Abstract
Zebrafish has been used as a powerful model system in biological and biomedical studies studying development and diseases. Comparative, functional, and developmental studies on zebrafish morphology require precise visualization of 3D morphological structures. Few methods that can visualize whole-volume of zebrafish tissues are available because optical bio-imaging methods are limited by pigmentation and hard tissues. To overcome these limitations, the 3D microstructures of a hypercholesterolemic zebrafish model are visualized using synchrotron X-ray micro-computed tomography (SR-μCT). The model spatial resolution ranged from sub- to several microns. The microstructures of various zebrafish organs are observed by combining high-contrast staining (osmium tetroxide and uranyl acetate) and embedding a protocol to enhance the image contrast of soft tissues. Furthermore, blood vessels are identified using a barium sulfate injection technique. The internal organs and cells, such as liver, intestine, oocytes, and adipocytes, of a hypercholesterolemic zebrafish are compared with those of normal organs and cells. The SR-μCT is useful for understanding the pathogenesis of circulatory vascular diseases by detecting the modifications in the 3D morphological structures of the whole body of the zebrafish. This bio-imaging technique can be readily used to study other disease models.
Introduction
Zebrafish (Danio rerio) is a useful vertebrate model system to investigate human diseases. This tiny fresh water fish has genetic similarity to human and contains the orthologues of more than 70% of human disease-related genes in its genome.1 Disease development is efficiently assisted by gene manipulation techniques and expression level control in zebrafish. In this process, the phenotypic outcome screening is highly facilitated by the zebrafish's high fecundity and rapid development from embryo to adult. To date, various disease models of zebrafish have already been developed in different fields, such as carcinogenesis, pathogenic infection, inflammation, immunological diseases, metabolic disorders, and hormonal and nutritional diseases. These models provide invaluable insight for disease pathogenesis and are also used for substance screening during drug development.
Although a zebrafish lives for 2–3 years and grows up to 6 cm in length, studies have focused only on the zebrafish's first several days after fertilization because its body is optically transparent. In this initial stage, screening abnormal phenotypes is easy because their internal organs and fluorescence labels for monitoring specific molecules of interest can be directly observed without any surgical treatment. This transparency is lost at about 14 days postfertilization (dpf) because pigmentation cells in the body are activated. Recently, a mutant strain that remains transparent throughout its life has been developed. However, its internal organs remain opaque, thus limiting the extent of the visible areas. In addition, its tissues' intrinsic scattering characteristics can also lead to a relatively poor image contrast in optical microscopy.
Recent advances in high-resolution X-ray micro-computed tomography (μCT) enable us to observe zebrafish models at later stages. In addition, the small size of zebrafish makes possible whole-body imaging at cell resolutions by μCT. Since the synchrotron μCT with X-ray beam of moderate energy (10–25 keV) is unimpeded by the pigmentation or tissue thicknesses encountered in zebrafishes at larval stages and beyond, it is well suited to the detailed observation of their phenotypes.2 This imaging modality provides 3D views of the internal and external morphological structures of the opaque test samples at a micrometer-scale resolution and is nondestructive. However, obtaining high-quality images of the soft tissues in bio samples using μCT is a challenging task compared with optical microscopy. Although the optical transparency of a developing zebrafish provides new opportunities to investigate the mechanisms governing the adipocyte biology, the zebrafish adipocytes still remain uncharacterized.3 Therefore, in this study, the zebrafish adipocytes are visualized using labeling lipids with high-atomic number elements, such as osmium tetroxide, uranyl acetate, and barium sulfate, to enhance the image contrast of the soft tissues.
A special type of μCT with synchrotron radiation X-ray beam as its source (SR-μCT) is utilized to examine the anatomical modifications of zebrafish induced by hypercholesterolemia. Hypercholesterolemia is one of the major risk factors for cardiovascular diseases (CVDs), and its clinical treatment is an essential strategy to prevent CVDs.4 Whole-body images of hypercholesterolemic zebrafish of 38 and 45 dpf exhibit lipid accumulation in perivascular fat, subcutaneous fat, and lipid droplets in the small intestine and the liver.
Materials and Methods
Feeding zebrafish
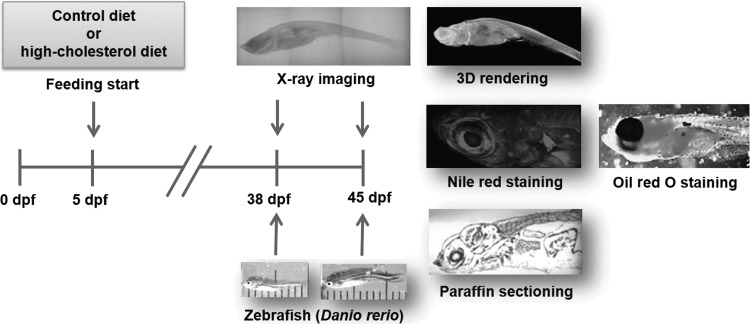
All experimental procedures complied with the act on life ethics and safety of the Ministry of Health and Welfare of South Korea. Wild-type zebrafish embryos were obtained through in vitro fertilization and natural spawning of adults maintained at 28.0°C±0.5°C under a 14 h:10 h light–dark cycle. The hypercholesterolemic zebrafish model was prepared by soaking the fish food (Baby meal; Jail Feed Corporation) in a diethyl ether solution containing cholesterol (Sigma-Aldrich) to achieve 4% (w/w) cholesterol in the food after evaporating the ether solution.5 The zebrafish larvae were fed twice a day, starting at the 5 dpf, with either control (normal) diet or high-cholesterol diet (HCD) for 33 or 40 days. Figure 1 shows a schematic of the experimental setup utilized to observe the hypercholesterolemic zebrafish.
FIG. 1.
