Summary
Invertebrate model systems are powerful tools for studying human disease owing to their genetic tractability and ease of screening. We conducted a mosaic genetic screen of lethal mutations on the Drosophila X-chromosome to identify genes required for the development, function, and maintenance of the nervous system. We identified 165 genes, most of whose function has not been studied in vivo. In parallel, we investigated rare variant alleles in 1,929 human exomes from families with unsolved Mendelian disease. Genes that are essential in flies and have multiple human homologs were found to be likely to be associated with human diseases. Merging the human datasets with the fly genes allowed us to identify disease-associated mutations in six families and to provide insights into microcephaly associated with brain dysgenesis. This bidirectional synergism between fly genetics and human genomics facilitates the functional annotation of evolutionarily conserved genes involved in human health.
Keywords: EMS mutagenesis, Drosophila X-chromosome, whole-exome sequencing, CRX, bull’s eye maculopathy, DNM2, Charcot-Marie-Tooth disease, ANKLE2, microcephaly
Introduction
Unbiased genetic chemical mutagenesis screens in flies have led to the discovery of the vast majority of genes in developmental signaling pathways (Nusslein-Volhard and Wieschaus, 1980). Most genes important to these pathways have now been shown to function as oncogenes or tumor suppressors (Pastor-Pareja and Xu, 2013). Similarly, in some areas of neurobiology, genetic screens in flies have led to the discovery of genes important to nervous system function including TRP channels, potassium channels, and pathways that affect diurnal rhythmicity. Subsequent studies have identified many diseases that are associated with mutations or deletions of human homologs (Bellen et al., 2010). However, our molecular understanding of neurological disorders such as neurodegenerative disease has mostly relied on reverse genetics (Lu and Vogel, 2009). Although some genes required for neuronal maintenance have been identified from genetic screens for viable mutations that exhibit shortened life span, electroretinogram defects, abnormal phototaxis and retinal histology defects, or temperature sensitive paralysis, no large-scale systematic screens to directly probe neurodegeneration have been carried out, reviewed in (Jaiswal et al., 2012). In addition, genes that cause lethality prevent the identification of numerous genes required for neuronal maintenance. We therefore implemented a genetic mosaic screen to identify essential genes required for neuronal maintenance on the X-chromosome.
One major limitation in chemical mutagenesis screens has been the inability to systematically identify an abundance of causative mutations. However, with the advent of numerous mapping tools and whole-genome sequencing (WGS), it should be possible to identify hundreds of causative mutations from a single mutagenesis experiment in which a multitude of phenotypes are scored in parallel for each mutation.
In humans, the study of Mendelian traits has led to the discovery of thousands of disease genes. Currently, identification of rare disease-causing mutations is rapidly evolving because whole-exome sequencing (WES) technologies are driving the process (Bainbridge et al., 2011; Lupski et al., 2013). However, the capability to detect rare variants in personal genomes has provided a diagnostic challenge. Traditionally, the identification of causative or associated genetic variation has relied on gene identification in families or patient cohorts followed by genetic studies in model organisms to define the function of the gene in vivo. Several studies have made use of phenotypic information in Drosophila to identify genes associated with human diseases or traits (Bayat et al., 2012; Neely et al., 2010). However, the large number of variants detected by WES with poorly defined phenotypic consequences makes it challenging to tie a specific variant/gene to a given disease phenotype. Yet, these rare variants have a strong contribution to disease (Lupski et al., 2011). The interpretation of such genome-wide variation is hindered by our lack of understanding of gene function for the majority of annotated genes in the human genome.
We identified mutations in 165 genes, most of which have not been characterized previously in vivo. We provide data that suggest this gene set can be utilized as a resource to study numerous disease causing genes. In addition, we present data that there is a fundamental difference between EMS screens and RNAi screens. Moreover, we show that fly genes with more than one homolog are much more likely to be associated with human genetic disorders. Finally, we demonstrate that merging datasets - genes identified in the fly screen and rare variant alleles in the human homologs in families with Mendelian disease - can assist in human disease gene discovery and provide biological insights into disease mechanisms.
Results
A mosaic genetic screen on the X-chromosome
To isolate mutations in essential genes that are required for proper development, function, and maintenance of the Drosophila nervous system, we performed an F3 adult mosaic screen on an isogenic (iso) y w FRT19A X-chromosome (Figures 1 and S1–S2). We mutagenized males using a low concentration of ethyl methane-sulfonate (EMS), established 31,530 mutagenized stocks, and identified 5,857 stocks that carry recessive lethal mutations. To identify a broad spectrum of mutations and isolate genes that affect multiple biological processes, we screened for numerous phenotypes that affect the nervous system. We also screened for seemingly unrelated phenotypes, such as wing and pigmentation defects. Genes that affect wing veins and notching have been shown to play roles in critical pathways that affect numerous organs, including the nervous system. To assess phenotypes in the tissues of interest, we induced mitotic clones in the thorax and wing with Ultrabithorax-flippase (Ubx-FLP) (Jafar-Nejad et al., 2005) and in the eye with eyeless-flippase (ey-FLP) (Newsome et al., 2000). We did not pursue mutations that caused cell lethality or showed no/minor phenotypes (Figure 1A). While these genes are clearly important, they are difficult to study and these mutants were not kept. We selected 2,083 lethal lines with interesting phenotypes for further characterization (Figure 1A and 1B).
Figure 1. Summary of the Drosophila X-chromosome screen.
(A) Pie chart and (B) bar graph of phenotypes scored in the screen. The numbers represent mutations in each phenotypic category. Note that one strain may show more than one phenotype in (B). (C–D) Examples of phenotypes observed in the notum. Homozygous wild-type bristles are marked by singed. Homozygous mutant bristles are marked by yellow (encircled by dotted line). Heterozygous bristles are wild-type for these two markers. (C) Wild-type. (D) Bristle [large bristles (macrochaetae) and small bristles (microchaetae)] loss. (E–I) Examples of ERG traces from mutant clones in the eye. A typical ERG has an on-transient (blue arrows), depolarization (orange line), and an off-transient (blue arrow head). ERGs were recorded in young (1–3 day old) and old (3–4 weeks old) flies for each genotype. (E) ERG of young or aged flies that show no obvious difference. (F) ERGs showing amplitude reduction in aged flies. (G) ERGs showing amplitude and on- and off-transient reduction in both young and aged mutants. (H) ERGs showing no or very small on-transient in both young and aged flies. (I) ERGs showing on- and off-transients that are either absent or very small in aged flies carrying mutant clones in eye. (J–M) Ultrastructural analysis using transmission electron microscopy (TEM) on young (two days old) and aged (three weeks old) mosaic flies. Red arrowheads indicate the rhabdomeres. (J) Young wild-type control eye: regular array of ommatidial structures with seven rhabdomeres surrounded by pigment (glia) cells. (K) Young mutant rhabdomeres showing intact structures. (L) Aged control eye tissue with intact rhabdomeres. (M) Aged mutant eye tissue with a strong degeneration of rhabdomeres. See also Figures S1, S2, S3.
In the Ubx-FLP screen, we assessed the number and size of mechanosensory organs (bristles) on the fly cuticle to identify genes required for neural development (Figure 1C–D and S2A–C) (Charng et al., 2014). We also screened for alterations in the color of bristles and cuticle to permit identification of genes involved in dopamine synthesis, secretion, metabolism, or melanization (Yamamoto and Seto, 2014) (Figure S2D). In addition, we selected mutations that affect wing morphogenesis to isolate genes that regulate core signaling pathways, including Notch, Wnt, Hedgehog, and BMP/TGF-β (Bier, 2005) (Figure S2E–J). Indeed, these pathways have been implicated in synaptic plasticity and neuronal maintenance in both fly and vertebrate nervous systems. In the ey-FLP screen, we assessed morphological defects in the eye and head to isolate genes involved in neuronal patterning, specification, and differentiation (Figure S2K–O). Moreover, we screened for mutations that cause glossy eye patches (Figure S2P) or mutations that cause a head overgrowth (Figure S2Q–S). Glossy eye phenotypes are associated with mitochondrial mutations (Liao et al., 2006), while head overgrowth is linked to genes in Hippo signaling, TOR signaling, intracellular trafficking, and cell polarity/adhesion, and these pathways are implicated in disorders such as autism, intellectual disability, and neurodegenerative diseases (Emoto, 2012; Saksena and Emr, 2009).
