Abstract
Cluster analysis faces two problems in high dimensions: first, the “curse of dimensionality” that can lead to overfitting and poor generalization performance; and second, the sheer time taken for conventional algorithms to process large amounts of high-dimensional data. We describe a solution to these problems, designed for the application of “spike sorting” for next-generation high channel-count neural probes. In this problem, only a small subset of features provide information about the cluster member-ship of any one data vector, but this informative feature subset is not the same for all data points, rendering classical feature selection ineffective. We introduce a “Masked EM” algorithm that allows accurate and time-efficient clustering of up to millions of points in thousands of dimensions. We demonstrate its applicability to synthetic data, and to real-world high-channel-count spike sorting data.
Keywords: spike sorting, high-dimensional, clustering algorithm
1 Introduction
Cluster analysis is a widely used technique for unsupervised classification of data. A popular method for clustering is fitting a mixture of Gaussians, often achieved using the Expectation-Maximization (EM) algorithm (Dempster et al., 1977) and variants thereof (Fraley & Raftery, 2002). In high dimensions however, this method faces two challenges (Bouveyron & Brunet-Saumard, 2012). First, the “curse of dimensionality” leads to poor classification, particularly in the presence of a large number of uninformative features; second, for large and high-dimensional data sets, the computational cost of many algorithms can be impractical. This is particularly the case where covariance matrices must be estimated, leading to computations of order , where p is the number of features; furthermore even a cost of can render a clustering method impractical for applications in which large high-dimensional data sets must be analyzed on a daily basis. In many cases however, the dimensionality problem is solvable at least in principle, as the features sufficient for classification of any particular data point are a small subset of the total available.
A number of approaches have been suggested for the problem of high-dimensional cluster analysis. One approach consists of modifying the generative model underlying the mixture of Gaussians fit to enforce low-dimensional models. For example the Mixture of Factor Analyzers (Ghahramani et al., 1996; McLachlan et al., 2003) models the covariance matrix of each cluster as a low rank matrix added to a fixed diagonal matrix forming an approximate model of observation noise. This approach can reduce the number of parameters for each cluster from to , and may thus provide a substantial improvement in both computational cost and performance. An alternative approach - which offers the opportunity to reduce the both the number of parameters and computational cost substantially below - is feature selection, whereby a small subset of informative features are selected and other non-informative features are discarded (Raftery & Dean, 2006). A limitation of global feature selection methods however, is that they cannot deal with the case where different data points are defined by different sets of features. One proposed solution to this consists of assigning each cluster a unique distribution of weights over all features, which has been applied to the case of hierarchical clustering (Friedman & Meulman, 2004).
The algorithm described below was developed to solve the problems of high-dimensional cluster analysis for a particular application: “spike sorting” of neurophysiological recordings using newly developed high-count silicon microelectrodes (Einevoll et al., 2012). Spike sorting is the problem of identifying the firing times of neurons from electric field signatures recorded using multisite microfabricated neural electrodes (Lewicki, 1998). In low-noise systems, such as retinal explants ex vivo, it has been possible to decompose the raw recorded signal into a sum of waveforms, each corresponding to a single action potential (Pillow et al., 2013; Marre et al., 2012; Prentice et al., 2011). For recordings in the living brain, noise levels are considerably higher, and an approach based on cluster analysis is more often taken. In a typical experiment, this will involve clustering millions of data points each of which reflects a single action potential waveform that could have been produced by one of many neurons. Historically, neural probes used in vivo have had only a small number of channels (usually 4), typically resulting in feature vectors of 12 dimensions which required sorting into 10-15 clusters. Analysis of “ground truth” shows that the data is quite well approximated by a mixture of Gaussians with different covariance matrices between clusters (Harris et al., 2000). Accordingly, in this low-dimensional case, traditional EM-derived algorithms work close to optimally, although specialized rapid implementation software is required to cluster the millions of spikes recorded on a daily basis (Harris et al., 2000 - 2013). More recent neural probes, however, contain tens to hundreds of channels, spread over large spatial volumes, and probes with thousands are under development. Because different neurons have different spatial locations relative to the electrode array, each action potential is detected on only a small fraction of the total number of channels, but the subset differs between neurons, ruling out a simple global feature selection approach. Furthermore, because spikes produced by simultaneous firing of neurons at different locations must be clustered independently, most features for any one data point are not simply noise, but must be regarded as missing data. Finally, due to the large volumes of data produced by these methods, we require a solution that is capable of clustering millions of data points in reasonably short running time. Although numerous extensions and alternatives to the simple cluster sorting method have been proposed: (Takahashi et al., 2003; Quian Quiroga et al., 2004; Wood & Black, 2008; Sahani, 1999; Lewicki, 1998; Gasthaus et al., 2008; Calabrese & Paninski, 2011; Ekanadham et al., 2013; Shoham et al., 2003; Franke et al., 2010; Carin et al., 2013), none have been shown to solve the problems created by high-count electrode arrays.
