 30: 3635–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak JA, Fuchs E. 2009. Isolation and culture of epithelial stem cells. Methods Mol Biol 482: 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al. . 2013. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Harris AR, Hinckley MR, Feldman SR, Fleischer AB, Coldiron BM. 2006. Incidence estimate of nonmelanoma skin cancer in the United States, 2006. Arch Dermatol 146: 283–287. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Goebeler M, Posern G, Feller SM, Seitz CS, Brocker EB, Rapp UR, Ludwig S. 2000. Ras-independent activation of the Raf/MEK/ERK pathway upon calcium-induced differentiation of keratinocytes. J Biol Chem 275: 41011–41017. [DOI] [PubMed] [Google Scholar]
- Schonthaler HB, Guinea-Viniegra J, Wculek SK, Ruppen I, Ximenez-Embun P, Guio-Carrion A, Navarro R, Hogg N, Ashman K, Wagner EF. 2013. S100A8–S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 39: 1171–1181. [DOI] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet 22: 356–360. [DOI] [PubMed] [Google Scholar]
- Seo HR, Kwan Y-W, Cho C-K, Bae S, Lee S-J, Soh J-W, Chung H-Y, Lee Y-S. 2004. PKCα induces differentiation through ERK1/2 phosphorylation in mouse keratinocytes. Exp Mol Med 36: 292–299. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Fleischmann A, Behrens A, Stingl L, Carroll J, Watt FM, Schlessinger J, Wagner EF. 2000. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102: 211–220. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708. [DOI] [PubMed] [Google Scholar]
- Smith CH, Barker JN. 2006. Psoriasis and its management. BMJ 333: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soza-Ried C, Bleul CC, Schorpp M, Boehm T. 2008. Maintenance of thymic epithelial phenotype requires extrinsic signals in mouse and zebrafish. J Immunol 181: 5272–5277. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Mann M. 2009a. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res 8: 5674–5678. [DOI] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. 2009b. Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. . 2012. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, Sibilia M, Wagner EF. 2003. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell 4: 879–889. [DOI] [PubMed] [Google Scholar]
- Zenz R, Eferl R, Kenner L, Florin L, Hummerich L, Mehic D, Scheuch H, Angel P, Tschachler E, Wagner EF. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature 437: 369–375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.