To isolate mutations that affect neuronal development, function, and maintenance in the visual system, we recorded electroretinograms (ERGs) in mutant eye clones in 3–4 week old flies (Figure 1E–I). By analyzing the on and off transients of ERGs (Figure 1H), one can assess photoreceptor synaptic activity and axon guidance. A loss or reduction in the amplitude of depolarization (Figure 1G) is typically associated with genes that play a role in phototransduction, loss of which typically causes retinal degeneration (Wang and Montell, 2007). To identify mutations that cause a progressive demise of neurons, we screened young and old animals for ERG defects (Figure 1F and 1I). Ultrastructural defects in the photoreceptor terminals of young and old flies were also examined in some mutants with strong ERG phenotypes (Figure 1J–M). Based on both the morphology screen and the ERG screen, we attempted to map 1,918 mutations (Figure S1 and S3).
Mutation identification
On the X-chromosome, complementation testing requires a genomic duplication on another chromosome to rescue male lethality. We selected 21 large (~0.5 Mb to ~2 Mb) duplications that cover ~95% of the X-chromosome (Cook et al., 2010), crossed them into the mutant backgrounds, and rescued the lethality of 1,385 mutations (Figure S3). This permitted mapping of the lethality to 26 cytological intervals of the X-chromosome. Complementation tests between mutants with similar phenotypes rescued by the same duplication allowed us to establish complementation groups. We grouped 450 mutations into 109 multiple allele complementation groups. The remaining 935 mutant strains include single alleles and a large number of mutations not yet been assigned to complementation groups. To map the genes, we first performed deficiency mapping and Sanger sequencing. This allowed identification of the locus for 63 complementation groups. For the remaining groups and single alleles, we performed WGS (Haelterman et al., 2014) and rescued the phenotypes with molecularly defined ~80 kb P[acman] duplications (Venken et al., 2010). By using both approaches, we were able to map 614 mutations to 165 genes, including 81 loci that have not been characterized in vivo (Tables 1 and S1) and are predicted to be involved in many diverse processes based on Gene Ontology analysis (Figure S2T–U).
Table 1. List of 165 fly genes and 259 corresponding human homologs identified from the screen.
Human genes associated with Mendelian disease are marked with an asterisk and bold type, the corresponding fly gene is shown in bold. See also Table S1, S5.
| Fly Gene | Human Homologs (*OMIM) |
|---|---|
| Aats-his | HARS*, HARS2* |
| AP-1γ | AP1G1, AP1G2, AP4E1* |
| ari-1 | ARIH1 |
| arm | CTNNB1* |
| Arp2 | ACTR2 |
| ATP7 | ATP7A*, ATP7B* |
| baz | PARD3, PARD3B |
| ben | UBE2N |
| br | - |
| Brms1 | BRMS1, BRMS1L |
| cac | CACNA1A*, CACNA1B CACNA1E |
| Cap | SMC3* |
| car | VPS33A |
| CDC45L | CDC45L |
| Cdk7 | CDK7 |
| CG11409 | - |
| CG11417 | ESF1 |
| CG11418 | MTPAP* |
| CG12125 | FAM73A, FAM73B |
| CG12991 | - |
| CG13365 | - |
| CG14442 | ZNF821 |
| CG14786 | LRPPRC* |
| CG15208 | C21orf2 |
| CG15896 | KIAA0391 |
| CG1597 | MOGS* |
| CG1703 | ABCF1 |
| CG1749 | UBA5 |
| CG17776 | - |
| CG17829 | HINFP |
| CG18624 | NDUFB1 |
| CG2025 | NRD1 |
| CG2918 | HYOU1 |
| CG3011 | SHMT1, SHMT2 |
| CG3149 | RFT1* |
| CG32649 | ADCK3*, ADCK4 |
| CG32694 | - |
| CG32795 | TMEM120A, TMEM120B |
| CG34401 | ZSWIM8 |
| CG3446 | NDUFA13* |
| CG3704 | GPN1 |
| CG3857 | SMG9 |
| CG4078 | RTEL1* |
| CG4165 | USP16 |
| CG42237 | PLA2G3, PROCA1 |
| CG42593 | UBR3 |
| CG42749 | - |
| CG4407 | FLAD1 |
| CG4542 | ALG8* |
| CG7065 | - |
| CG7358 | ZC3H13 |
| CG8184 | HUWE1* |
| CG8636 | EIF3G |
| CG8949 | WAC |
| CG9650 | BCL11A, BCL11B, ZNF296 |
| Chc | CLTC, CLTCL1 |
| CkIIbeta | CSNK2B |
| CkIα | CSNK1A1, CSNK1A1L |
| comt | NSF |
| COQ7 | COQ7 |
| Crag | DENND4A, DENND4B, DENND4C |
| Cyp4d2 | CYP4V2* |
| DAAM | DAAM1, DAAM2 |
| dlg1 | DLG1, DLG2, DLG3*, DLG4 |
| Dlic | DYNC1LI1, DYNC1LI2 |
| dor | VPS18 |
| dsh | DVL1, DVL2, DVL3 |
| Dsor1 | MAP2K1*, MAP2K2* |
| dwg | MZF1, ZSCAN22 |
| Efr | SLC35B4 |
| egh | - |
| eIF2B-ε | EIF2B5* |
| elav | ELAVL1, ELAVL2, |
| ELAVL3, ELAVL4 | |
| ewg | NRF1 |
| fh | FXN* |
| fliI | FLII |
| flw | PPP1CB |
| fs(1)h | BRD2, BRD3, BRD4, BRDT |
| Fur2 | PCSK5, PCSK6 |
| Gtp-bp | SRPR |
| hfw | - |
| Hlc | DDX56 |
| hop | JAK1, JAK2*, JAK3*, TYK2* |
| Hr4 | NR6A1 |
| Hsc70-3 | HSPA5 |
| Inx2 | - |
| kdn | CS |
| l(1)1Bi | MYBBP1A |
| l(1)G0156 | IDH3A |
| l(1)G0222 | ANKLE2 |
| l(1)G0230 | ATP5D |
| l(1)G0255 | FH* |
| l(1)G0334 | PDHA1*, PDHA2 |
| Marf | MFN1, MFN2* |
| Mcm6 | MCM6* |
| mew | ITGA3*, ITGA6*, ITGA7* |
| mRNA-cap | RNGTT |
| mRpL38 | MRPL38 |
| mRpS25 | MRPS25 |
| mRpS30 | MRPS30 |
| mst | MSTO1 |
| mus101 | TOPBP1 |
| mxc | NPAT |
| Myb | MYB*, MYBL1, MYBL2 |
| mys | ITGB1, ITGB2*, ITGB4*, ITGB5, ITGB6, ITGB7, ITGB8 |
| N | NOTCH1*, NOTCH2*, |
| NOTCH3*, NOTCH4 | |
| nej | CREBBP*, EP300* |
| Nmd3 | NMD3 |
| nonC | SMG1 |
| Nrg | CHL1, L1CAM*, NFASC, NRCAM |
| Nup93-1 | NUP93 |
| oc | CRX*, OTX1, OTX2* |
| para | SCN1A*, SCN2A*, SCN3A, SCN4A*, SCN5A*, SCN7A, SCN8A*, SCN9A*, SCN10A*, SCN11A* |
| parvin | PARVA, PARVB, PARVG |
| pck | CLDN12 |
| Pgd | PGD* |
| phl | ARAF, BRAF*, RAF1* |
| PI4KIIIalpha | PI4KA |
| por | PORCN* |
| pot | - |
| PpV | PPP6C |
| Prosα4 | PSMA7, PSMA8 |
| Psf3 | GINS3 |
| rap | FZR1 |
| Rbcn-3A | DMXL1, DMXL2 |
| Rbcn-3B | WDR7, WDR72* |
| Rbf | RB1*, RBL1, RBL2 |
| Rhp | RHPN1, RHPN2 |
| RpII215 | POLR2A |
| RpS5a | RPS5 |
| Sas10 | UTP3 |
| schlank | CERS1, CERS2, CERS3*, CERS4, CERS5, CERS6 |
| scu | HSD17B10* |
| Sec16 | SEC16A, SEC16B |
| sgg | GSK3A, GSK3B |
| shi | DNM1, DNM2*, DNM3 |
| sicily | NDUFAF6* |
| skpA | SKP1 |
| Smox | SMAD2, SMAD3* |
| smr | NCOR1, NCOR2 |
| SNF1A | PRKAA1, PRKAA2 |
| sno | SBNO1, SBNO2 |