Here we introduce a “Masked EM” algorithm to solve the problem of high-dimensional cluster analysis, as found in the spike-sorting context. The algorithm works in two stages. In the first stage, a “mask vector” is computed for each data point via a heuristic algorithm, encoding a weighting of each feature for every data point. This stage may take advantage of domain-specific knowledge, such as the topological constraint that action potentials occupy a spatially contiguous set of recording channels. In the case that the majority of masks can be set to zero, the number of parameters per cluster and running time can both be substantially below . We note that the masks are assigned to data points, rather than clusters, and need only be computed once at the start of the algorithm. The second stage consists of cluster analysis. This is implemented using a mixture of Gaussians EM algorithm, but with every data point replaced by a virtual mixture of the original feature value, and the fixed subthreshold noise distribution weighted by the masks. The use of this virtual mixture distribution avoids the splitting of clusters due to arbitrary threshold crossings. At no point is it required to generate samples from the virtual distribution, as expectations over it can be computed analytically.
2 The Masked EM Algorithm
2.1 Stage 1: mask generation
The first stage of the algorithm consists of computing a set of “mask vectors” indicating which features should be used to classify which data points. Specifically, the outcome of this algorithm is a set of vectors mn with components, mn,i ∈ [0, 1]. A value of 1 indicates that feature i is to be used in classifying data point xn, a value of 0 indicating it is to be ignored, and intermediate values corresponding to partial weighting. We refer to features being used for classification as “unmasked”, and features being ignored as “masked” (i.e. concealed). As described below, masked features are not simply set to zero, but are ignored by replacing them with a virtual ensemble of points, drawn from a distribution of subthreshold noise.
The use of masks provides two major advantages over a standard mixture of Gaussians classification: first, it overcomes the curse of dimensionality, because assignment of points to classes is no longer overwhelmed by the noise on the large number of masked channels; second, it allows the algorithm to run in time proportional to rather than . Because a small number of features may be unmasked for each data point, this can allow computational costs substantially below . The way masks are chosen can depend on the application domain and would typically follow a heuristic method. A simple approach that can work in general is to compute masks based on a standard deviation of each feature:
| (1) |
This strategy smoothly interpolates between a mask of zero for features below a lower threshold, and a mask of 1 for features above a higher threshold; such smooth interpolation avoids the artificial creation of discrete clusters when variables cross a fixed boundary. In the case of spike sorting, a slightly more complex procedure is used to derive the masks, which takes advantage of the topological constraint that spikes must be distributed across continuous groups of recording channels (Section 3.2). In practice, we have found that good performance can be obtained for a range of masking parameters, provided the majority of non-informative features have a mask of 0, and that features that are clearly suprathreshold are given a mask of 1 (Section 3.1).
Once the masks have been computed, an additional set of quantities is pre-computed before the main EM loop starts. Specifically, the subthreshold noise mean for feature i, νi is obtained by taking the mean of feature i whenever that particular feature is masked, i.e. mn,i = 0, and analogously, the noise variance for feature i, :
where .
2.2 Stage 2: clustering
The second stage consists of a maximum-likelihood mixture of Gaussians fit, with both the E and M steps modified by replacing each data point xn with a virtual ensemble of points distributed as
| (2) |
where mn,i ∈ [0, 1] is the mask vector component associated to xn,i for the nth spike. Intuitively, any feature below a noise threshold is replaced by a ‘virtual ensemble’ of the entire noise distribution. The noise on each feature will be modelled as independent univariate Gaussian distributions for each i, which we shall refer to as the noise distribution for feature i. This is, of course, a simplification, as the noise may be correlated. However for tractability, ease of implementation and as we shall later show, efficacy, this approximation suffices.
The algorithm maximizes the expectation of the usual log likelihood over the virtual distribution:
The masked EM algorithm therefore acts as if it were passed an ensemble of points, with each data point replaced by an infinite sample, corresponding to different possibilities for the noisy masked variables. This solves the curse of dimensionality by “disenfranchising” each data point’s masked features, disregarding the value that was actually measured, and replacing it by a virtual ensemble that is the same in all cases, and thus does not contribute to cluster assignment.
