Abstract
Purpose
To evaluate if the differential exchange rates with bulk water between amine and amide protons can be exploited using chemical exchange saturation transfer magnetic resonance (CEST-MR) to monitor the release of glutamate induced by carboxypeptidase G2 (CPG2), an enzyme utilized in cancer gene therapy.
Procedures
The CEST properties of glutamate (amine) and the CPG2 substrate (amide), 3,5-difluorobenzoyl-L-glutamate (3,5-DFBGlu), were evaluated at 11.7T, 37°C and varying pH. The ability of CEST-MR to monitor CPG2-mediated release of glutamate was assessed in extracts of CPG2-expressing cancer cells and purified solution of CPG2.
Results
The addition of CPG2 to a solution containing 3,5-DFBGlu led to a marked and progressively increasing CEST effect (+3ppm), concomitant with the time-dependent release of glutamate induced by CPG2.
Conclusion
CEST-MR affords the detection of CPG2 activity in vitro and supports the translation of CEST-MRI to assess CPG2-based gene therapy in vivo.
Keywords: CEST MRI, Carboxypeptidase G2, Gene therapy, GDEPT
Introduction
Chemical exchange saturation transfer magnetic resonance imaging (CEST-MRI) exploits the direct chemical exchange of metabolite protons with the bulk tissue water, enabling the detection of specific tissue metabolites at concentrations that are otherwise below the detection limit of conventional 1H MRI [1]. Recently, CEST MRI has been successfully utilized to acquire high temporal and spatial resolution images of glutamate (Glu) in the human brain, where the neurotransmitter is present at mM concentration [2]. Glutamate demonstrates fast amine exchange rates, giving rise to a strong CEST effect at a resonance offset of +3 ppm from the water frequency (GluCEST), which is dependent on glutamate concentration and pH.
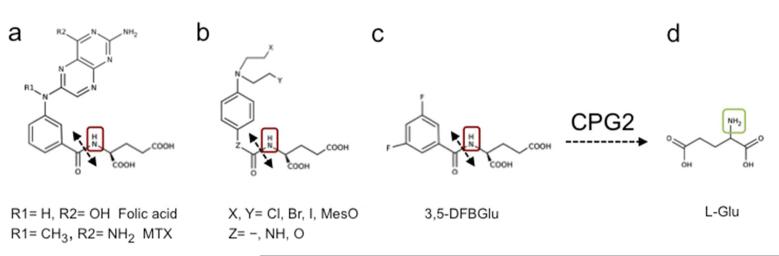
The bacterial enzyme carboxypeptidase G2 (CPG2) (EC 3.4.17.11) is a zinc-dependent enzyme which mediates the release of the C-terminal glutamate residue from a wide range of N-acylating moieties, including peptidyl, aminoacyl, benzoyl, benzyloxycarbonyl, folyl and pteroyl groups (Fig. 1). This unique ability of CPG2 has been exploited to activate carboxyl, phenol or aniline mustard prodrugs, in which the DNA alkylating chain has been synthetically deactivated through N-substitution of L-glutamate [3]. CPG2 underpins the gene-directed enzyme prodrug therapy (GDEPT) strategy due to enter Phase I clinical evaluation at the Royal Marsden Hospital (UK) in 2013 [4]. In GDEPT, the gene encoding the prodrug-activating enzyme is targeted selectively to the tumor prior to administration of the prodrug, resulting in the activation of the cytotoxic drug specifically in the tumor. The success of CPG2-based GDEPT relies on the careful optimization of the timing of injection of the prodrug following expression of the transgene and the generation of sufficient concentrations of CPG2 in the tumor. Non-invasive imaging strategies to monitor the prodrug-activating enzyme activity and its bio-distribution would thus be invaluable in guiding the successful translation of this promising therapeutic approach to the clinic [5].
Figure 1.
Carboxypeptidase G2 (CPG2)-mediated hydrolysis of the glutamate moiety of nitrogen mustard prodrugs, and the 19F MRS in vivo reporter probe 3,5-difluorobenzoyl-L-glutamate (3,5-DFBGlu).
In this study, we have exploited the differential exchange rates of amine and amide functional groups [2], to dynamically monitor CPG2 activity through the “activation” of the concentration-dependent GluCEST signal induced by the CPG2-mediated release of glutamate.