| Sp1 | SP7*, SP8, SP9 |
| stim | STIM1*, STIM2 |
| svr | CPD |
| tay | AUTS2 |
| temp | PTAR1 |
| TH1 | NELFCD |
| tko | MRPS12 |
| trr | KMT2C, KMT2D* |
| ubqn | UBQLN1, UBQLN2*, UBQLN3, UBQLN4, UBQLNL |
| Upf1 | UPF1 |
| Upf2 | UPF2 |
| Usf | USF1, USF2 |
| Usp7 | USP7 |
| vnd | NKX2-2, NKX2-8 |
| Vps26 | VPS26A, VPS26B |
| wapl | WAPAL |
| wds | WDR5, WDR5B |
| wus | DNAJC22 |
| Ykt6 | YKT6 |
| Zpr1 | ZNF259 |
| βCop | COPB1 |
| β-Spec | SPTB*, SPTBN1, SPTBN2*, SPTBN4 |
| δCOP | ARCN1 |
Chemical mutagenesis versus RNAi screens
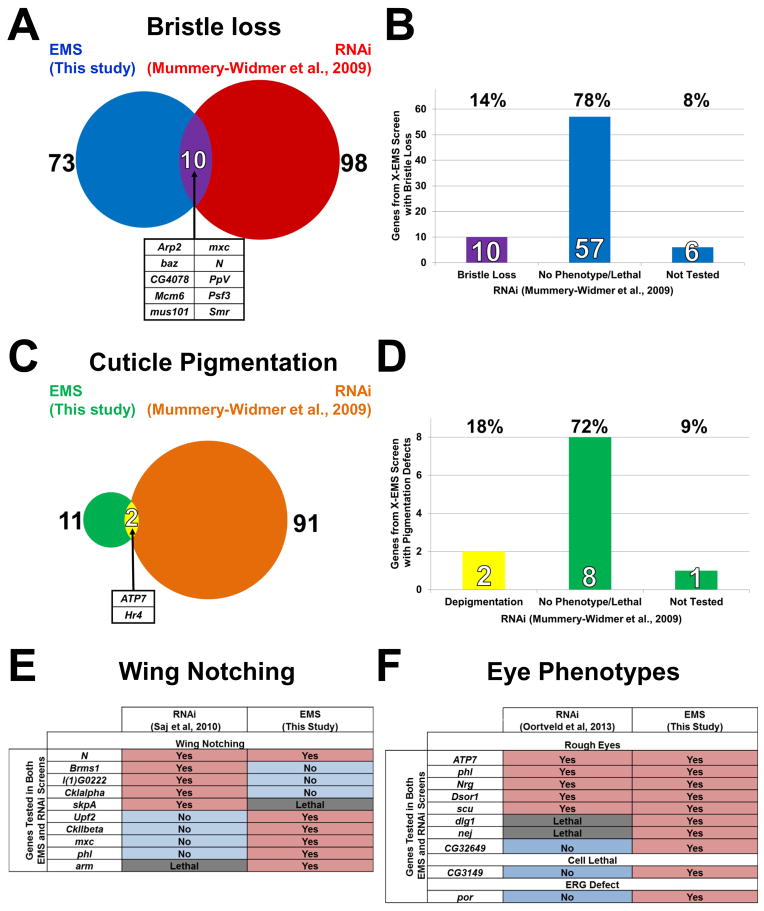
Two of the phenotypes that we screened, bristle development and depigmentation, allow a direct comparison between this screen and a genome-wide RNAi screen (Mummery-Widmer et al., 2009). This RNAi screen covered ~80% of all X-chromosome protein coding genes. Interestingly, only 14% of the genes we identified in the bristle screen were also isolated in the RNAi screen (Figure 2A–B). Similarly, only 18% of the genes that we identified from the pigmentation screen were also identified in the RNAi screen (Figure 2C–D). Conversely, we did not identify the vast majority of genes that were identified by RNAi. In addition, a comparison of our gene list and those of two RNAi screens for wing margin (Saj et al., 2010) and eye morphological defects (Oortveld et al., 2013), show that these screens also identified very different sets of genes (Figure 2E–F). In summary, chemical screens identify a distinctive set of genes when compared to RNAi based screens.
Figure 2. Comparison of results from this EMS screen and previous RNAi screens.
(A) Venn diagram and (B) bar graph showing overlap between two screens for bristle loss defects. The genes that were identified in the EMS screen were also screened by RNAi (Mummery-Widmer et al., 2009) and 10 caused a bristle loss whereas 57 showed no phenotype or caused lethality. (C) Venn diagram and (D) bar graph showing overlap between two screens for pigmentation defects (this screen and the RNAi screen of Mummery-Widmer). (E) Comparison of the results of these screens for wing notching defects. (F) Comparison of the results of these two screens for eye morphological defects.
Links to Human Diseases based on Online Mendelian Inheritance in Man (OMIM)
We next sought to determine if the 165 genes we identified in flies could enhance the understanding of human disease associated genes. Strikingly, 93% (153) of the fly genes isolated have homologs in humans (Tables 1, S1; Figure 3A). This is a strong enrichment (χ2 = 129, p < 0.001) for evolutionarily conserved genes between humans and flies when compared to the whole fly genome as only 48% of all fly genes have human homologs (Figure 3B). Moreover, the human homologs of 31% (48/153) of the identified fly genes have been associated with human disease in OMIM, 79% (38/48) of which exhibit neurological signs and symptoms (Figure 3A; Table S1). Of the genes that are conserved but not yet associated with Mendelian diseases with neurological symptoms, 65 genes have potential relationships to neurologic diseases (Figure 3A; Table S2). Therefore, the essential genes that we identified in this screen are highly conserved and many of their homologs have already been implicated in human disorders, showing that the screening strategy is effective.
Figure 3. Essential fly genes associated with more than one human homolog are more likely to be linked to human diseases.
(A) Classification of genes identified in the screen based on human homologs and associated diseases. (B) Classification of the whole fly genome according to the same criteria as in (A). (C–D) Relationship between the number of human homologs per fly gene and their association with human diseases for genes identified in the screen (C) and the whole fly genome (D). (E) The number of human homologs per fly gene and their enrichment in OMIM associated human diseases. (F) Relationship between the number of human homologs per fly gene and lethality in flies. (G) Relationship between genes associated with lethality in flies and OMIM associated human diseases. See also Table S2.
Data analysis revealed a striking difference in the number of genes associated with disease depending on the number of human homologs for each fly gene. Fly genes that have a single human homolog have many fewer disease genes represented in the OMIM database than those that have more than one homolog. There is a two-fold enrichment (χ2 = 10.7, p < 0.001) of fly genes with more than one human homolog associated with diseases in the OMIM database compared to fly genes that have only one human homolog, 47% versus 22% (Figure 3C). This prompted us to assess if the bias is conserved for all fly genes. We found that a similar bias holds throughout the genome. Fly genes with more than one human homolog are more likely to be associated with diseases in the OMIM database than those with a single homolog, 40% versus 20% (χ2 = 386, p < 0.001) (Figure 3D and Extended Experimental Procedures). Indeed, 57 fly genes with more than one human homolog account for 100 diseases in the OMIM database (1.75 diseases per fly gene), an 8-fold enrichment when compared to fly genes with a single homolog (0.22 diseases per fly gene) (Figure 3E). This enrichment is not simply due to an absolute increase in the total number of human homologs because evolutionarily conserved genes that have more than one homolog are three times more enriched for OMIM diseases, 0.62 versus 0.22 diseases per human gene (Figure 3E). The difference between 1.75 and 0.62 is due to the number of homologs. Indeed, there are on average ~3 human homologs for every fly gene that has more than one human homolog (data not shown). These data suggest that evolutionary gene duplication with divergence and further specialization of gene function may allow tolerance of mutation and viability versus lethality.