Experimental scheme for observing zebrafishes fed with control or high-cholesterol diet (HCD). When zebrafish larvae start free feeding (5 days postfertilization [dpf]), normal diet or HCD starts, and feeding is continued for 33 or 40 days. Lower figures show representative photographs of 38 and 45 dpf juvenile zebrafishes. The total body lengths of the 38 and 45 dpf models are about 10 and 13 mm, respectively. Barium sulfate was injected into the vessels to observe the blood vessels of a zebrafish sample. Zebrafish was fixed in formalin and stained with osmium tetroxide and uranyl acetate. The fish was then embedded in Spurr's resin for X-ray microscopy and scanned in the resin block. Upper figures show a typical phase-contrast X-ray image of a 38 dpf zebrafish and its 3D rendering image. Nile red and Oil red O staining methods and two-photon microscopy technique were performed to identify adipocytes or lipid droplets in zebrafish models and to compare with synchrotron X-ray micro-computed tomography (SR-μCT) images. Paraffin sectioning (10 μm thickness) and EVG staining were also carried out to compare PSR-μCT images with histological sectioning images.
In vivo labeling and imaging of zebrafish adipocytes
In this study, we examined the SR-μCT images of the normal and hypercholesterolemic zebrafishes. The bright parts in the X-ray images presumably indicate lipid droplets or adipocytes. To prove this presumption, the SR-μCT images were compared with the corresponding Nile red images.
Nile red (Sigma-Aldrich) was dissolved in acetone at 500 mg/mL and stored in a dark environment at −20°C. The vessel containing live unanesthetized zebrafish was supplemented with Nile red to a final concentration of 5 μg/mL egg water. The vessel was then placed in the dark for 30 min. Afterward, zebrafish was anesthetized with 0.4% tricaine (Sigma-Aldrich) and mounted in 7% methylcellulose. Optical images were captured through Leica two-photon microscope (TPM) (TCS SP5II MP; Leica Microscopy Systems, GMBH) with a 10×objective lens (Leica Microscopy Systems, GMBH) and a laser (1100 nm). The acquired TPM images were analyzed and processed using LAS AF 2.7 software (Leica Microscopy Systems, GMBH).
Injection of barium sulfate
Barium sulfate (Sigma-Aldrich) and gelatin (from cold-water fish skin; Sigma-Aldrich) were used as contrast agents to obtain high-contrast images of the circulatory vascular system. Barium sulfate was subsequently filtered (pore size 5 μm; Millex-SV; Millipore) to perfuse microvessels.6 The 15% barium sulfate and 2% gelatin were then suspended in phosphate-buffered saline (PBS) solution.7 Zebrafishes were anesthetized with 0.4% tricaine (Sigma-Aldrich). To inject barium sulfate into the bloodstream of zebrafish, a procedure in which retro-orbital injection was given into venous sinus was adopted and monitored. This injection method can minimize mortality and allows the efficient delivery of barium sulfate/gelatin solution into the bloodstream.8 A heated barium sulfate/gelatin solution was injected into zebrafish, which was subsequently placed on ice for 20 min to promote suspension gelling within the vasculature. Zebrafishes were fixed with 2% glutaraldehyde and 2% paraformaldehyde for 24 h at 4°C. The samples used for SR-μCT were prepared in a similar manner as with the transmission electron microscopy (TEM) sample preparation. Briefly, barium sulfate was injected into a blood vessel of the anesthetized zebrafish. The zebrafish model was euthanized and soaked in a fixing agent and transferred to the staining solutions of osmium tetroxide and uranyl acetate solutions in tandem to enhance the image contrast of the soft tissues. The sample was then dehydrated and finally infiltrated with Spurr's resin to solidify.
Contrast stains and sample preparation
All materials used for the contrast staining and sample preparation were purchased from Sigma-Aldrich. The staining process was performed by placing the fixed zebrafishes in 1% osmium tetroxide for 24 h (osmium tetroxide penetrates into tissues by diffusion). Zebrafishes were removed and washed with sodium cacodylate buffer (0.1 M, pH 7.4) solution. The samples were subsequently postfixed with 0.5% uranyl acetate for 24 h. This process was followed by further washing, as previously described. The specimens were sequentially dehydrated using ethanol (30%, 50%, 70%, 100%, and 100%) for 1 h and then infiltrated with 0%, 25%, 50%, and 75% Spurr's resin in propylene oxide for 2 h each. After the overnight infiltration with 100% Spurr's resin at room temperature, each sample was embedded in the Spurr's resin using a 200 μL heat-sealed pipette tip and then incubated at 60°C for 24 h to rigidify the resin.
Experimental setup and data acquisition
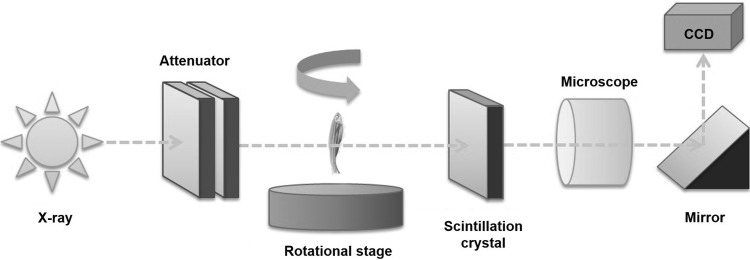
All SR-μCT experiments were performed at the biomedical imaging (BMI) beamline of the Pohang Light Source-II (PLS-II) in Pohang, Korea. The new BMI beamline was recently constructed, and regular user services started on September 2013. A white beam emitted by a multipole wiggler source of the PLS-II 3 GeV storage ring was filtered through 1 mm graphite, after passing through double multilayer monochromator, resulting in a photon energy of 10–50 keV. The optimal monochromatic X-ray energy for tomographic scanning was experimentally determined to be ∼24 keV. Figure 2 shows a schematic layout of the BMI beamline. The detector was positioned at 20 cm from the test sample for phase-contrast imaging. The monochromatic beam transmitted through the sample was detected by a detector (Andor Zyla) with 2560×2160 pixels, on which a YAG:Ce (30 μm thick) scintillation crystal was attached. The X-ray image of the charge-coupled device (CCD) camera on the scintillator was converted into a visible image. The image was then magnified using two different 4× (for 45 dpf zebrafish imaging) and 10× (for 38 dpf zebrafish imaging) microscopic objective lenses. The field of view (FoV) of these two lenses has horizontal and vertical sizes of 4.2×3 and 1.7×1.4 mm2, respectively. The corresponding effective pixel size is 1.6×0.65 μm. The specimen embedded in a resin was mounted on a sample holder. An air-bearing rotary stage was used for tomographic scanning. To reconstruct a tomographic image, 360 projection images were obtained from 0° to 180° at intervals of 0.5°.