Materials and Methods
All studies were performed at 500 MHz on a 11.7T Bruker Avance (Bruker Instruments) vertical bore magnet equipped with a 5mm BBO probe. Magnetic field homogeneity was optimized on solutions in which 10% D2O was added to provide a lock signal. Experiments were subsequently performed unlocked without D2O.
CEST NMR
Glutamic acid (monosodium salt) was obtained from Sigma Aldrich. The in vivo reporter molecule of CPG2 activity, 3,5-difluorobenzoyl-L-glutamate (3,5-DFBGlu) was custom-synthesized as previously described [6]. Z spectra of solutions of 10 mM 3,5-DFBGlu, and the products of its CPG2-mediated cleavage, 3,5-difluorobenzoic acid (3,5-DFBA) and glutamate, were acquired at 37°C in phosphate buffered saline at pH 5,6,7 and 8, using a series of spectra (16 averages, TR=14 s) with the saturation frequency at different offset frequency incremented by 0.2 ppm from −5 ppm to 5 ppm from the water resonance. The frequency selective saturation was achieved using continuous wave presaturation with 3sec duration and γB1/2π ~ 155 Hz. Spectra were apodized with a 3 Hz exponential line broadening. Peak areas were measured using TopSpin3.0 (Bruker) and z spectra and MTR asymmetry plot were generated in Excel (Microsoft).
Dynamic detection of CPG2 activity in cell extracts
The generation of β-gal (control) and stCPG2(Q)3-expressing WiDr stable cell lines used in this study has been described previously [7]. The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (Gibco) at 37°C in a 5% CO2 atmosphere. The dense cell monolayers were then extracted as described previously[8]. Briefly, cells were first washed twice with PBS, followed by 5 mL of extraction buffer (250mM TrisHCl, 10% (v/v) glycerol, 1% (v/v) Triton X-100, pH 7) added to the flask. Cells were lysed for 5 min at room temperature. Cell extracts were then spun down in a microfuge (18000g) to remove cell debris. 50μl of extract was then added to 500μl of a 12mM solution of 3,5-DFBGlu in 100mM phosphate buffer, containing 260μM ZnCl2, at pH 7and maintained at 37°C. Acquisition of a series of 4 spectra (4 averages, TR=17s) was initiated immediately after the sample was positioned in the spectrometer, with either no saturation, or selective saturation at +3ppm, +2ppm, or +1ppm. This sequence was repeated over a period of 3 hours. The frequency selective saturation was achieved using continuous wave presaturation with 5sec duration and γB1/2π ~ 280Hz. Each spectrum was apodised with a 3Hz exponential line broadening function. Peak areas were calculated using TopSpin3.0 (Bruker) and fitted as (Msat(t)−M0)/M0 in Excel.
Detection of CPG2 activity in purified solution of CPG2
10mU of purified CPG2 (EC 3.4.17.11, Sigma Aldrich, 1U cleaves ~ 2μmol 3,5-DFBGlu.min−1 [6]) or vehicle solution were added to a 5mM solution of 3,5-DFBGlu (in 100mM phosphate buffer containing 260μM ZnCl2, at pH 7 and maintained at 37° C). 1H MRS spectra (16 averages, TR=14s) were acquired two hours later (to ensure total conversion of 3,5-DFBGlu into glutamate) with either no saturation, or selective saturation at −3ppm or +3 ppm, using continuous wave presaturation with 3sec duration and γB1/2π ~ 155 Hz. Each spectrum was apodised with a 3Hz exponential line broadening function. The GluCEST signal was calculated as MTRassym = (M−3ppm − M+3ppm) /M0, where M−3ppm and M+3ppm represent the bulk water signals acquired with selective RF saturation at ±3ppm resonance offset from the water frequency.