Since all of the mutations we isolated cause homozygous lethality, we analyzed the correlation between lethality, the number of human homologs, and their links to OMIM diseases for the entire fly genome. The number of essential genes in Drosophila has been estimated to be approximately 5,000 (Benos et al., 2001). Currently only ~2,000 essential genes in FlyBase have transposable elements or EMS/X-ray-induced mutations (Marygold et al., 2013), representing about 40% of all essential fly genes. The proportion of essential genes varies with evolutionary conservation: an estimated 11% of the genes that do not have human homologs are essential, whereas 38% of the genes that have a single human homolog are essential (χ2 = 354, p <0.001) (Figure 3F). Finally, an estimated 61% of the fly genes with more than one human homolog are essential. These data show that fly genes that have more than one human homolog are more likely to cause lethality when mutated. Finally, human homologs of essential genes in Drosophila are more likely to be associated with human genetic diseases (χ2 = 88, p <0.001) (Figure 3G). Therefore, we conclude that genes that are essential in flies and have multiple human homologs are the most likely to be associated with human diseases, potentially due to gene duplication and redundancy.
Combining fly and human mutant screen datasets to identify disease genes
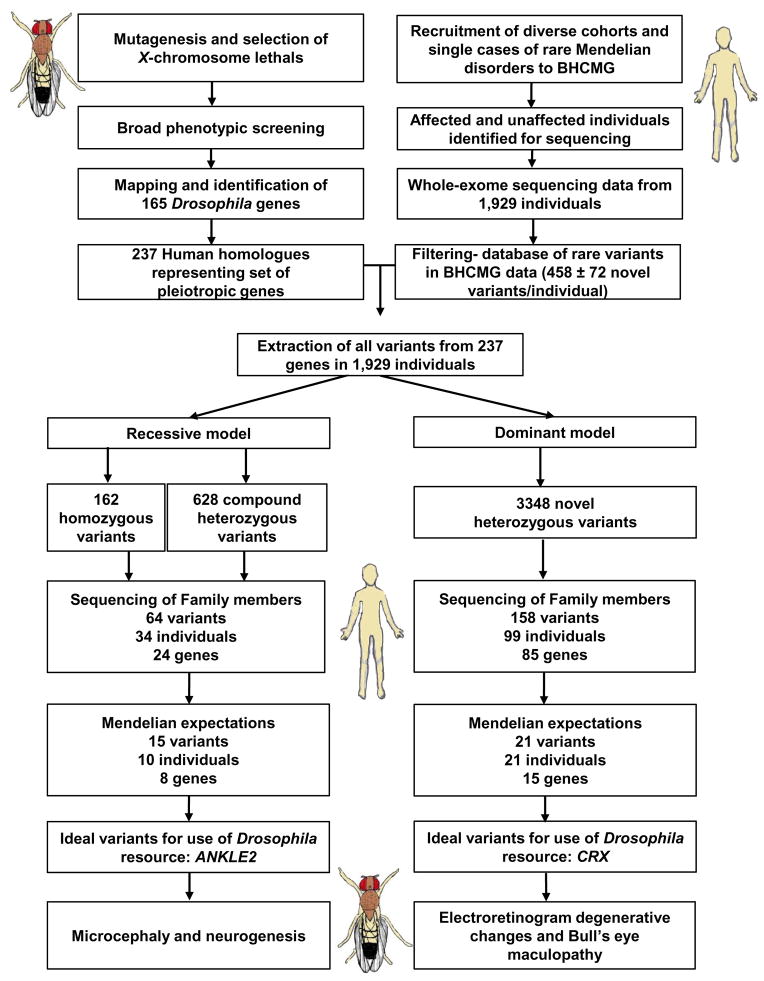
We next utilized the fly gene dataset uncovered from the forward genetic screen in combination with a human exome dataset, to identify new human disease genes. We undertook a systematic search of all the variants in the human homologs of the genes identified from the Drosophila screen within WES data generated from undiagnosed cases of Mendelian diseases. This included 1,929 individuals in the Baylor-Hopkins Centers for Mendelian Genomics (BHCMG) (Figure 4).
Figure 4. Flowchart for discovery and functional studies of disease genes using the Drosophila resource and human exome data.
BHCMG uses next-generation sequencing, to discover the genetic basis of as many Mendelian diseases as possible (Bamshad et al., 2012). The study population includes singleton cases with sporadic disease, single families, and when possible, larger cohorts of affected individuals with a range of rare Mendelian phenotypes. A wide range of disorders are under investigation (http://www.mendelian.org/). In general, patients are recruited when a Mendelian disease seems highly likely, and all reasonable efforts at a molecular diagnosis have failed. Due to the rare nature of the phenotypes, information from other patients or additional biological information from model organisms is required to fulfill the burden of proof for gene/disease association in such cases. For this reason, our Drosophila resource of mutant genes was integrated with our human exome variant and Mendelian phenotype (Hamosh et al., 2013) databases, and the combination approach was used to solve some of the cases.
We analyzed 237 out of the 259 (Table 1) homologs of fly genes identified through the X-chromosome screen as they were validated at the time of analysis. We included all 237 genes, regardless of whether they were previously identified to be associated with Mendelian diseases in OMIM, to avoid any bias. We filtered out variants reported as having greater than 1% allele frequency in databases of control individuals (See Extended Experimental Procedures). Under the assumption of a recessive model dataset, we included all variants that met these criteria and were homozygous or had two heterozygous variants affecting the same gene. The latter set was not tested for cis or trans orientation of the variants prior to analysis. A dominant model included heterozygous variants. These were filtered even more stringently for allele frequency such that only variants that had not been observed in the control datasets were studied (Table S3).
To explore potential associations with disease, we prioritized variants for segregation analysis within families (Figure 4). We performed Sanger sequencing or explored segregation in families for 64 variants in 24 genes within 34 individuals in the recessive dataset and found that 15 variants in 8 genes within 10 individuals fulfilled Mendelian expectations for recessive inheritance. Likewise, for the dominant dataset, we explored the segregation for 158 variants in 85 genes within 99 individuals. We found 22 variants in 15 genes within 21 individuals that fulfilled Mendelian expectations of a dominantly inherited disorder in the family under investigation. Interestingly, 22/31 individuals in which the variant met Mendelian expectations had a neurological disease.
As a proof-of-principle, we report six patients/families with mutations in three genes. In addition, we identified 25 other individuals in which the variant in the homolog of the fly gene met Mendelian expectation. Some of these individuals were found to have candidate variants in multiple genes, some had too few living relatives for further study, and for others, studies are ongoing. Therefore, a systematic search of variants within the genes identified in the Drosophila screen was able to identify and prioritize a subset of variants with Mendelian inheritance in the family that could be studied. Amongst these, we found examples of known disease genes (DNM2),, a novel disease association to a known disease gene (CRX), and novel candidate genes for disease (ANKLE2).
DNM2 and Charcot-Marie-Tooth Neuropathy
Examination of a homolog of Drosophila shibire (shi), the gene that encodes Dynamin, led to a molecular diagnosis for two individuals with heterozygous mutations in DNM2 (Figure S4A and B). Both patients were diagnosed with a distal symmetric polyneuropathy consistent with Charcot-Marie-Tooth disease (CMT) (See Supplemental Text). Mutations in DNM2 are associated with CMT Type 2M (OMIM# 606482), an axonal form primarily affecting neurons (Figure S4C). Patient 1, the proband in Figure S4A, presented at age 12 with hand tremor, calf cramps, lower limb paresthesias, and difficulty with heel walking. She is a member of a family with three generations of neuropathy (Figure S4A), and the heterozygous G358R variant co-segregated with CMT (Figure S4A). Patient 2, the proband in Figure 4B, (currently 88 years) presented at age 40 with lower extremity weakness. His nerve conduction studies showed low amplitudes and borderline slowed velocities. He carries an E341K mutation in DNM2 (Figure S4D). In addition to DNM2, WES revealed a variant in another CMT gene, LRSAM1 in this patient (Figure S4B). Interestingly, dominant as well as recessive mutations in LRSAM1 can cause CMT2P (OMIM#614436). Hence, either one or a combination of both genes may cause CMT in this family. While some clinical features of the probands made diagnosis difficult, the phenotypes of these cases were ultimately consistent with CMT type 2.