FIG. 2.
Schematic of the experimental set-up for SR-μCT experiment performed at 6C biomedical beamline of PLS. Mono beam was attenuated by a graphite attenuator. Beam propagated 23 cm from test sample to a CdWO4 scintillation crystal that was magnified by a 4× or 10× objective lens. Images were captured using a charge-coupled device (CCD) camera. Sample was rotated on a sample stage, and projection images were captured from 0° to 180° at intervals of 0.5°.
Image analysis
In a raw projection image, all data along the synchrotron X-ray beam pathway are overlapped. Therefore, the local information is hardly discernible. To retrieve the local information, an image reconstruction process is required. The 3D morphological structure of the sample was numerically reconstructed from the tomographic projection images using Octopus 8.5 software (in CT). For the 3D reconstruction, the sizes of the projection images were reduced by 2× binning. The 3D volume image of the specimen was then obtained by applying a filtered back-projection algorithm to the projection images. The cross-sectional images were stacked to produce a 3D image. The stacked images were rendered as a 3D structure using Amira® 5.3.3 image-analysis software (Visualization Sciences Group).
Oil red O staining
Control and hypercholesterolemic zebrafish models were euthanized with 0.4% tricaine (Sigma-Aldrich). They were fixed in 2% glutaraldehyde and 2% paraformaldehyde for 24 h at 4°C. After fixation, they were washed with PBS and infiltrated with a graded series of propylene glycol (Fisher Scientific) and stained with 0.5% Oil red O (Sigma-Aldrich) in 100% propylene glycol. The Oil red O-stained samples were washed with propylene glycol and observed by a microscope (Zeiss Axiovert 200) with a 2.5× objective lens.
Sample preparation for histological analysis
Control and hypercholesterolemic zebrafishes were euthanized with 0.4% tricaine (Sigma-Aldrich). They were fixed in 2% glutaraldehyde and 2% paraformaldehyde for 24 h at 4°C. After fixation, they were embedded in paraffin according to the standard protocol and were histologically processed. Slices of 10 μm were deparaffined and stained with elastic van gieson (EVG, Merck). The EVG-stained samples were observed by a microscope (Zeiss Axiovert 200) with a 40× phase-contrast objective lens.
Sample preparation for TEM imaging
Temporal variations of lipid droplets in the zebrafish models were observed with a TEM. Zebrafish samples were fixed in 2% glutaraldehyde and 2% formaldehyde, postfixed in 2% osmium tetroxide, dehydrated in a graded ethanol series up to 100%, and embedded in LX-112 resin. They were sectioned using Ultramicrotome (MT-X; RMC). The sliced samples were stained with 2% uranyl acetate and lead citrate. Their images were recorded by using TEM (JEOL JEM-1011).
Results
Whole-body imaging of hypercholesterolemic zebrafish
We collected the 38 and 45 dpf female zebrafishes for the SR-μCT study. The widths of the 38 dpf control and hypercholesterolemic zebrafishes are around 1 mm, which is well fitted to the SR-μCT FoV under the high-spatial resolution mode with a 10× lens (FoV: 1.7×1.4 mm, pixel size: 0.65 μm). Whole-body images of the control and hypercholesterolemic zebrafish models were obtained using SR-μCT.
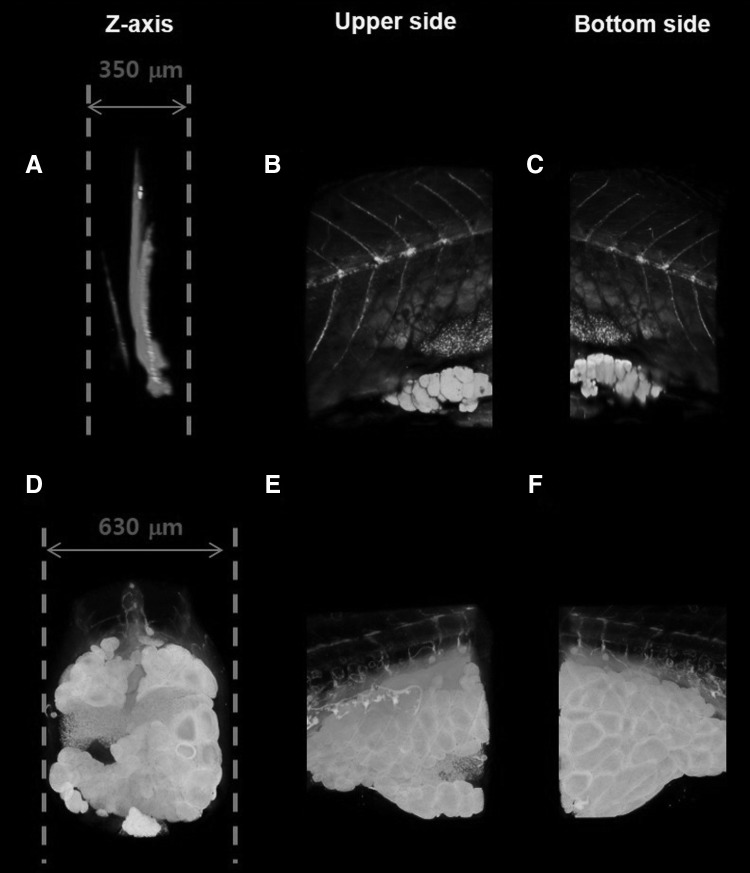
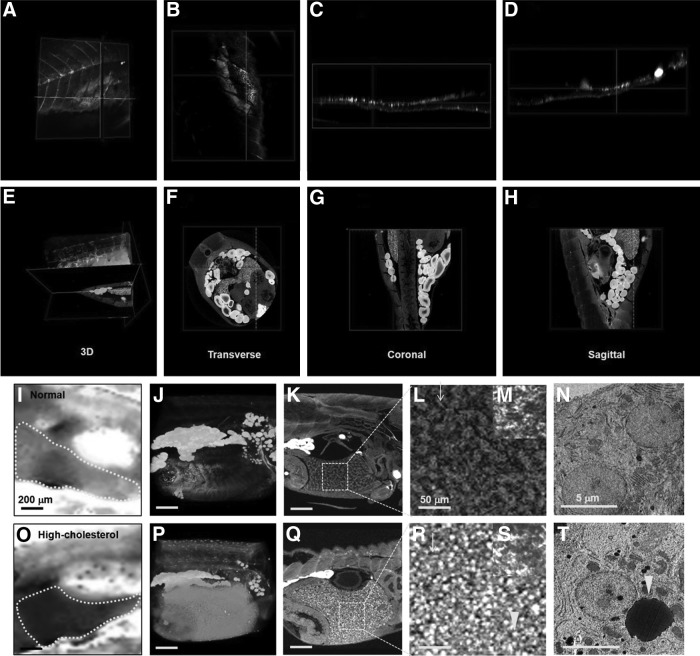