Results
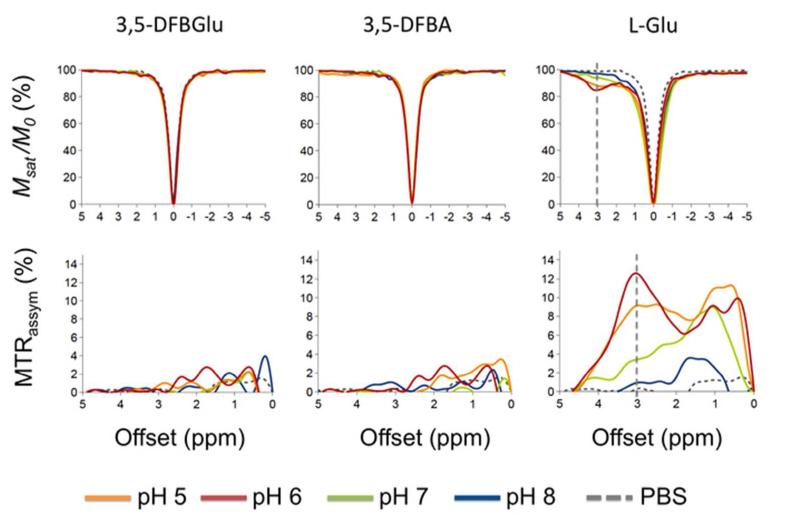
Fig. 2 shows z-spectra and the magnetization transfer ratio asymmetry (MTRassym) of the water 1H NMR signal as a function of saturation offset frequency for 10mM solutions of the imaging reporter probe 3,5-DFBGlu, and the products of its CPG2-mediated cleavage, 3,5-DFBA and glutamate, at varying pH representative of the physiological range associated with tumors (pH 5-8). Glutamate demonstrates a sharp CEST peak in the z spectrum at +3ppm offset from the water resonance at lower pH as previously demonstrated [2], whilst 3,5-DFBA, and most importantly, 3,5-DFBGlu, did not demonstrate any detectable CEST effect over this range of pH.
Figure 2.
CEST z-spectra acquired at 11.7 T of 10 mM 3,5-DFBGlu, 3,5-difluorobenzoic acid (3,5-DFBA), and glutamate (L-Glu) in PBS at varying pH and at 37 °C. Msat and M0 represent the bulk water signals acquired with and without selective RF saturation, respectively. Inserts show corresponding MTRassym plots, defined as MTRassym = (M−Δω − M+Δω) /M0 , where M−Δ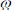 and M+Δω represent the bulk water signals acquired with selective RF saturation at ±Δω resonance offset from the water frequency.
and M+Δω represent the bulk water signals acquired with selective RF saturation at ±Δω resonance offset from the water frequency.
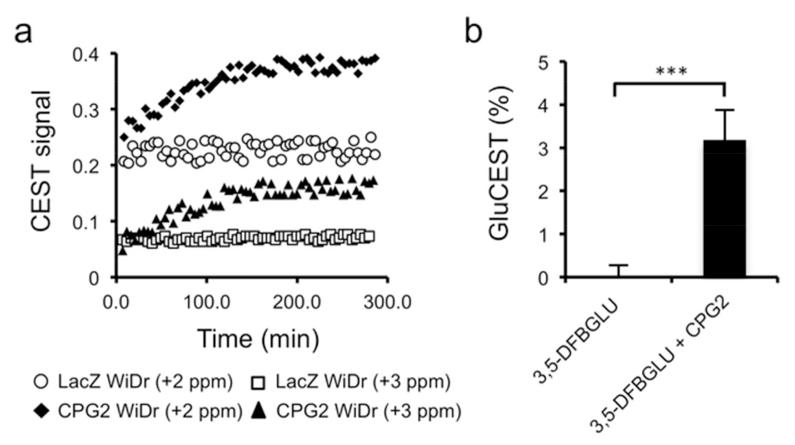
The ability of CEST MR to detect CPG2 activity was subsequently evaluated using extracts of stCPG2(Q)3 WiDr colon carcinoma cells, genetically-engineered to express CPG2, and control WiDr cells, genetically engineered to express β-galactosidase (LacZ WiDr). The addition of 3,5-DFBGlu to the extracts of CPG2-expressing WiDr cells caused a time dependent increase of the CEST effect at +3ppm (Fig. 3a). No change in the CEST effect was observed following addition of 3,5-DFBGlu to the non-CPG2 expressing LacZ WiDr control cell extracts. CPG2 activity was also detected by the change in CEST signal at +2ppm with improved sensitivity, corroborating the z spectrum of 3,5-DFBGlu and glutamate at pH 7 (Fig. 2). Finally, a ~3% GluCEST signal (+3ppm) was detected two hours following the addition of 10mU purified CPG2 to a solution of 5mM 3,5-DFBGlu (Fig. 3b). No GluCEST signal was detected following the addition of CPG2 enzyme buffer alone.
Figure 3.