Before running the EM algorithm, the following quantities are also computed, which will greatly speed up computation of the modified M and E-steps:
2.3 M-step
For the M-step, replacing x with the virtual ensemble requires computing the expectation with respect to of the mean and the covariance of each cluster. For simplicity, we henceforth focus on a “hard” EM algorithm in which each data point xnis fully assigned to a single cluster, although a “soft” version can be easily derived. We denote by the set of data point indices assigned to the cluster with index k. It is straightforward to show that:
| (3) |
| (4) |
Note that this is the same formula as the classical M-step using, but with xn,i replaced by the expected value yn,i of the virtual distribution , plus a correction term to Σi,j corresponding to the covariance matrix ηn,i of . Computation of these quantities can be carried out very efficiently as we can decompose (μk)i and (Σk)ij as follows:
| (5) |
where denotes the set of points within cluster k for which feature i is fully masked. Note that if all data points in a cluster have feature i masked, then (μk)i = νi, the noise mean, and (Σk)ii = , the noise variance.
2.4 E-step
In the E-step, we compute the responsibility of each cluster for each point, defined as the probability that point n came from cluster k, conditional on its feature values. The responsiblity is computed via Bayes theorem from πnk, the log-likelihood of point n under the Gaussian model for cluster k. In the masked EM algorithm, we compute πnk as its expected value over the virtual distribution . Thus, the algorithm acts as if each data point is replaced by an infinite ensemble of points drawn from the distribution of , which must all be assigned the same cluster label. Explicitly,
| (6) |
The final term of Equation 6 corresponds to the expectation of the Mahalanobis distance of from cluster k; it can be shown that
This leads to the original E-step for the EM algorithm, but with yn,i substituted for xn,i, plus a diagonal correction term .
2.5 Penalties
Automatically determining the number of clusters in a mixture of Gaussians requires a penalty function that penalises over-fitting by discouraging models with a large number of parameters. Commonly used penalization methods include the Akaike Information Criterion (AIC) (Akaike, 1974) and Bayes Information Criterion (BIC) (Schwarz, 1978):
where κ is the number of free parameters in the statistical model, and L is the maximum of the likelihood for the estimated model and N is the number of data points.
For the classical mixture of Gaussians fit, the number of parameters κ is equal to where K is the number of clusters and p is number of features. The elements of the first term in κ correspond to the number of free parameters in a p × p covariance matrix, a p-dimensional mean vector, and a single weight for each cluster. Finally, 1 is subtracted from the total because of the constraint that the weights must sum to 1 for a mixture model.
For masked data the estimation of the number of parameters in the model is more subtle. Because masked features are replaced by a fixed distribution that does not vary between data points, the effective degrees of freedom per cluster is much smaller than . We therefore define a cluster penalty for each cluster C that depends only on the average number of unmasked features corresponding to that cluster. Specifically, let be the effective number of unmasked features for data point n (i.e. sum of the weights over the features). Define , where the three terms correspond to the number of free parameters in an r × r covariance matrix, mean vector of length r and a mixture weight, respectively.
Our estimate of the effective number of parameters is thus:
| (7) |
2.6 Implementation
The algorithm was implemented in custom C++ code, based on previously released open-source software for fast mixture of Gaussians fitting termed “KlustaKwik” (Harris et al., 2000 - 2013). Because we require the algorithm to run in reasonable time on large numbers of high-dimensional data points, several approximations are made to give faster running times without significantly impacting on performance. These include not only hard classification but also a heuristic that eliminates the great majority of E-step iterations; a split-and-merge feature that changes the number of clusters dynamically if this increases the penalized likelihood; and an additional uniform distributed mixture component to catch outliers. The software can be downloaded from https://github.com/klusta-team/klustakwik (Harris et al., 2013).