Figure 3 shows typical TPM images (Nile red images) and the corresponding SR-μCT images of a 38 dpf hypercholesterolemic zebrafish. The TPM images show the presence of neutral lipids in the zebrafish model. However, the whole body of a zebrafish is difficult to observe because of the technical limitations of the TPM technique. Figure 3C shows that the TPM image of the far side of the lens (bottom side) is somewhat distorted. However, the internal organs are clearly identified (presented with pseudo-coloring of each organ in Figure 3D–F in the SR-μCT images). This visibility is attributed to the staining of heavy metal compounds with osmium tetroxide and uranyl acetate, which bind to the lipid organic molecules of lipids, proteins, and nucleic acids, resulting in the image contrast enhancement of soft tissues. In addition, blood vessels became detectable because of the injection of barium sulfate. The combination of heavy metal staining and SR-μCT technique facilitated the assessment of the morphological characteristics and lipid distribution. By utilizing the absorption and phase-contrast effects, the major organs and tissues are readily identified in the SR-μCT images with a spatial resolution of about 0.65 μm. Especially, the blood vessels were clearly identified by injecting barium sulfate. In this study, the high SR-μCT spatial resolution combined with the high-contrast staining method allows us to visualize the microstructures of various zebrafish organs by enhancing the image contrast of the soft tissues. Therefore, the internal organs and cells, such as the intestine, liver, oocytes, and adipocytes, of the normal and hypercholesterolemic zebrafishes can be evidently compared.
FIG. 3.
Comparison of SR-μCT images and corresponding two-photon microscope (TPM) images of a hypercholesterolemic zebrafish (38 dpf). (A) Three-dimensional image of HCD zebrafish reconstructed from images scanned along the z-axis. Zebrafish was stained with Nile red and imaged with two-photon laser scanning fluorescence microscopy. Nile red staining reveals neutral lipid in zebrafish model. (D) Three-dimensional volume rendering image reconstructed from projection images captured by SR-μCT. (B, E) Upper-side images of 3D reconstructed images (A, B), respectively. (C, F) Bottom-side images of 3D reconstructed images (A, B), respectively.
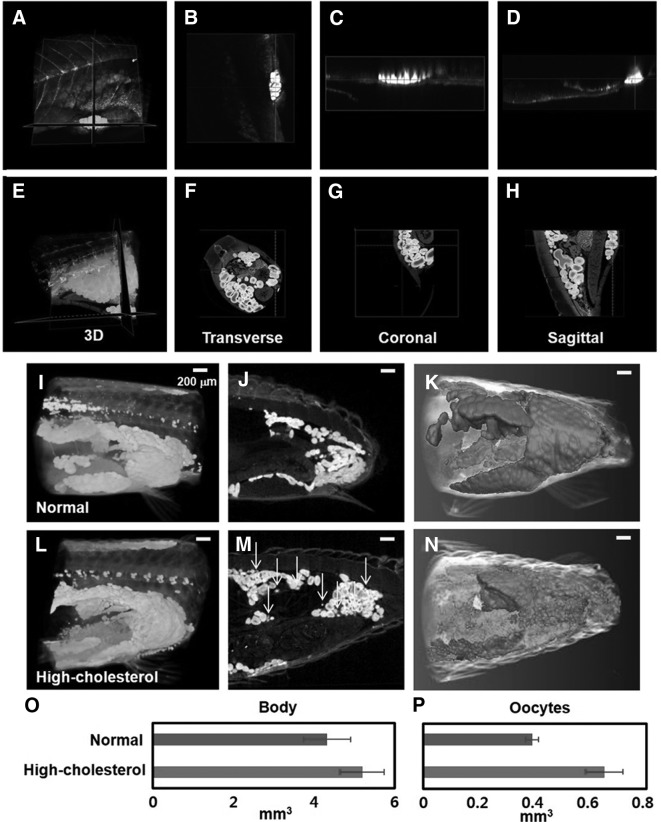
Oocytes of hypercholesterolemic zebrafishes
Figure 4A–D show that the zebrafish oocytes are not distinctively observed in the TPM images. However, the morphological variations in the oocytes from the zebrafish disease models are more evidently observed in the SR-μCT images (Fig. 4E–H) than in the TPM images. Figure 4I and L show the typical 3D volume-rendered PSR-μCT images of the normal and hypercholesterolemic zebrafish models. Various stages of oocyte maturation are simultaneously observed because the oogonium maturation is asynchronous. However, the normal zebrafishes have a relatively large number of mature eggs in the ovary and oviduct compared with the hypercholesterolemic zebrafishes. Figure 4J and M show that the sagittal sections of the 3D reconstructed image exhibit structural differences between the oocytes of the normal and hypercholesterolemic zebrafish models. The arrows in Figure 4M indicate the immature oocytes (stage I oocytes). Immature oocytes are ∼10–50 μm in physical dimension. Three-dimensional reconstruction image (Fig. 4N) shows immature oocytes more clearly, compared to Figure 4M. The volumetric changes in the normal and hypercholesterolemic female zebrafishes are assessed. As shown in Figure 4O, the net volumes of the body of the normal and hypercholesterolemic zebrafish models do not show statistically significant differences. However, the net volume of the oocytes in the hypercholesterolemic female zebrafish is about 66% larger than that from the control group (Fig. 4P). From these results, we can conclude that female infertility is induced by the disorders in the lipid metabolism through HCD.