CRX and Bull’s Eye Maculopathy
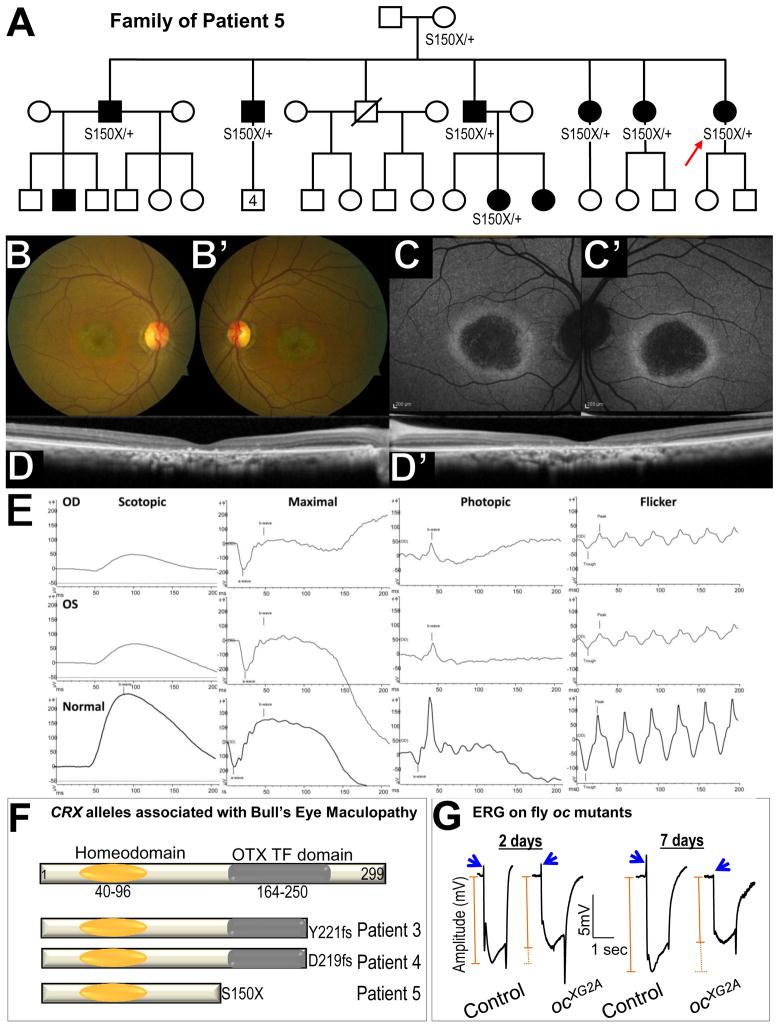
Examination of one of the human homologs of Drosophila ocelliless (oc, CRX in humans) led to the identification of three cases of bull’s eye maculopathy associated with dominant CRX alleles. oc encodes a homeobox transcription factor that regulates photoreceptor development (Vandendries et al., 1996). Identifying cases of bull’s eye maculopathy, a late-onset slowly progressive retinal disorder, with CRX alleles was surprising because CRX is typically associated with much more severe childhood vision loss seen in dominant cone-rod dystrophy, Leber congenital amaurosis, and autosomal dominant retinitis pigmentosa (OMIM #120970, #613829).
The three cases of bull’s eye maculopathy included two individuals with no family history of retinal disease (Patients 3 and 4) and one multigenerational pedigree (Patient 5 [p.S150X])(Figure 5A). The affected individuals in the family of Patient 5 developed symptoms at age 50 (range 28–63 years), and three family members with the S150X mutation had minimal symptoms at initial evaluation between the age of 55–60. Despite having near normal vision, ophthalmologic exam in the retina of these individuals revealed advanced bull’s eye maculopathy with foveal sparing explaining the modest effect on vision.
Figure 5. Mutations in CRX cause bull’s eye maculopathy.
(A) Pedigree of the family of Patient 5 (red arrow) with multiple individuals with bull’s eye maculopathy. The S150X mutation in CRX was identified in 7 patients. (B–D) Clinical phenotypes of Patient 5. (B–B′) Fundus photography show fine granularity in the outer retina and speckled glistening deposits arranged in a ring around the macula. Peripheral fundi appear unaffected. (C–C′) Autofluorescence images reveal a bull’s eye phenotype with hypo-fluorescent macula surrounded by a hyper-autofluorescent ring, suggesting a continuously atrophic macular area. (D–D′) Optical coherence tomography shows central loss of the outer nuclear layer, ellipsoid line, external limiting membrane, and retinal pigment epithelium atrophy corresponding to area of hypo-autofluorescence in (C–C′). (E) ERG of the proband: Electroretinographic traces showed implicit time delay and amplitude reduction in both scotopic and especially photopic responses in keeping with generalized cone-rod dysfunction. (F) Structure of CRX protein and mutations in Patients 3–5. (G) ERG of control and oc mutant clone in 2 days and 7 day (in light) old adult flies. Blue arrows indicate on transient in ERG. On-transients are lost in 7 days old flies. The orange line indicates the amplitude of ERG.
Patient 5 exhibits retinal abnormalities (Figure 5B–B′), abnormal autofluorescence in the fundus (Figure 5C–C′), aberrant Optical Coherence Tomography (OCT, Figure 5D–D′) and electroretinograms (Figure 5 E), all consistent with bull’s eye maculopathy. The three new alleles are all encoding predicted truncations of the OTX transcription factor domain (Figure 5F).
Functional analysis of homozygous oc mutant clones reveal that the ERGs in young animals are nearly normal (Figure 5G) but defective in 7 day old flies, including reduced amplitude and loss of on-transients (Figure 5G, blue arrows). This suggests that the photoreceptors become impaired over time. In summary, the defects in flies and humans show similarities.
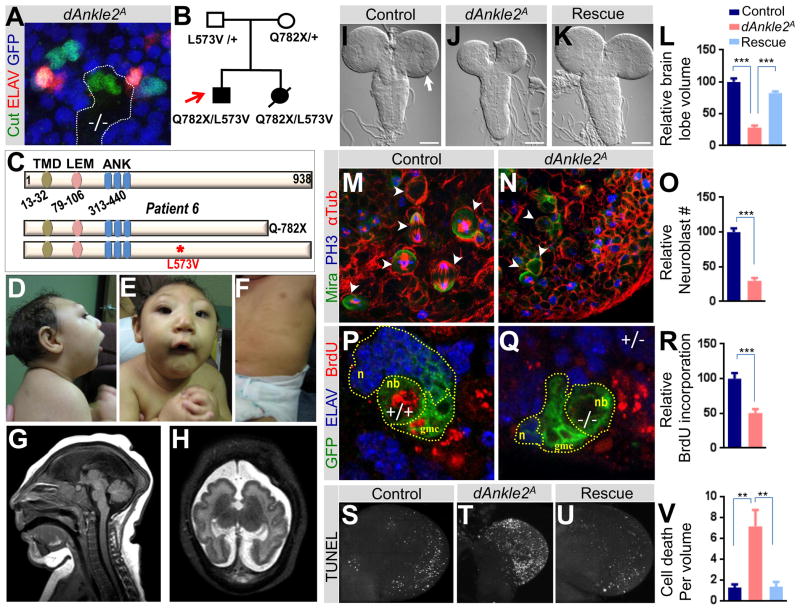
ANKLE2 and microcephaly
The Drosophila screen identified a mutation in l(1)G0222, the homolog of ANKLE2 (dAnkle2) (Table 1). The mutation causes a loss of thoracic bristles and underdevelopment of the sensory organs in clones (Figure 6A). The human WES data identified variants in ANKLE2 in a family with apparent recessive microcephaly (Figure 6B and C). The proband, Patient 6, has an extreme small head circumference, a low sloping forehead, ptosis, small jaw, multiple hyper- and hypopigmented macules over all areas of his body, and spastic quadriplegia (Figure 6D–H; Supplemental Text, Clinical case histories). During his first year of life, he had unexplained anemia, and glaucoma. At 3 years, he had onset of seizures, and at 5.5 years, his weight was 10.7 kg (−4 standard deviations (SD)), length 83.8 cm (−6 SD) and fronto-occipital circumference 38.2 cm (−9 SD).
Figure 6. ANKLE2 and microcephaly.