3 Evaluation
3.1 Mixture of Gaussians
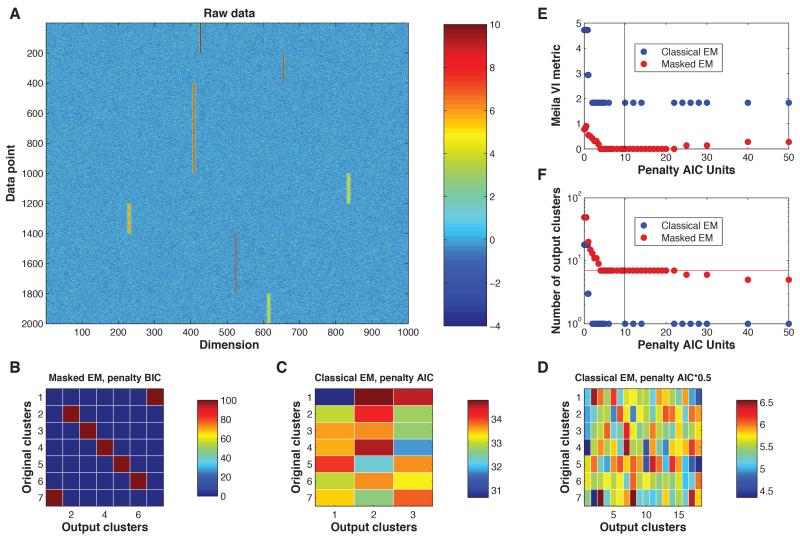
We first demonstrate the efficacy of the Masked EM algorithm using a simple data set synthesized from a high-dimensional mixture of Gaussians. The data set consisted of 20000 points in 1000 dimensions, drawn from 7 separate clusters. The means were chosen by centering probability density functions of gamma distributions on certain chosen features. All covariance matrices were identical, namely a Toeplitz matrix with all the diagonal entries 1 and off-diagonal entries that decayed exponentially with distance from the diagonal. Figure 1A shows this data set in pseudocolor format.
Figure 1.
Simulated Data. A: Subsampled raw data. B: Confusion matrix in percentages for Masked EM with α = 2, β = 3, and BIC penalty (equivalent to 10*AIC for 20000 points). C: Confusion matrix in percentages for Classical EM for an AIC penalty. D: Confusion matrix in percentages for Classical EM for a penalty of 0.5 AIC. E: VI metric measure of performance of both algorithms using various values for penalty, where the black vertical line indicates BIC. F: The number of clusters obtained for various values of penalty, where the black vertical line indicates BIC.
Figure 1B shows a confusion matrix generated by the Masked EM algorithm on this data, with the modified BIC penalty and masks defined by equation 1, indicating perfect performance. By contrast, Figure 1C shows the result of classical EM, in which many clusters have been erroneously merged; the results for AIC penalty are shown since using a BIC penalty yielded only a single cluster. To verify that this is not simply due to an inappropriate choice of penalty, we reran with the penalty term linearly scaled by various factors. Figure 1D shows the results of a penalty 0.5 AIC that gave more clusters than the ground truth data. Even in this case, however, the clusters produced by classical EM contained points from a mixture of the original clusters, and could not be corrected even by manual post-hoc cluster merging. To systematically evaluate the effectiveness of both algorithms, we measured performance using the Variation of Information (VI) metric (Meilă, 2003), for which a value of 0 indicates a perfect clustering. Both algorithms were tested for a variety of different penalties measured in multiples of AIC (Figure 1E,F). Whereas the Masked EM algorithm was able to achieve a perfect clustering for a large range of penalties around BIC, the classical EM algorithm was only able to produce a poorer value of 1.83 (corresponding to the poor result of merging all the points into a single cluster).
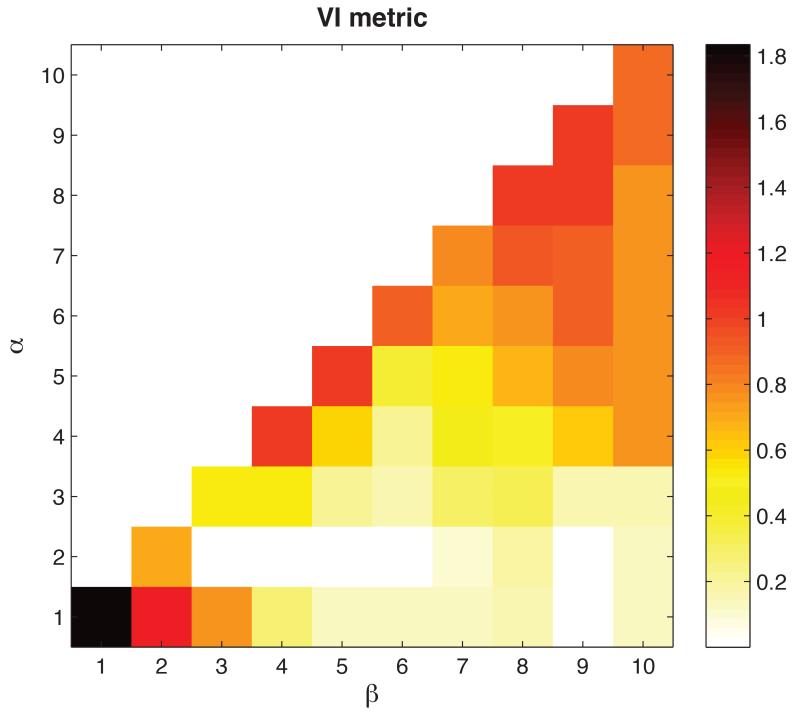
Figure 2 shows how clustering performance depends on the precise choice of masking parameters α and β defined in Equation (1), using BIC penalty. Good performance did not require a single precise parameter setting, but could be obtained with a range of parameters with α ≈ 2 and 3 ≤ β ≤ 7. The results illustrate the benefits of using a double threshold approach in preference to a single threshold: performance when α = β is markedly worse than when β > α.