FIG. 4.
Comparison of Nile red and SR-μCT images of oocyst of a hypercholesterolemic zebrafish (38 dpf). (A–H) Images of Nile red (A–D) and SR-μCT (E–H) of a hypercholesterolemic zebrafish (38 dpf). (B–D, F–H) Multi-view sections of a zebrafish model. (A–D) TPM (Nile red) images exhibit technical limitation in the whole-body imaging of zebrafish. (I, L) Three-dimensional volume-rendered SR-μCT images of normal (I) and hypercholesterolemic zebrafish models (45 dpf) (J). (J, M) Sagittal images of 3D reconstructed images of (I, L), showing structurally different oocytes of the normal and hypercholesterolemic zebrafishes, respectively. (K, N) Tomographic reconstruction of 3D structure of oocytes in the normal and hypercholesterolemic zebrafishes. (M) Arrows indicate immature oocytes (stage I oocytes). High-cholesterolemic zebrafish model exhibits a large number of immature oocytes. (O, P) Comparison of total volume of the body and oocytes in the normal and hypercholesterolemic zebrafish models (45 dpf) represented in (I–N), respectively (n=3 animals/group). Scale bar: 200 μm.
Lipid droplets in the liver
In this study, the bright parts in the SR-μCT images are assumed to indicate lipid. To prove this presumption, the SR-μCT images are compared with the corresponding Nile red images. Although lipid droplets in the liver of a hypercholesterolemic zebrafish are detected in the captured Nile red images (Fig. 5A–D), however, they are not so clearly discriminated. We suppose that the image contrast of TPM images of lipid droplets is largely deteriorated by the surround thick tissues and pigmentation. However, the SR-μCT and heavy metal staining method employed in this study clearly reveals lipid droplets in the liver of hypercholesterolemic zebrafish models (Fig. 5E–H).
FIG. 5.
Comparison of Nile red, Oil red O stained images, paraffin-sectioned images, SR-μCT images, and transmission electron microscopy (TEM) images of livers of normal and hypercholesterolemic zebrafishes (38 dpf). The lipid droplets in the liver of a hypercholesterolemic zebrafish are well detected in the SR-μCT images. (A–D) TPM images of a hypercholesterolemic zebrafish. (A) Three-dimensional reconstruction image. (B–D) Multi-view section images of (A). Nile red staining method somewhat exhibit lipid droplets in the liver of hypercholesterolemic zebrafish, but not so distinctive. (E–H) SR-μCT images of a hypercholesterolemic zebrafish. (E) Three-dimensional reconstruction image. (F–H) Multi-view section images of (E). The combination of SR-μCT and heavy-metal staining method reveals lipid droplets clearly in the liver of the hypercholesterolemic zebrafish. (I, O) Oil red O stained images of the normal and hypercholesterolemic zebrafish models, respectively. Each boundary of the liver is delineated by a dotted line. Lipid droplets in the liver of the hypercholesterolemic zebrafish model are dyed deeper red, compared to the normal model. (J, P) Three-dimensional volume-rendered images of normal-diet zebrafish (J) and HCD zebrafish (P). (K, Q) Sagittal images of 3D reconstructed images (J, P). (L, R) Highly magnified views of dotted boxes in (K, Q). Arrows indicate sinusoid. (M, S) Images of paraffin-sectioned liver stained with EVG of normal and hypercholesterolemic zebrafishes. (N, T) TEM images of the liver of the normal and hypercholesterolemic zebrafishes. Arrowhead indicates a lipid droplet. Scale bars: (I–K, O–Q) 200 μm, (L, M, R, S) 50 μm, (N, T) 5 μm.
The Oil red O image in Figure 5O shows that more lipid droplets are present in the liver of the high-cholesterolemic zebrafish model compared with that of the normal zebrafish model (Fig. 5I). Figures 5J and P show the typical 3D of the normal and hypercholesterolemic zebrafishes. Figures 5K and Q are transverse sectional images of Figure 5J and P, respectively. Figures 5L and R show that the sinusoidal organization and the lipid contents of the livers of the normal and hypercholesterolemic zebrafishes are evidently different. The intensity level in the livers of the hypercholesterolemic zebrafishes is higher than that of normal zebrafishes.
To find the reason for high intensity of SR-μCT images, the histological images of the liver of the 38 dpf control and hypercholesterolemic zebrafishes are compared. Paraffin sectioning (10 μm thickness) and EVG staining were performed to compare the present SR-μCT images with the histological sectioning images. These slice images provide only 2D information, and the sample preparation procedure may induce a variety of stresses, which influence the morphological structures of the tissues. In the paraffin-sectioned images, the sinusoids and hepatocytes in the livers of the normal and hypercholesterolemic zebrafishes do not exhibit any noticeable difference (Fig. 5G, H). This is attributed to the fact that lipid components of zebrafish tissues were melted by xylene in the pretreatment process of paraffin section samples. However, based on the results observed in TEM images (Fig. 5T), we found that bright spots in SR-μCT images represent the lipid droplets inside hepatocytes of the hypercholesterolemic model. These results demonstrate that SR-μCT is a useful and powerful tool for detecting lipid droplets in zebrafish CVD models.