(A) dAnkle2 mutant clone of the peripheral nervous system in the thorax of a fly. In wild-type tissue (GFP, shown in blue), sensory organs are comprised of four cells marked by Cut (green), one of which is a neuron marked by ELAV (red). In the mutant clone (−/−, non-blue), the number of cells per sensory organ is reduced to two and does not contain a differentiated neuron. (B) Pedigree of the family of Patient 6 (red arrow) with a severe microcephaly phenotype. Both affected individuals inherited variants from both parents in ANKLE2. (C) Structure of ANKLE2 protein and mutations in Patient 6. Abbreviations: transmembrane domain (TMD), LAP2/emerin/MAN1 domain (LEM), ankyrin repeats (ANK). (D–E) Clinical phenotypes of the proband with a severe sloping forehead, microcephaly, and micrognathia. (F) Scattered hyperpigmented macules on the trunk. (G) Sagittal brain MRI of the proband in infancy with severe microcephaly, agenesis of the corpus callosum, and a collapsed skull with scalp ruggae. (H) Axial brain MRI showing polymicrogyria-like cortical brain malformations. (I–L) Third instar larval brain of (I) control (y w FRT19Aiso), scale bar indicates 100 microns (J) dAnkle2 mutant, and (K) dAnkle2 mutant in which the human ANKLE2 cDNA is ubiquitous expressed (Rescue). Note that brain lobe (arrow in I) size is reduced in dAnkle2 mutant (J) and the phenotype is rescued by ANKLE2 expression (K). Relative brain lobe volume of control, dAnkle2 and rescue using 3D confocal images is quantified in (L). (M–O) Larval CNS neuroblasts (arrowheads) in control and dAnkle2 mutant. Neuroblasts are marked by Miranda (Mira, green), chromosomes in dividing cells are marked by Phospo-Histone3 (PH3, blue), and spindles in dividing cells are marked by α-Tubulin (αTub, red). Relative number of neuroblasts in control and dAnkle2 is shown in (O). (P–R) BrdU incorporation (red) in control (P) and dAnkle2 mutant clones (Q) marked by GFP (green, dotted lines) in larval brains. Differentiated neurons are marked by ELAV (blue). Neuroblast (nb), ganglion mother cells (gmc), and neurons (n) are marked. Quantification of relative BrdU incorporation is shown in (R). (S–V) TUNEL assay in third instar larval brain lobes of (S) control, (T) dAnkle2 mutant, and (U) Rescue. Quantification of TUNEL positive cells/volume (cell death) is shown in (V). In Figures L, O, R and V, *** indicates a p-value < 0.001 and ** indicates a p-value < 0.01. See also Table S4, Figure S5.
Brain MRI in the newborn period demonstrated a low forehead, several scalp ruggae, and mildly enlarged extra-axial space with communication between the posterior lateral ventricles and the mesial extra-axial space. Other brain abnormalities included a simplified gyral pattern, mildly thickened cortex, small frontal horns of the lateral ventricles with mildly enlarged posterior horns of the lateral ventricles, and agenesis of the corpus callosum. The brainstem and cerebellum appeared relatively normal (Figure 6G and H). A younger sister born a year later had severe microcephaly, spasticity, and similar hyper- and hypopigmented macules over all areas of her body. She died 24 hours after delivery from cardiac failure associated with poor contractility, although the basis for this was not known.
WES data of the proband, his affected sister, and both parents revealed four candidate genes that meet Mendelian expectation and are expressed in the CNS (Table S4). Table S4 shows the variants with their scores from four predictions programs (Liu et al., 2011). ANKLE2 was prioritized as a good candidate. To assess if dAnkle2 is involved in CNS development, we examined the brains of Drosophila mutant larvae. Brain size in early third instar larval stages is similar to that of controls (Figure S5A). However, later in third larval stage, the brain becomes progressively smaller than control larvae (Figure S5A and Figure 6I and J). To confirm that dAnkle2 is an ortholog of human ANKLE2, we ubiquitously expressed human ANKLE2 in mutant flies and observed rescue of lethality and the small brain phenotype (Figure 6K–L). These data indicate that ANKLE2 is implicated in CNS development and its molecular function is evolutionarily conserved.
To explore the cause of the small brain phenotype in dAnkle2 mutants we assessed defects in processes which can cause small brain phenotypes: mitosis, asymmetric cell division, and apoptosis (Rujano et al., 2013). The number of neuroblasts, marked by Miranda (Ceron et al., 2001). is severely reduced in late third instar brain lobes (Figure 6M–O and S5B–C). In the few neuroblasts that undergo division, the spindles are properly oriented towards the polarity axis (Figure S5D and E). In addition, centriole duplication, impaired in many primary human microcephaly syndromes (Kaindl et al., 2010), is not affected in dAnkle2 mutants (Figure S5F and G). Hence, loss of dAnkle2 causes a severe reduction in neuroblast number but does not seem to affect asymmetric division or centriole number.
To assess proliferation in the CNS, we induced mitotic clones of dAnkle2 in the brain and labeled them with Bromodeoxyuridine (BrdU)(Figure 6P–R). As shown in Figure 6R, BrdU incorporation is strongly reduced in mutant clones when compared to wild clones, indicating that cell proliferation is severely impaired. In addition, the mutant clones (Figure 6Q,) that contain a neuroblast and its progeny, the ganglion mother cells and neurons, contain many fewer cells than wild type clones (Figure 6P). Finally, we observe a dramatic increase in apoptotic cells marked by TUNEL in the dAnkle2 mutant brain lobes (Figure 6S, T, and V). This cell death is rescued by the expression of the human cDNA encoding ANKLE2 (Figure 6U–V). Therefore, defects in proliferation and excessive apoptosis are both contributing to the loss of CNS cells in dAnkle2.
Discussion
Here we describe the generation of a large set of chemically-induced lethal mutations on the Drosophila X-chromosome that were screened for predominantly neurological phenotypes in adult mosaic flies. The mutations were assigned to complementation groups, mapped, and sequenced to associate as many genes as possible with specific phenotypes. We identified and rescued the lethality associated with mutations in 165 genes using a variety of mapping and sequencing methods. These mutations are available through the Bloomington Drosophila Stock Center and provide a valuable resource to study the function of human genes in Drosophila especially since 93% of the genes are evolutionarily conserved in human.
This mutant collection contains 21 genes associated with human diseases for which no mutations were previously available. The fly mutants thus immediately enables the study of the basic molecular mechanism of 26 human diseases, including Leigh syndrome (CG14786/LRPPRC, l(1)G0334/PDHA1, and sicily/NDUFAF6), congenital disorders of glycosylation (CG1597/MOGS, and CG3149/RFT1), Usher syndrome (Aats-his/HARS), Friedereich’s ataxia (fh/FXN), and amyotrophic lateral sclerosis (ubqn/UBQLN2). Based on the gene list from the Drosophila screen, we explored a database of 1,929 human exomes from a Mendelian disease resource of patients with rare diseases. We examined the personal genomes for rare variants of the fly homologs and prioritized a subset of human rare variant alleles for segregation analysis. We report six families with distinct diseases in which the variants segregate and are likely responsible for causing the associated Mendelian disease.
The approach described here provides a valuable resource to study the function of many disease genes in different tissues. We propose that the screen strategy be expanded to the autosomes, and a number of guiding principles should be considered based on this study. First, the use of low concentrations of EMS is important as it minimizes the number of second site lethal and visible mutations (Haelterman et al., 2014). Second, screening for lethal mutations has major advantages as 93% of the isolated genes that are essential for viability are conserved, whereas only 48% of all Drosophila genes have evolutionarily conserved human homologs. Third, the isolation of lethal mutations also greatly facilitates genetic mapping. Fourth, screening for many different phenotypes casts a broader net and permits isolation of mutations in many different genes, a strategy that is also used in mice (White et al., 2013). Fifth, analyzing different phenotypes revealed that mutations in the majority of the genes cause more than one phenotype, consistent with extensive pleiotropism.
Comparison of the gene list identified from our EMS screen and several RNAi screens have shown that these approaches reveal very distinct sets of genes. There are multiple reasons that may lead to this difference. For example, since our screen was aimed at identifying mutations that cause lethality, we have not screened for genes that are non-essential. Thus, a number of genes that are non-essential but cause morphological defects are missed in our screen. On the other hand, RNAi may not be efficient or cause off targeting effects (Green et al., 2014; Mohr, 2014). Regardless of the methods that are being used, rescue experiments and independent validation are critical to determine that the phenotype one observes is due to loss of the gene of interest when performing a genetic screen.