Figure 2.
The effect of varying α and β in Equation (1) for the simulated 1000-dimensional dataset with 7 clusters. The plot shows a pseudocolor representation of performance measured using the Meila VI metric for various values of α and β using BIC penalty.
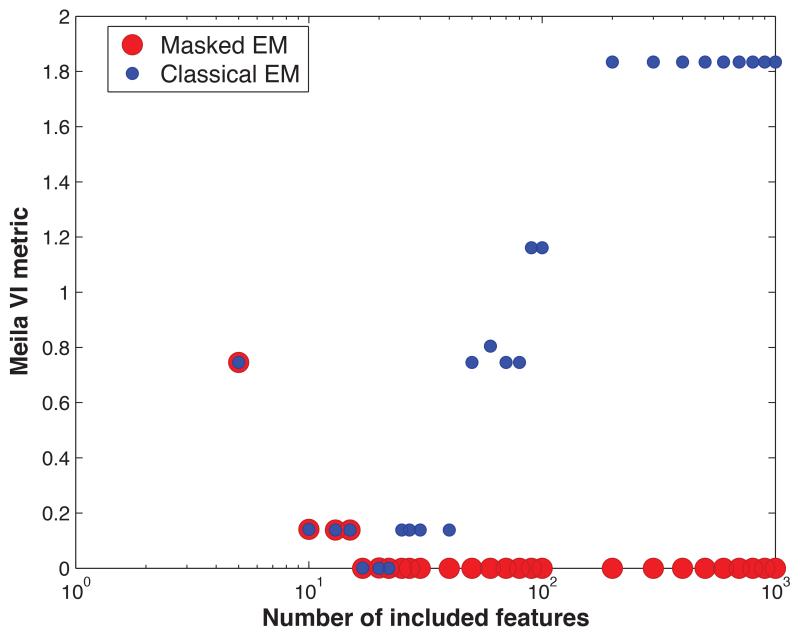
Finally, in order to explore in more detail how the classical and masked EM algorithm deal with increasing dimensionality, we varied the number of features input to the algorithms. First, we sorted the features in rough order of relevance, according to mean value of that feature over all input patterns. Both algorithms were then run on subsets of the most relevant p features, for varying values of p. Performance was quantified with the VI metric (Figure 3); in order to ensure differences between algorithms were not simply due to differences in penalty strategy, we also permitted post-hoc manual merging of clusters that were overspilt. With less than 17 features, both algorithms performed badly. For 17 to 22 features, both algorithms perform perfectly; however as more uninformative features were added, the performance of the classical, but not masked EM algorithm started deteriorating. The performance of the masked algorithm remained good for all dimensionalities tested.
Figure 3.
The effect of increasing the number dimensions on the quality of cluster analysis. The dimensions are added in order of relevance relative to the 7 ground truth clusters of the simulated mixture of Gaussians dataset.
3.2 Spike sorting
To test the performance of the Masked EM algorithm on our target application of high-channel-count spike sorting requires a ground truth data set. Previous work established the performance of the classical EM algorithm for low-channel spike sorting with ground truth obtained by simultaneous recordings of a neuron using not only the extracellular array, but also an intracellular method using a glass pipette that unequivocally determined firing times (Harris et al., 2000). Because such dual recordings are not yet available for high-count electrodes, we created a simulated ground truth we term “hybrid datasets”. In this approach, the mean spike waveform on all channels of a single neuron taken from one recording (the “donor”) is digitally added onto a second recording (the “acceptor”) made with the same electrode in a different brain. Because the “hybrid spikes” are linearly added to the acceptor traces, this simulates the linear addition of neuronal electric fields, and recreates many of the challenges of spike sortingsuch as the variability of amplitudes and waveforms of the hybrid spike between channels, and overlap between the digitally added hybrid with spikes of other neurons in the acceptor data set (Harris et al., 2000). Furthermore, to simulate the common difficulty caused by of amplitude variability in bursting neurons, the amplitude of the hybrid spike was varied randomly between 50% and 100% of its original value. The hybrid datasets we consider were constructed from recordings in rat cortex kindly provided by Mariano Belluscio and György Buzsáki, using a 32-channel probe with a zig-zag arrangement of electrodes and minimum 20μm spacing between neighbouring contacts. Three principal components were taken from each channel resulting in 96-dimensional feature vectors.