Lipid droplets in the small intestine
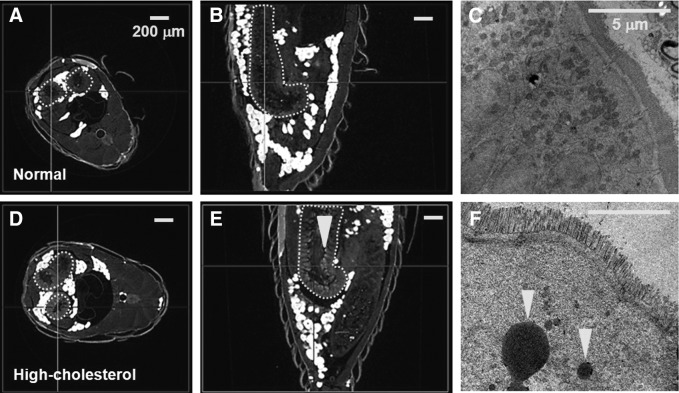
The small intestine is the primary site of dietary lipid absorption in mammals. In the previous study, to visualize intestinal lipids in the hypercholesterolemic zebrafish, fluorescently labeled lipids, lipophilic dyes, and fluorescent fusion proteins are used to monitor subcellular events without surgical or other invasive procedures.9 All of the transverse (Fig. 6A, D) and sagittal section images (Fig. 6B, E) show that the intensity of intestine of hypercholesterolemic zebrafishes (45 dpf) is higher than that of normal zebrafishes. From the previous study, we can conjecture that the high intensity of hypercholesterolemic zebrafish intestine is caused by HCD. In addition, TEM images of intestine of the normal and hypercholesterolemic zebrafishes (Fig. 6C, F) also support that higher intensity of hypercholesterolemic model results from the lipid droplets in the epithelial cells of small intestine. These results show that the use of SR-μCT would be helpful to enhance the understanding on the metabolism of intestinal lipid by detecting the modifications of lipid contents in 3D whole-body structures of zebrafish.
FIG. 6.
Cross-sectional views of SR-μCT images of the normal and hypercholesterolemic zebrafishes (45 dpf). (A, D) Transverse section images of the normal and hypercholesterolemic zebrafishes. (B, E) Sagittal section images of the normal and hypercholesterolemic zebrafishes. (A, B, D, E) The dotted lines represent boundaries of the small intestine. (E, F) Arrowheads indicate lipid droplets in the small intestine of the normal and hypercholesterolemic zebrafishes. Scale bars: (A, B, D, E) 200 μm, (C, F) 5 μm.
Lipid accumulation around blood vessels and under the skin
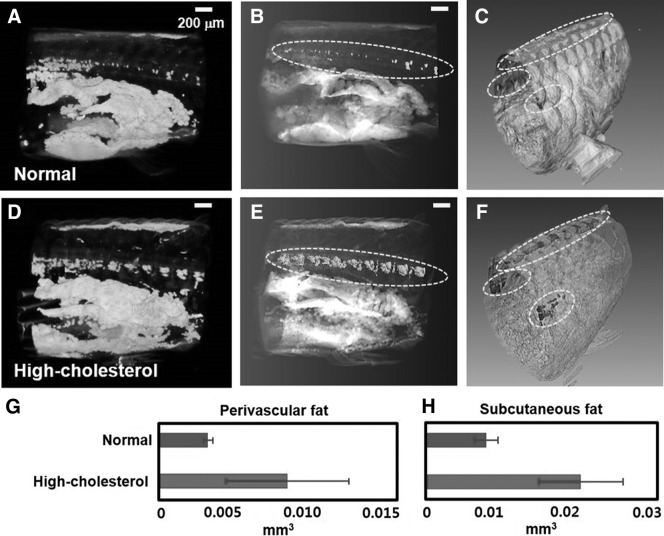
White adipose tissues are located at distinct depots in adult zebrafishes. Adipocytes have a unilocular lipid droplet. In the slightly inflated abdomen of the hypercholesterolemic zebrafish, adipose tissues expand into the regions surrounding the intestine and liver. In the SR-μCT images, adipocytes are hardly distinguishable from oocytes because their colors (image contrast) and sizes are similar. However, the perivascular fat and subcutaneous fat are distinct from the oocytes because of their site-related differences. Subcutaneous fat are generally located at distinct depots in adult zebrafishes and accumulate around the outer skin of a zebrafish. Perivascular fat are located around blood vessels. Especially, perivascular fat near the spine are clearly distinguished from other adipocytes and oocytes. Therefore, the volumetric changes of perivascular fat (near the spine, represented by dotted lines in Figures 7B and 7E and subcutaneous fat, represented by dotted lines in Figures 7C and 7F) in the body of the normal and hypercholesterolemic female zebrafishes are assessed in this study (Fig. 7). The internal anatomy of the 45 dpf hypercholesterolemic female zebrafish exhibits the accumulation of perivascular fat and subcutaneous fat. The net volumes of the normal and hypercholesterolemic zebrafish models are nearly similar. However, the mean volume of the perivascular fat and subcutaneous fat of the hypercholesterolemic female zebrafishes is about 167% and 161% larger than those of the control models, respectively (Fig. 7G, H).
FIG. 7.
Comparison of lipid accumulation in the normal (A) and hypercholesterolemic zebrafish models (D) (45 dpf). (B, E) Tomographic reconstruction of 3D structure of the perivascular fat in the dotted lines of images (A, D). (C, F) Tomographic reconstruction of 3D structure of subcutaneous fat in the dotted lines of images (A, D). (G, H) Comparison of the perivascular fat and subcutaneous fat of the normal and hypercholesterolemic zebrafish models shown in (A, D), respectively. Bar graphs indicate the total volumes of the subcutaneous fat and the perivascular fat (n=3 animals per group).
Discussion
Atherosclerosis and its complications are the main causes of death in developed countries. One of the major risks for atherosclerosis is hypercholesterolemia.10 Hypercholesterolemia is a well-established cardiovascular risk factor, and its clinical treatment is essential to prevent CVDs.4 A hypercholesterolemia disease model was established by feeding a zebrafish with cholesterol-enriched diet.5 The zebrafish model was observed to well respond to the diet, and gradually became obese. Hypercholesterolemic zebrafishes (obese zebrafish) tend to have higher percentage of body fat, compared with normal zebrafishes. Adipocytes serve as the major sites of fat storage and as the regulators of energy balance and inflammation.11 Therefore, we can better understand the adipocyte-associated diseases by using the information about percentage of body fat and distribution of adipocytes.