It is interesting to note that from our screen, essential fly genes with two or more homologs in human have a significantly higher likelihood of being associated with Mendelian diseases than those that only have a single human homolog (Figure 3). This suggests that gene duplications of essential genes and subsequent evolutionary divergence may lead to genes that are partially redundant and more likely to be disease associated. Hence, when analyzing human exomes, it would seem more productive to start with homologs of evolutionarily conserved essential Drosophila genes that have two or more human homologs. In addition to these relationships to Mendelian traits, 17% (26/153) of the fly genes that have human homologs have been identified in GWAS (genome wide association studies) for neurological disorders (Table S5). Hence, the collection of mutations described here may permit us to study genes for complex traits
We uncovered a genetic basis in a few cases for which the gene was previously known. For example, the study of DNM2 revealed previously studied phenotypes associated with mutations in the gene (CMT, Figure S4). In another case we observed that mutations in a gene caused unexpected phenotypes. Indeed, we identified three families with bull’s eye maculopathy, a condition that is much milder and with a later age of onset than conditions typically associated with CRX truncations such as Leber congenital amaurosis (leading to blindness before a year of life) and cone rod dystrophy (a condition with onset in the first or second decade). Interestingly, other truncating alleles have been reported both N- and C-terminal to the OTX transcription factor domain in patients with these severe phenotypes. CRX mutations can produce variable phenotypes (Huang et al., 2012). However, bull’s eye maculopathy has not been associated with CRX. Our data suggest that some symptoms may manifest at older ages and the true phenotypic spectrum of CRX mutations includes late-onset mild retinopathy.
We identified deleterious alleles in ANKLE2 in two individuals in a family affected by severe microcephaly. In flies, we observed severe defects in neuroblast proliferation and excessive apoptosis in the third instar larval brain of dAnkle2 mutants. This knowledge, combined with the observation that expression of human ANKLE2 in dAnkle2 mutants rescues lethality, brain size, and apoptosis, provide strong evidence that ANKLE2 is responsible for the microcephaly in the family. Moreover, ANKLE2 has been shown to physically and genetically interact with VRK1 in C. elegans and vertebrates (Asencio et al., 2012), and loss of fly VRK1 (also known as ballchen (ball) or nhk-1 in flies) also causes a small brain phenotype in third instar larvae (Cullen et al., 2005). It is therefore interesting to note that mutations in VRK1 also cause microcephaly in patients (Figure S5H) (Gonzaga-Jauregui et al., 2013).
The pattern of brain abnormalities and microcephaly in our patient with ANKLE2 mutations is somewhat similar to patients with autosomal recessive CLP1 mutations. CLP1 encodes an RNA kinase involved in tRNA splicing (Karaca et al., 2014; Schaffer et al., 2014). The Clp1 homozygous kinase-dead mouse exhibits microcephaly that worsens with age due to apoptosis. Hence, apoptosis may be a common denominator in these forms of microcephaly.
Phenotypic information of Drosophila mutants allows researchers to understand the potential in vivo function of their human homologs. The cases of oc/CRX and dAnkle2/ANKLE2 are examples in which some direct phenotypic comparisons are possible between the fly mutant and human conditions. However, one of the major drawbacks of comparing phenotypes in different species is that a comparison between different tissues and organs is not always obvious. How do we relate wing vein defects or a rough eye with the phenotypes observed in human genetic diseases? Numerous strategies have been outlined by Lehner (Lehner, 2013) and one of the most compelling strategies is based on orthologous phenotypes or phenologs (McGary et al., 2010). Genes tend to work in evolutionarily conserved pathways, allowing the direct transfer from genotype–phenotype relations between species. For example, mutations in a subset of genes that function in mitochondrial quality control cause a high incidence of muscle mitochondrial defects in adult flies and Parkinson’s disease (PD) in humans (Jaiswal et al., 2012), suggesting that new genes that affect muscle mitochondria in adult flies make good candidates for PD. Indeed, it may well be that phenotypic similarities between fly and man will be the exception rather than the rule. Regardless, we provide evidence that the use of unbiased screens in the fly and the resulting genetic resources will provide opportunities to prioritize human exome variants and to explore the underlying function of these and many other disease-causing genes in vivo.
Experimental Procedures
Fly strains
The strains used in this study including the mutations and duplications and deletion strains used for mapping are described in flybase (Marygold et al., 2013) (see also Extended Experimental Procedures).
Isogenization and Mutagenesis
Isogenization of y w FRT19A chromosome was performed using standard genetic crosses. Mutagenesis was performed by feeding isogenized y w FRT19A iso males with sucrose solution containing low concentration (7.5–10mM) of EMS as described (Bokel, 2008). After recovery from mutagenesis, these males were mated en masse with Df(1)JA27/FM7c Kr>GFP virgin females for 3 days. In the F1 generation, y w mut* FRT19A/FM7c Kr>GFP (mut* indicates the EMS-induced mutation) virgins were collected and 33,887 individual females were crossed with FM7c Kr>GFP males to establish independent balanced stocks. 5,859 lines carried lethal mutations and the remaining stocks were discarded.
Complementation and Mapping
Lines that exhibited a strong morphological and/or ERG phenotype were subjected initially to duplication mapping. Subsequently, lines that were rescued by the same duplication and exhibit similar phenotypes were crossed inter se to establish complementation groups based on lethality. Complementation groups were further fine mapped using deficiencies that cover the region of interest.
Gene identification
When a complementation group was mapped to a small region (~30–300 kb, varies depending on available resources), we searched for publically available lethal mutations that map to the same region using FlyBase (Marygold et al., 2013). We performed complementation tests using >1 mutant allele when possible. For complementation groups that complemented all available lethal mutations in the region, we performed Sanger sequencing using standard methods. To expedite gene identification we also used Illumina based whole-genome sequencing technology (Haelterman et al., 2014)
Ethics Statement
Informed consent was obtained prior to participation from all subjects or parents of recruited subjects under an Institutional Review Board approved protocol at BCM.
Study Subjects
The analysis of 1,929 exomes from BHCMG described was performed in a database from the WES of over 160 separate phenotypic cohorts. The sequencing data included family-based studies in which both affected and unaffected family members were sequenced, single individuals with unique phenotypes, as well as larger cohorts of up to 50–60 cases with the same phenotype. Selection of subjects was performed by a phenotypic review committee based on the likelihood of the Mendelian inheritance for the disease phenotype.
Whole-Exome Capture, Sequencing and Data analysis
All of the subjects enrolled in the BHCMG underwent WES using methods previously described (Lupski et al., 2013) (Extended Experimental Procedures). Produced sequence reads were mapped and aligned to the GRCh37 (hg19) human genome reference assembly using the HGSC Mercury analysis pipeline (http://www.tinyurl.com/HGSC-Mercury/). Variants were determined and called using the Atlas2 suite to produce a variant call file (VCF). High-quality variants were annotated using an in-house developed suite of annotation tools (Bainbridge et al., 2011).
ANKLE2 construct and transgenesis
Human ANKLE2 cDNA was cloned into pUASTattB (Bischof et al., 2007) tagged vectors (N-terminal FLAG) using In-Fusion HD Cloning Kit (Clontech) and vector was linearized with NotI and XhoI. The construct was inserted in VK33 (Venken et al., 2006)
Supplementary Material
Highlights.
A Drosophila resource of mutants affecting adult neural development and maintenance
Essential fly genes with multiple human homologs are often associated with disease
Variants in human homologs of these genes are associated with Mendelian disease
ANKLE2 is associated with small brain size and microcephaly in fly and man
Acknowledgments
We thank Y. Chen, C. Benitez, X. Shi, S. Gibbs, A. Jawaid, H. Wang, Y. Q. Lin, D. Bei and L. Wang, Yuchun He and Hongling Pan for technical support, Y-N. Jan, T. Kaufman, C. Doe, U Banergee, J. Olson, K. Cook and D. Bilder for reagents, J. Shulman, J. Zallen, E. Seto and H. Y. Zoghbi for critical reading of this manuscript. This study was supported by the National Institutes of Health (NIH) 1RC4GM096355-01 (HJB and RCh), U54HG006542 (BHCMG), R01NBS058529 (JRL), 5P30HD024064 (Confocal microscopy at the Intellectual and Developmental Disabilities Research Center), K23NS078056 (WW) 5R01GM067858 (HS), T32 NS043124-11(HS), and 5K12GM084897 (HS), K08NS076547(MW), EY021163 (RA), EY019861 (RA), and EY019007 (RA and SHT). Additional support: Nakajima Foundation and the Jan and Dan Duncan Neurological Research Institute at Texas Children’s Hospital (SY) National Science Centre Poland (DEC-2012/06/M/NZ2/00101) (WW), Houston Laboratory and Population Science Training Program in Gene-Environment Interaction from the Burroughs Wellcome Fund (Grant No. 1008200) (BX), NSF DMS# 1263932 (ZL), Bogazici Univeristy Research Found (09B101P) (EB) Research to Prevent Blindness to the Department of Ophthalmology, Columbia University (RA, SHT). HJB is a Howard Hughes Medical Institute Investigator and received funds from the Robert and Renee Belfer Family Foundation, the Huffington Foundation, and Target ALS.