Detection of CPG2-mediated cleavage of 3,5-DFBGlu with CEST-MR. (a) Evolution of the CEST signal over time, following the addition of 10mM 3,5-DFBGlu to extracts of WiDr cells engineered to express CPG2 (CPG2 WiDr) or β-galactosidase (LacZ WiDr), at pH 7 and 37°C. CEST signal is expressed as (M0 − Msat(t))/M0, where M0 and Msat(t) are the bulk water signal acquired without and with saturation (at +2ppm and +3ppm) at a time t after addition of 3,5-DFBGlu. (b) GluCEST signal from a 5mM solution of 3,5-DFBGlu measured 2h following the addition of 10mU of purified CPG2 or enzyme buffer. The GluCEST signal was calculated as MTRassym = (M−3ppm − M+3ppm) /M0 , where M−3ppm and M+3ppm represent the bulk water signals acquired with selective RF saturation at ±3ppm resonance offset from the water frequency. (*** p<0.005, Student’s 2-tailed unpaired t-test with a 5% level of significance, n=3)
Discussion and Conclusions
This study demonstrates the potential of CEST-MRI to monitor CPG2 activity via its sensitivity to the CPG2-mediated release of glutamate, resulting in the“activation”of GluCEST contrast (+3ppm). We have previously demonstrated the utility of 19F MRS, in combination with the imaging reporter 3,5-DFBGlu, to monitor CPG2 activity in vivo using human tumor xenografts derived from CPG2-expressing WiDr cells, exploiting the 1.4ppm 19F MRS chemical shift change upon CPG2 mediated cleavage of 3,5-DFBGlu [6]. Although the 19F MRS approach provides invaluable information on the level of CPG2 activity in the tumor, the inherent lack of sensitivity of conventional MRS precluded the assessment of the heterogeneous distribution of enzyme activity within the tumor, an important prognostic factor for successful therapy. We also investigated if the increased signal provided by hyperpolarized 13C MRS could be utilized to detect and potentially provide CPG2 activity maps. While CPG2 activity was successfully detected in vitro, we concluded that the relatively short T1 of the 13C within 3,5-DFBGlu would only afford a short 13C signal enhancement time window, making the translation of the approach in vivo challenging [9].
GluCEST MRI is an attractive method for metabolic imaging of glutamate since it utilizes the bulk signal from water protons, providing a signal enhancement of ~700 fold, enabling the indirect detection of glutamate concentration at higher temporal and spatial resolution compared to its conventional direct detection by 1H MRS [2]. The release of glutamate from the amido, carbonyl or ureido bonds is the hallmark of CPG2 activity. As a result, the approach could be applied to substrates of CPG2, including natural substrates such as folic acid, MTX (CPG2 is approved for use in MTX overdose), the noninvasive imaging reporter 3,5-DFBGlu, and, most interestingly, to the arsenal of prodrugs designed for GDEPT using CPG2. The change in glutamate concentration measured with GluCEST MRI could provide a noninvasive biomarker of CPG2 activity, in addition to acting as a surrogate for prodrug activation and the concentration of activated drug in the tumor. The information provided on the active drug is of particular interest in the clinical setting, as the extremely short half-life of the active drug precludes the direct measure of its intra-tumoral concentration [3].
The activity of cytosine deaminase (CD), another prodrug activating enzyme utilized in GDEPT, has been previously monitored using CEST MRI in vitro [10]. CD mediates the conversion of the prodrug 5-fluorocytosine (5-FC) or cytosine into the conventional chemotherapy agent 5-fluorouracil (5-FU) and uracil, respectively, through the removal of a CEST active amine group. CD activity thus leads to a reduction of the CEST signal (negative contrast). CPG2 activity offers more favorable characteristics than CD towards CEST MRI, since it is sensitive to CPG2-mediated glutamate production (positive contrast).
Glutamate metabolism and diffusion out of the extracellular space may potentially cause limitations for the translation of the approach in vivo. The success of CPG2 detection with CEST MRI will thus depend on the intra-tumoral CPG2 activity levels, CEST sensitivity and careful optimization of the CEST MRI acquisition time. In the forthcoming clinical trial, CPG2 will be targeted to the tumor using a replication-competent adenoviral vector. These vectors drive the infected cell to express CPG2 whilst replicating. The vector is designed to target and infect tumor cells following intravenous delivery, and to release both CPG2 and CPG2 armed virus into the extracellular space. Preclinically, the use of such vectors results in CPG2 concentrations of up to 100 units/g of tumor [11]. Since CPG2 has favorable kinetics, this excess of enzyme is able to cleave the prodrug and release glutamate in the tumor within minutes of prodrug administration, as we demonstrated using 19F MRSI [6]. Elevated levels of glutamate, typically between 5-10mM, have been detected in a range of human solid tumors [12]. This, coupled with the slightly acidic pH associated with tumors, means that the endogenous pool of glutamate (both extra and intracellular) will provide a significant baseline GluCEST signal, and the necessary threshold for detection of CPG2-mediated glutamate release in the mM range [6].