For the spike data, masks were generated using a generalization of Equation (1) that took into account the topology of the electrode array. Data were first high-pass filtered (500Hz cutoff), then spikes were detected and masks were generated using a two-threshold floodfill algorithm. The floodfill algorithm finds spatiotemporally connected sets S of samples (t, c) (where t is time and c is channel number), for which the filtered signal exceeds a lower threshold α for every point in each set, and each set contains at least one sample where the filtered signal exceeds an upper threshold β. The values of α and β were set as 2 and 4.5 times the standard deviation of the filtered signal, which was estimated robustly as a scaled median absolute deviation. For each spike, a mask for channel c was defined as maxt:(t,c)∈Sθ(t, c), where . Spikes were resampled and aligned to a mean spike time estimated as . Finally, feature vectors were extracted from the resampled filtered spike waveforms by principal component analysis channel by channel. For each channel the first 3 principal components were used to create the feature vector, hence for a C-channel dataset, each spike was given a 3C-dimensional feature vector. The mask vector for was obtained for each feature as maxtθ(t, c) computed on the corresponding channel. The data set contained 138572 points of 96 dimensions, and running 1500 iterations of the clustering algorithm on it took 10 hours on a single core of a 2.4GHz Intel Xeon L5620 computer running Scientific Linux 5.5. The data we analyzed is publicly available at https://github.com/klusta-team/hybrid_analysis.
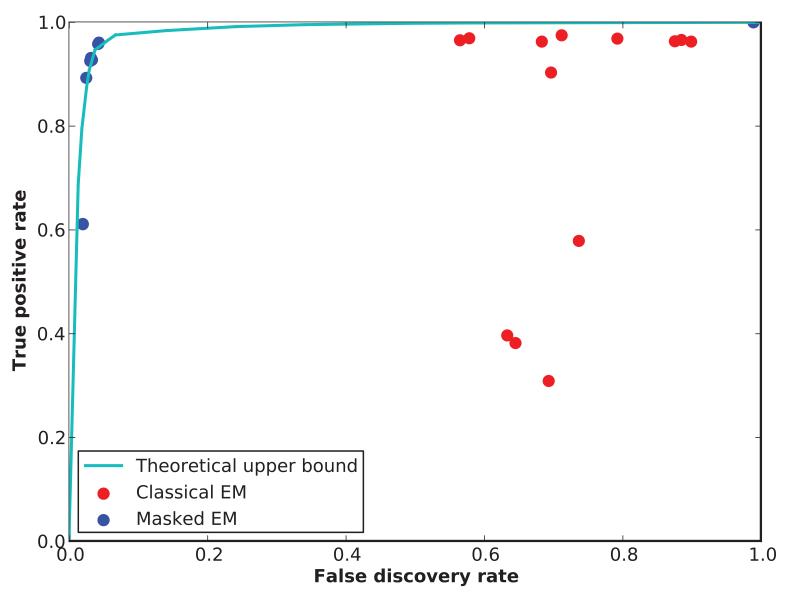
To evaluate the performance of the masked EM algorithm on this data set, we first identified the cluster with the highest number of true positive spikes, and used it to compute a false discovery rate, , and a true positive rate , where FP denotes the number of false positive, TP, the number of true positive, FN, the number of false negative spikes. This performance was compared against a theoretical upper bound obtained by supervised learning. The upper bound was obtained by using a quadratic support vector machine (SVM) (Cortes & Vapnik, 1995) trained using the ground truth data, with performance evaluated by 20-fold cross-validation. In order to ensure we estimated maximal performance, the SVM was run over a large range of parameters such as margin, class weights, as well as including runs in which only features relevant to hybrid cell were included. The theoretical optimum performance was estimated as a receiver operating characteristic (ROC) curve, by computing the convex hull of FP and TP rates for all SVM runs.
Figure 4 shows the performance of the Masked EM algorithm and classical EM algorithm on the hybrid data set, set against the theoretical optimum estimated by the SVM. While the Masked EM algorithm performs at close to the estimated upper bound, the classical EM algorithm is much poorer. To verify that this poorer performance indeed resulted from a curse of dimensionality, we re-ran the classical EM algorithm on only the subset of features that were unmasked for the hybrid spike (9 out of 96 features). As expected, the upper-bound performance was poorer in this case, but the classical EM algorithm performed close to the theoretical upper bound. This indicates that the classical algorithm fails in high-dimensional settings, whereas the Masked EM algorithm performs well.