The optical transparency of zebrafishes at developing stage provides new opportunities for investigating the governing mechanisms of the adipocyte biology. However, zebrafish adipocytes are still unrevealed yet. Edward et al. observed adipose depots in subcutaneous layers along the dorsal and ventral midlines and near the base of other fins in adults older than 28 dpf. They visualized zebrafish adipocytes under in vivo condition by labeling neutral lipid droplets with Nile red.3 Although they mentioned that they observed smaller neutral lipid droplets in the liver and intestinal epithelia, the corresponding data were not included in their article. We suppose that the image contrast of fluorescent images of the lipid droplets is weakened by the surrounding thick tissues and pigmentation.
In this study, Nile red staining patterns in the TPM images and heavy metal staining patterns in the SR-μCT images were compared with the collocation arrangement. In addition, bio-imaging methods were developed to visualize the zebrafish adipocytes by labeling neutral lipid droplets with heavy metals. Approximately 10 to 25 keV X-ray beams penetrated the zebrafish model stained with a variety of contrast agents that have high atomic numbers, such as osmium and uranium.12–15 The osmium tetroxide staining method was employed to enhance the image contrast of μCT images. The K-shell energy of osmium (73.9 keV) and its tissue-binding properties allows osmium tetroxide to become a natural candidate for X-ray contrast staining.16 As osmium tetroxide chemically reacts with carbon-double bonds,17 it readily stains lipid-containing structures, such as plasma membrane and other lipid-rich structures.18–21 Therefore, in this study, the high-spatial resolution of SR-μCT combined with the high-contrast staining method makes us to visualize the microstructures of various zebrafish organs by enhancing image contrast of the soft tissues. In this study, we observed the increase of lipid accumulation in the whole body of hypercholesterolemic zebrafishes. Especially, based on the different osmium staining effects, lipid droplets are evidently observed in the livers and small intestines of the hypercholesterolemic zebrafishes. Fat depots in the liver can cause adverse cardiometabolic risks by affecting glucose and lipid metabolism.22 Perivascular fat has influence on the coronary atherosclerosis, cardiac function, and hemodynamic features.22 Therefore, the ectopic fat depots may contribute to better understanding about the link between body compositions and cardiometabolic risks.23
Conventional μCT system nondestructively acquires the 3D images of a thick tissue. However, its spatial resolution is insufficient to observe sinusoids. Therefore, applying the μCT system to hepatic biliary, arterial, and venous circulatory systems is difficult.24,25 Considering that the synchrotron radiation μCT has a spatial resolution at the submicron level, it can be used to observe sinusoids and their alterations in 3D space.26–28 In this study, the structural modifications in the sinusoids of the pathologically injured livers are detected through SR-μCT.
SR-μCT does not require an intricate dissection process and needs only formalin fixation and staining preparation. Therefore, the morphological variations in the oocytes from the zebrafish disease models are more evidently observed in the SR-μCT images. In this study, we can conclude that female infertility is induced by the disorders in the lipid metabolism through HCD. However, this study does not reflect the entire lifetime of the zebrafish models. Therefore, to overcome this limitation, further studies on zebrafishes from different age groups are necessary in the near future.
Combining with the contrast staining method, SR-μCT provides quantitative, high-resolution, and high-contrast images of 3D soft tissues without destroying the test samples. The obtained images are helpful for comparative, functional, and quantitative morphological studies. For example, the present imaging technique can be utilized effectively and uniquely in the Zebrafish Phenome Project,2 by probing the functions of vertebrate genes through phenotyping of mutants. In addition, this technique can be further extended to the bio-imaging experiments of other small laboratory animals suffering from other diseases.
Conclusion
X-ray μCT is a nondestructive bio-imaging technique that provides information about the 3D internal morphological structures of bio samples. However, direct application of this technique to soft tissues is difficult to their low X-ray attenuation. In this study, we developed a method for visualizing oocytes and adipocytes in zebrafishes by adopting heavy-metal staining. Especially, the lipid accumulations around blood vessels, under the skin, in the small intestine and the liver of zebrafish disease models are distinctly distinguished by combining the SR-μCT and the contrast staining method using osmium tetroxide, uranyl acetate, and barium sulfate. Using the contrast staining method, the SR-μCT provides quantitative, high-spatial resolution and high-contrast images of 3D soft tissues without destroying the test samples. The obtained images are helpful for comparative, functional, and quantitative morphological studies. The biological applications of the SR-μCT imaging technique to hypercholesterolemic zebrafish models have a strong potential for in vivo studies aiming to reveal the basic HCD mechanisms. In addition, this technique would be further extended to the bio-imaging modalities for observing other organs/tissues of small laboratory animals suffering from other diseases.
Acknowledgments
The authors are grateful to Ui-Ryong Kang and Jaeung Sim for their technical assistance, Sung-Hee Park for breeding and maintaining the zebrafishes, Sujung Kim for the valuable help in the TEM imaging experiments performed at the TEM laboratory of the POSTECH Biotech Center, and Gyu Seop Jo for the valuable help in the TPM imaging experiments performed at the POSTECH in vivo Microscopy Laboratory of the POSTECH Biotech Center. The authors are also grateful to the staff and supporting groups of the 6C X-ray medical imaging beamline of the Pohang Light Source (Pohang, Korea) for their assistance during the experiments. This study was supported by the National Research Foundation of Korea (NRF) of the Korean government (MSIP; Grant No. 2008-0061991).
Authors' Contributions
Conceived and designed the experiments: E.S.S. Performed the experiments: E.S.S. Analyzed the experimental data: E.S.S. Contributed analysis tools: J.H.L. and S.J.S. Wrote the article: E.S.S., S.J.L., J.H.L., and S.J.S.