Footnotes
Accession numbers
Data have been reported to dbGAP, and a list of accession numbers is available in Table S6. The project accession ID at dbGAP is phs000711.v1.p1 and additional details are available at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000711.v1.p1
Author Contributions
Thorax and wing screen SY and WC. Eye and ERG screen MJ, BX, KZ and VB. Fly mapping SY, MJ, WC, BX, KZ, VB, HS, NH, GD, TL, KC, UG, AM, KS and RCh. Bioinformatic analysis: SY, MJ, MW, YW and ZL. Whole-exome sequencing: TG, SJ, DM, EBo, RG and JL. Human genome analysis SY, MJ, WC, TG, EK, WW, LV, JD, TH, HS, NH, GD, TL, KC, UG and MW. Clinical data and segregation analysis: EK, DP, YP, MS and EBaSP, YX, ST and RA SP, GM, RCl, CC and WD. Fly experiments on oc mutant: MJ, and dAnkle2 mutant : MJ, NL, WC. Designed the study and wrote the manuscript: SY, MJ, JL, MW and HB. SY and MJ contributed equally.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asencio C, Davidson IF, Santarella-Mellwig R, Ly-Hartig TB, Mall M, Wallenfang MR, Mattaj IW, Gorjanacz M. Coordination of kinase and phosphatase activities by Lem4 enables nuclear envelope reassembly during mitosis. Cell. 2012;150:122–135. doi: 10.1016/j.cell.2012.04.043. [DOI] [PubMed] [Google Scholar]
- Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Shendure JA, Valle D, Hamosh A, Lupski JR, Gibbs RA, Boerwinkle E, Lifton RP, Gerstein M, Gunel M, et al. The Centers for Mendelian Genomics: a new large-scale initiative to identify the genes underlying rare Mendelian conditions. Am J Med Genet A. 2012;158A:1523–1525. doi: 10.1002/ajmg.a.35470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat V, Thiffault I, Jaiswal M, Tetreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci. 2010;11:514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benos PV, Gatt MK, Murphy L, Harris D, Barrell B, Ferraz C, Vidal S, Brun C, Demaille J, Cadieu E, et al. From first base: the sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 2001;11:710–730. doi: 10.1101/gr.173801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokel C. EMS screens : from mutagenesis to screening and mapping. Methods Mol Biol. 2008;420:119–138. doi: 10.1007/978-1-59745-583-1_7. [DOI] [PubMed] [Google Scholar]
- Ceron J, Gonzalez C, Tejedor FJ. Patterns of cell division and expression of asymmetric cell fate determinants in postembryonic neuroblast lineages of Drosophila. Developmental biology. 2001;230:125–138. doi: 10.1006/dbio.2000.0110. [DOI] [PubMed] [Google Scholar]
- Charng WL, Yamamoto S, Bellen HJ. Mechanisms underlying Drosophila peripheral nervous system and neuromuscular junction development share features with human neurodegenerative diseases. Curr Opin Neurobiol. 2014 doi: 10.1016/j.conb.2014.03.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook RK, Deal ME, Deal JA, Garton RD, Brown CA, Ward ME, Andrade RS, Spana EP, Kaufman TC, Cook KR. A new resource for characterizing X-linked genes in Drosophila melanogaster: systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics. 2010;186:1095–1109. doi: 10.1534/genetics.110.123265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Brittle AL, Ito T, Ohkura H. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol. 2005;171:593–602. doi: 10.1083/jcb.200508127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K. Signaling mechanisms that coordinate the development and maintenance of dendritic fields. Curr Opin Neurobiol. 2012;22:805–811. doi: 10.1016/j.conb.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Lotze T, Jamal L, Penney S, Campbell IM, Pehlivan D, Hunter JV, Woodbury SL, Raymond G, Adesina AM, et al. Mutations in VRK1 Associated With Complex Motor and Sensory Axonal Neuropathy Plus Microcephaly. JAMA Neurol. 2013 doi: 10.1001/jamaneurol.2013.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11:222–223. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- Haelterman N, Jiang L, Li S, Bayat V, Ugur B, Tan KL, Zhang K, Bei D, Xiong B, Charng WL, et al. Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Research. 2014 doi: 10.1101/gr.174615.114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Sobreira N, Hoover-Fong J, Sutton VR, Boehm C, Schiettecatte F, Valle D. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34:566–571. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Xiao X, Li S, Jia X, Wang P, Guo X, Zhang Q. CRX variants in cone-rod dystrophy and mutation overview. Biochem Biophys Res Commun. 2012;426:498–503. doi: 10.1016/j.bbrc.2012.08.110. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Jaiswal M, Sandoval H, Zhang K, Bayat V, Bellen HJ. Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu Rev Genet. 2012;46:371–396. doi: 10.1146/annurev-genet-110711-155456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaindl AM, Passemard S, Kumar P, Kraemer N, Issa L, Zwirner A, Gerard B, Verloes A, Mani S, Gressens P. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–383. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M, Campbell IM, et al. Human CLP1 Mutations Alter tRNA Biogenesis, Affecting Both Peripheral and Central Nervous System Function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- Liao TS, Call GB, Guptan P, Cespedes A, Marshall J, Yackle K, Owusu-Ansah E, Mandal S, Fang QA, Goodstein GL, et al. An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function. Genetics. 2006;174:525–533. doi: 10.1534/genetics.106.061705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Vogel H. Drosophila models of neurodegenerative diseases. Annual review of pathology. 2009;4:315–342. doi: 10.1146/annurev.pathol.3.121806.151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, Kovar CL, Wang M, Hawes AC, Reid JG, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary KL, Park TJ, Woods JO, Cha HJ, Wallingford JB, Marcotte EM. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr SE. RNAi screening in Drosophila cells and in vivo. Methods. 2014;68:82–88. doi: 10.1016/j.ymeth.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Oortveld MA, Keerthikumar S, Oti M, Nijhof B, Fernandes AC, Kochinke K, Castells-Nobau A, van Engelen E, Ellenkamp T, Eshuis L, et al. Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet. 2013;9:e1003911. doi: 10.1371/journal.pgen.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Xu T. Dissecting social cell biology and tumors using Drosophila genetics. Annu Rev Genet. 2013;47:51–74. doi: 10.1146/annurev-genet-110711-155414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujano MA, Sanchez-Pulido L, Pennetier C, le Dez G, Basto R. The microcephaly protein Asp regulates neuroepithelium morphogenesis by controlling the spatial distribution of myosin II. Nat Cell Biol. 2013;15:1294–1306. doi: 10.1038/ncb2858. [DOI] [PubMed] [Google Scholar]
- Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Durrenberger M, Paro R, Boutros M, Merdes G. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell. 2010;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Saksena S, Emr SD. ESCRTs and human disease. Biochem Soc Trans. 2009;37:167–172. doi: 10.1042/BST0370167. [DOI] [PubMed] [Google Scholar]
- Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Silhavy JL, Xue Y, Kayserili H, Yasuno K, et al. CLP1 Founder Mutation Links tRNA Splicing and Maturation to Cerebellar Development and Neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandendries ER, Johnson D, Reinke R. orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol. 1996;173:243–255. doi: 10.1006/dbio.1996.0020. [DOI] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Popodi E, Holtzman SL, Schulze KL, Park S, Carlson JW, Hoskins RA, Bellen HJ, Kaufman TC. A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics. 2010;186:1111–1125. doi: 10.1534/genetics.110.121285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Seto ES. Dopamine dynamics and signaling in Drosophila: an overview of genes, drugs and behavioral paradigms. Exp Anim. 2014;63:107–119. doi: 10.1538/expanim.63.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.