Figure 4.
Performance of the masked and classical EM algorithms for a spike sorting data. Blue and red points indicate performance of multiple runs of the masked and classical EM algorithms with different penalty parameter settings. Cyan curve indicates optimal possible performance, estimated as the convex hull of supervised learning results obtained from a support vector machine with quadric kernel.
4 Discussion and Conclusions
We have introduced a method for high-dimensional cluster analysis, applicable to the case where a small subset of the features is informative for any data point. Unlike global feature selection methods, both the number and the precise set of unmasked features can vary between different data points. Both the number of free parameters and computational cost scale with the number of unmasked features per data point, rather than the total number of features. This approach was found to give good performance on simulated high-dimensional data, and in our target application of neurophysiological spike sorting for large electrode arrays.
A potential caveat of allowing different features to define different clusters is the danger of artificial cluster splitting. If simply a hard threshold were used to decide whether a particular feature should be used for a particular cluster or data point, this could lead to a single cluster being erroneously split in two, according to whether or not the threshold was exceeded by noisy data. The Masked EM algorithm overcomes this problem with two approaches. First, because the masks are not binary but real-valued, crossing a threshold such as that in Equation (1) leads to smooth rather than discontinuous changes in responsibilities; second, because masked features are replaced by a virtual distribution of empirically measured subthreshold data, the assignment of points with masked features is close to that expected if the original subthreshold features had not been masked. With these approaches in place, we found that erroneous cluster splitting was not a problem in simulation or in our target application.
In the present study, we have applied the masking strategy to a single application of unsupervised classification using a hard EM algorithm for mixture of Gaussians fitting. However the same approach may apply more generally whenever only a subset of features are informative for any data point, and when the expectation of required quantities over the modelled subthreshold distribution can be analytically computed. Other domains in which this approach may work therefore include not only cluster analysis with soft EM algorithms or different probabilistic models, but also model-based supervised classification.
Table 1.
Mathematical Notation
| Notation | |
|---|---|
| Dimensions (number of features) | p |
| Data | xn,i, point n, feature i |
| Masks | mn,i ∈ [0,1] |
| Cluster label | k |
| Total number of clusters | K |
| Mixture weight, cluster mean, covariance | ωk, μk, Σk |
| Probability density function of the multivariate Gaussian distribution | φ (x|μk, Σk) |
| Total number of data points | N |
| Number of points for which feature i is masked | |
| Noise mean for feature i | v i |
| Noise variance for feature i | |
| Virtual features (random variable) | |
| Mean of virtual feature | |
| z n,i | |
| Variance of virtual feature | |
| Log likelihood of in cluster k | π nk |
| Set of data points assigned to cluster k | |
| Subset of for which feature i is fully masked | |
References
- Akaike Hirotugu. A New Look at the Statistical Model Identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723. [Google Scholar]
- Bouveyron Charles, Brunet-Saumard Camille. Model-based clustering of high-dimensional data: A review. Computational Statistics & Data Analysis. 2012 [Google Scholar]
- Calabrese Ana, Paninski Liam. Kalman filter mixture model for spike sorting of non-stationary data. Journal of Neuroscience Methods. 2011;196(1):159–169. doi: 10.1016/j.jneumeth.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Carin L, Wu Q, Carlson D, Lian Wenzhao, Stoetzner C, Kipke D, Weber D, Vogelstein J, Dunson D. Sorting Electrophysiological Data via Dictionary Learning & Mixture Modeling. 2013. [DOI] [PubMed] [Google Scholar]
- Cortes Corinna, Vapnik Vladimir. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- Dempster Arthur P, Laird Nan M, Rubin Donald B. Maximum likelihood from incomplete data via the EM algorithm. Journal of the Royal Statistical Society. Series B (Methodological) 1977:1–38. [Google Scholar]