Disclosure Statement
The authors have no competing interests to declare.
References
- 1.Langheinrich U. Zebrafish: a new model on the pharmaceutical catwalk. Bioessays 2003;25:904–912 [DOI] [PubMed] [Google Scholar]
- 2.Cheng KC, Xin X, Clark DP, La Riviere P. Whole-animal imaging, gene function, and the Zebrafish Phenome Project. Curr Opin Genet Dev 2011;21:620–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edward JF, III, Chad MT, John FR. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J Lipid Res 2009;50:1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mannarino MR, Ministrini S, Pirro M. Nutraceuticals for the treatment of hypercholesterolemia. Eur J Intern Med 2014;25:592–599 [DOI] [PubMed] [Google Scholar]
- 5.Stoletov K, Fang L, Choi SH, Hartvigsen K, Hansen LF, Hall C, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res 2009;104:952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myojin K, Taguchi A, Umetani K, Fukushima K, Nishiura N, Matsuyama T, et al. Visualization of intracerebral arteries by synchrotron radiation microangiography. AJNR Am J Neuroradiol 2007;28:953–957 [PMC free article] [PubMed] [Google Scholar]
- 7.Duvall CL, Taylor WR, Weiss D, Guldberg RE. Quantitative microcomputed tomography analysis of collateral vessel development after ischemic injury. Am J Physiol Heart Circ Physiol 2004;287:H302–H310 [DOI] [PubMed] [Google Scholar]
- 8.Pugach EK, Li P, White R, Zon L. Retro-orbital injection in adult zebrafish. J Vis Exp 2009;e1645 DOI: 10.3791/1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters JW, Anderson JL, Bittman R, Pack M, Farber SA. Visualization of lipid metabolism in the zebrafish intestine reveals a relationship between NPC1L1-mediated cholesterol uptake and dietary fatty acid. Chem Biol 2012;19:913–925 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sozen E, Karademir B, Yazgan B, Bozaykut P, Ozer NK. Potential role of proteasome on c-jun related signaling in hypercholesterolemia induced atherosclerosis. Redox Biol 2014;2:732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dru I, Kirsten CS. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev Dyn 2010;239:3013–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betz O, Wegst U, Weide D, Heethoff M, Helfen L, Lee WK, et al. Imaging applications of synchrotron X-ray phase-contrast microtomography in biological morphology and biomaterials science. I. General aspects of the technique and its advantages in the analysis of millimetre-sized arthropod structure. J Microsc 2007;227(Pt 1):51–71 [DOI] [PubMed] [Google Scholar]
- 13.Mizutani R, Takeuchi A, Uesugi K, Takekoshi S, Osamura RY, Suzuki Y. X-ray microtomographic imaging of three-dimensional structure of soft tissues. Tissue Eng Part C Methods 2008;14:359–363 [DOI] [PubMed] [Google Scholar]
- 14.Metscher BD. MicroCT for developmental biology: a versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn 2009;238:632–640 [DOI] [PubMed] [Google Scholar]
- 15.Metscher BD. MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 2009;9:11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiernan JA: Histological and Histochemical Methods: Theory and Practice. Pergamon Press, Oxford, England, 1990 [Google Scholar]
- 17.Ribi W, Senden TJ, Sakellariou A, Limaye A, Zhang S. Imaging honey bee brain anatomy with micro-X-ray-computed tomography. J Neurosci Methods 2008;171:93–97 [DOI] [PubMed] [Google Scholar]
- 18.Bozzola JJ, Russell LD: Electron Microscopy: Principles and Techniques for Biologists. Jones and Bartlett Publishers, Boston, 1992 [Google Scholar]
- 19.Di Scipio F, Raimondo S, Tos P, Geuna S. A simple protocol for paraffin-embedded myelin sheath staining with osmium tetroxide for light microscope observation. Microsc Res Tech 2008l;71:497–502 [DOI] [PubMed] [Google Scholar]
- 20.Raimondo S, Fornaro M, Di Scipio F, Ronchi G, Giacobini-Robecchi MG, Geuna S. Chapter 5 methods and protocols in peripheral nerve regeneration experimental research: part II-morphological techniques. Int Rev Neurobiol 2009;87:81–103 [DOI] [PubMed] [Google Scholar]
- 21.Hanker JS, Deb C, Wasserkrug HL, Seligman AM. Staining tissue for light and electron microscopy by bridging metals with multidentate ligands. Science 1966;152:1631–1634 [DOI] [PubMed] [Google Scholar]
- 22.Lim S, Meigs JB. Links between ectopic fat and vascular disease in humans. Arterioscler Thromb Vasc Biol 2014;34:1820–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim S, Meigs JB. Ectopic fat and cardiometabolic and vascular risk. Int J Cardiol 2013;169:166–176 [DOI] [PubMed] [Google Scholar]
- 24.Masyuk TV, Ritman EL, LaRusso NF. Hepatic artery and portal vein remodeling in rat liver: vascular response to selective cholangiocyte proliferation. Am J Pathol 2003;162:1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Op Den Buijs J, Bajzer Z, Ritman EL. Branching morphology of the rat hepatic portal vein tree: a micro-CT study. Ann Biomed Eng 2006;34:1420–1428 [DOI] [PubMed] [Google Scholar]
- 26.Meuli R, Hwu Y, Je JH, Margaritondo G. Synchrotron radiation in radiology: radiology techniques based on synchrotron sources. Eur Radiol 2004;14:1550–1560 [DOI] [PubMed] [Google Scholar]
- 27.Plouraboue F, Cloetens P, Fonta C, Steyer A, Lauwers F, Marc-Vergnes JP. X-ray high-resolution vascular network imaging. J Microsc 2004;215(Pt 2):139–148 [DOI] [PubMed] [Google Scholar]
- 28.Heinzer S, Krucker T, Stampanoni M, Abela R, Meyer EP, Schuler A, et al. Hierarchical microimaging for multiscale analysis of large vascular networks. Neuroimage 2006;32:626–636 [DOI] [PubMed] [Google Scholar]