The combined excellent temporal resolution of GluCEST MRI protocols as demonstrated by Cai and colleagues [2], and the high levels of CPG2 intra-tumoral activity, should provide a sufficient temporal imaging window during which the contribution of glutamate metabolism and diffusion into the extracellular space is minimized.
Conclusion
This study thus encourages the in vivo assessment of GluCEST-MRI for imaging CPG2 activity.
Acknowledgements
We acknowledge the support received for The Institute of Cancer Research CR-UK and EPSRC Cancer Imaging Centre, in association with the MRC and Department of Health (England) grants C1060/A10334, NHS funding to the NIHR Biomedical Research Centre, The Wellcome Trust grant #091763Z/10/Z, the Christopher’s Smile charity, Cancer Research UK grants C309/A8274 and C107/A10433, the Lewis Trust Ltd and the Head & Neck Trust.
The authors thank Geoffrey Payne for his RF pulse expertise, and Simon Walker-Samuel and David Collins for critically appraising the manuscript.
Footnotes
Conflict of Interest Statement
The authors declare that they have no conflict of interest.
References
- 1.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn Reson Med. 2011;65:927–48. doi: 10.1002/mrm.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai K, Haris M, Singh A, et al. Magnetic resonance imaging of glutamate. Nat Med. 2012;18:302–6. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niculescu-Duvaz D, Niculescu-Duvaz I, Springer CJ. Design of prodrugs for suicide gene therapy. Methods Mol Med. 2004;90:161–202. doi: 10.1385/1-59259-429-8:161. [DOI] [PubMed] [Google Scholar]
- 4.Hedley D, Ogilvie L, Springer C. Carboxypeptidase-G2-based gene-directed enzyme-prodrug therapy: a new weapon in the GDEPT armoury. Nat Rev Cancer. 2007;7:870–9. doi: 10.1038/nrc2247. [DOI] [PubMed] [Google Scholar]
- 5.Penet M-F, Chen Z, Li C, Winnard P, Bhujwalla Z. Prodrug enzymes and their applications in image-guided therapy of cancer: tracking prodrug enzymes to minimize collateral damage. Drug Deliv and Transl Res. 2012;2:22–30. doi: 10.1007/s13346-011-0052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamin Y, Smyth L, Robinson SP, et al. Noninvasive detection of carboxypeptidase G2 activity in vivo. NMR Biomed. 2011;24:343–50. doi: 10.1002/nbm.1597. [DOI] [PubMed] [Google Scholar]
- 7.Marais R, Spooner RA, Stribbling SM, et al. A cell surface tethered enzyme improves efficiency in gene-directed enzyme prodrug therapy. Nat Biotechnol. 1997;15:1373–7. doi: 10.1038/nbt1297-1373. [DOI] [PubMed] [Google Scholar]
- 8.Schepelmann S, Hallenbeck P, Ogilvie LM, et al. Systemic gene-directed enzyme prodrug therapy of hepatocellular carcinoma using a targeted adenovirus armed with carboxypeptidase G2. Cancer Res. 2005;65:5003–8. doi: 10.1158/0008-5472.CAN-05-0393. [DOI] [PubMed] [Google Scholar]
- 9.Jamin Y, Gabellieri C, Smyth L, et al. Hyperpolarized (13)C magnetic resonance detection of carboxypeptidase G2 activity. Magn Reson Med. 2009;62:1300–4. doi: 10.1002/mrm.22049. [DOI] [PubMed] [Google Scholar]
- 10.Liu G, Liang Y, Bar-Shir A, et al. Monitoring enzyme activity using a diamagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. J Am Chem Soc. 2011;133:16326–9. doi: 10.1021/ja204701x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schepelmann S, Ogilvie LM, Hedley D, et al. Suicide gene therapy of human colon carcinoma xenografts using an armed oncolytic adenovirus expressing carboxypeptidase G2. Cancer Res. 2007;67:4949–55. doi: 10.1158/0008-5472.CAN-07-0297. [DOI] [PubMed] [Google Scholar]
- 12.Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–25. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]