- Einevoll Gaute T, Franke Felix, Hagen Espen, Pouzat Christophe, Harris Kenneth D. Towards reliable spike-train recordings from thousands of neurons with multielectrodes. Current Opinion in Neurobiology. 2012;22(1):11–17. doi: 10.1016/j.conb.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanadham Chaitanya, Tranchina Daniel, Simoncelli Eero P. A unified framework and method for automatic neural spike identification. Journal of Neuroscience Methods. 2013 doi: 10.1016/j.jneumeth.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley Chris, Raftery Adrian E. Model-based clustering, discriminant analysis, and density estimation. Journal of the American Statistical Association. 2002;97(458):611–631. [Google Scholar]
- Franke Felix, Natora Michal, Boucsein Clemens, Munk Matthias HJ, Obermayer Klaus. An online spike detection and spike classification algorithm capable of instantaneous resolution of overlapping spikes. Journal of computational neuroscience. 2010;29(1-2):127–148. doi: 10.1007/s10827-009-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman Jerome H, Meulman Jacqueline J. Clustering objects on subsets of attributes (with discussion) Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2004;66(4):815–849. [Google Scholar]
- Gasthaus Jan, Wood Frank, Gorur Dilan, Teh Yee W. Dependent Dirichlet process spike sorting. Pages 497-504 of: Advances in Neural Information Processing Systems. 2008 [Google Scholar]
- Ghahramani Zoubin, Hinton Geoffrey E, et al. The EM algorithm for mixtures of factor analyzers. University of Toronto; 1996. Tech. rept. Technical Report CRG-TR-96-1. [Google Scholar]
- Harris Kenneth D, Henze Darrell A, Csicsvari Jozsef, Hirase Hajime, Buzsáki György. Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. Journal of Neurophysiology. 2000;84(1):401–414. doi: 10.1152/jn.2000.84.1.401. [DOI] [PubMed] [Google Scholar]
- Harris Kenneth D., Kadir Shabnam N., Goodman Dan F.M. KlustaKwik. 2000 - 2013 https://sourceforge.net/projects/klustakwik/files/
- Harris Kenneth D., Kadir Shabnam N., Goodman Dan F.M. Masked KlustaKwik. 2013 https://klusta-team.github.com/klustakwik.
- Lewicki Michael S. A review of methods for spike sorting: the detection and classification of neural action potentials. Network: Computation in Neural Systems. 1998;9(4):R53–R78. [PubMed] [Google Scholar]
- Marre Olivier, Amodei Dario, Deshmukh Nikhil, Sadeghi Kolia, Soo Frederick, Holy Timothy E, Berry Michael J. Mapping a complete neural population in the retina. The Journal of Neuroscience. 2012;32(43):14859–14873. doi: 10.1523/JNEUROSCI.0723-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan GJ, Peel David, Bean RW. Modelling high-dimensional data by mixtures of factor analyzers. Computational Statistics & Data Analysis. 2003;41(3):379–388. [Google Scholar]
- Meilă Marina. Pages 173-187 of: Learning theory and kernel machines. Springer; 2003. Comparing clusterings by the variation of information. [Google Scholar]
- Pillow Jonathan W, Shlens Jonathon, Chichilnisky EJ, Simoncelli Eero P. A Model-Based Spike Sorting Algorithm for Removing Correlation Artifacts in Multi-Neuron Recordings. PLoS ONE. 2013;8(5):e62123. doi: 10.1371/journal.pone.0062123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice Jason S, Homann Jan, Simmons Kristina D, Tkačik Gašper, Balasubramanian Vijay, Nelson Philip C. Fast, scalable, Bayesian spike identification for multi-electrode arrays. PLoS ONE. 2011;6(7):e19884. doi: 10.1371/journal.pone.0019884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quian Quiroga R, Nadasdy Z, Ben-Shaul Y. Unsupervised spike detection and sorting with wavelets and superparamagnetic clustering. Neural computation. 2004;16(8):1661–1687. doi: 10.1162/089976604774201631. [DOI] [PubMed] [Google Scholar]
- Raftery Adrian E, Dean Nema. Variable selection for model-based clustering. Journal of the American Statistical Association. 2006;101(473):168–178. [Google Scholar]
- Sahani M. Ph.D. thesis. Caltech; 1999. Latent variable models for neural data analysis. [Google Scholar]
- Schwarz Gideon. Estimating the dimension of a model. The annals of statistics. 1978;6(2):461–464. [Google Scholar]
- Shoham Shy, Fellows Matthew R, Normann Richard A. Robust, automatic spike sorting using mixtures of multivariate¡ i¿ t¡/i¿-distributions. Journal of neuroscience methods. 2003;127(2):111–122. doi: 10.1016/s0165-0270(03)00120-1. [DOI] [PubMed] [Google Scholar]
- Takahashi Susumu, Anzai Yuichiro, Sakurai Yoshio. Automatic sorting for multi-neuronal activity recorded with tetrodes in the presence of overlapping spikes. Journal of Neurophysiology. 2003;89(4):2245–2258. doi: 10.1152/jn.00827.2002. [DOI] [PubMed] [Google Scholar]
- Wood Frank, Black Michael J. A nonparametric Bayesian alternative to spike sorting. Journal of Neuroscience Methods. 2008;173(1):1–12. doi: 10.1016/j.jneumeth.2008.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]