Abstract
Local protein interactions (“molecular context” effects) dictate amino acid replacements and can be described in terms of site-specific, energetic preferences for any different amino acid. It has been recently debated whether these preferences remain approximately constant during evolution or whether, due to coevolution of sites, they change strongly. Such research highlights an unresolved and fundamental issue with far-reaching implications for phylogenetic analysis and molecular evolution modeling. Here, we take advantage of the recent availability of phenotypically supported laboratory resurrections of Precambrian thioredoxins and β-lactamases to experimentally address the change of site-specific amino acid preferences over long geological timescales. Extensive mutational analyses support the notion that evolutionary adjustment to a new amino acid may occur, but to a large extent this is insufficient to erase the primitive preference for amino acid replacements. Generally, site-specific amino acid preferences appear to remain conserved throughout evolutionary history despite local sequence divergence. We show such preference conservation to be readily understandable in molecular terms and we provide crystallographic evidence for an intriguing structural-switch mechanism: Energetic preference for an ancestral amino acid in a modern protein can be linked to reorganization upon mutation to the ancestral local structure around the mutated site. Finally, we point out that site-specific preference conservation naturally leads to one plausible evolutionary explanation for the existence of intragenic global suppressor mutations.
Keywords: molecular evolution, ancestral proteins, amino acid replacements
Introduction
Molecular evolution can be described in terms of modifications in protein (or nucleic acid) sequences that result in changes in relevant molecular properties that may ultimately impact organismal fitness (Bershtein et al. 2006; Nowak 2006; Sikosek and Chan 2014). Evolutionary sequence modifications are, in most cases, single mutations. For evolution by natural selection to occur, a certain number of these single mutations (at least one) must be accepted by a functional protein. Expressed in terms of the Maynard–Smith’s sequence space concept (Smith 1970), functional proteins must form continuous networks in sequence space with nodes connected by acceptable single-mutation steps. There is considerable interest in understanding the structural, functional, and energetic factors that determine the basic evolutionary moves in protein sequence space, that is, the set of acceptable single mutations.
Molecular context is required to understand site-specific amino acid replacements. For instance, an amino acid is more likely to occur in a given position if it generates stabilizing interactions or positive contributions. Such context-related effects are widely recognized in the literature and described using a variety terms: Propensities, preferences, forming tendencies, and so on. We shall use the term “preference” here. Amino acid preferences can be estimated from statistical analyses of amino acid occurrence (“statistical” preferences; Chou and Fasman 1974; Richardson JS and Richardson DC 1988) and also from experimental stability measurements, such as mutational effects on stability (“thermodynamic” or “energetic” preferences; Kim and Berg 1993; Smith et al. 1994; Myers et al. 1997). Good correlations between statistical and energetic preferences for amino acids in different types of secondary structures have been reported (Pace and Scholtz 1988; Kim and Berg 1993; Smith et al. 1994).
Preferences for amino acids in secondary structures can be viewed as average or typical values that describe some relevant general trends. More fully, from a rigorous point of view, any particular site in any protein has an associated preference scale that may differ from the preference scales at other sites in the same protein and also from the scales at corresponding sites in other homologs. Site-specific energetic preferences can be assessed by determining the stability effects of mutations at the site and may show a high level of discrimination between the different amino acids. For instance, if packing at a buried site is optimized for a given hydrophobic residue, replacement with another hydrophobic amino acid of similar, but smaller, size is likely to be destabilizing due to less efficient packing (Godoy-Ruiz et al. 2005). This presumably small difference in energetic preference may have consequences since the stabilities of natural proteins are marginal, that is, only slightly above the evolutionary stability threshold for purifying selection (Taverna and Goldstein 2002; Bershtein et al. 2006; Godoy-Ruiz et al. 2006; Bloom et al. 2007; Tokuriki et al. 2007; Sikosek and Chan 2014), and even a moderately destabilizing mutation could bring stability below the threshold and thus prevent proper protein folding or facilitate protein degradation with the overall result that organismal fitness would be compromised and the mutation would be rejected—in the absence of drift.
The above reasoning does not imply that the amino acid present at any particular site is necessarily the residue at the top of the energetic preference ranking. In fact, several scenarios may explain the acceptance and fixation of a less energetically preferred residue at a given site. For instance, a previously stabilizing mutation in another region of the protein structure (a so-called compensating or permissive mutation; Weinreich et al. 2006; Bloom et al. 2007; Ortlund et al. 2007; Wyganowski et al. 2013) could enhance protein stability in such a way that the mutation to the less energetically preferred amino acid does not violate the stability threshold for purifying selection. Subsequently, the less preferred amino acid could persist at the site if its presence brings about some functional changes that translate into enhanced organismal fitness. Note, however, that, even in this case, energetic preferences play a fundamental role, as they determine the evolutionary trajectory leading to the acceptance of the less preferred amino acid.
Overall, there can be little doubt about the relevance of site-specific energetic preferences and, indeed, site-specific effects have been included in many efforts to model molecular evolutionary processes (Halpern and Bruno 1998; Lartillot and Philippe 2004; Le et al. 2008; Wang et al. 2008; Rodrigue et al. 2010; Tamuri et al. 2012; Bloom 2014). On the other hand, it is not at all clear whether the preferences themselves are conserved or change substantially during evolution. We may expect energetic preferences at a given site in a protein to be determined by the interactions of amino acids at that position with their neighboring sites (and also with those at distant sites if electrostatic charge–charge interactions are relevant). Since residues at interacting sites change during evolution, it is conceivable that preferences at each given position also change. Indeed, recent computational analyses (Pollock et al. 2012) support the notion that preferences change after mutation in the direction of making the new amino acid more acceptable over time, an adjustment that is referred to as the “evolutionary Stokes shift.” However, Fersht and coworkers (Serrano et al. 1993) found that effects on stability of mutations separating barnase and binase (85% sequence identity) were independent and additive. More recently, experimental stability studies (Ashenberg et al. 2013) supported evolutionary conservation of amino acid preferences for six mutations in nucleoproteins from four different strains of influenza A virus by claiming to disfavor strong changes in amino acid preferences during evolution. Yet, this general implication of the mutational data for different nucleoproteins has been called into question (Pollock and Goldstein 2014). It is therefore unresolved whether the preferences for different amino acids at each site in a protein change during evolution or remain essentially constant. Needless to say, this is a crucial issue that bears on the methodologies used for phylogenetic analysis and the description and modeling of molecular evolutionary processes. For instance, if preferences are conserved to a substantial extent, models that assume independent evolution at different protein sites are reasonable. On the other hand, if site-specific preferences change strongly, widespread amino acid coevolution must be explicitly included in molecular evolution models. At a more fundamental level, the recent controversy on the evolutionary rates of preference change (Pollock et al. 2012; Ashenberg et al. 2013; Pollock and Goldstein 2014) highlights our limited understanding of one of the fundamental steps in evolution: The replacement of amino acids in proteins.
The availability of large numbers of protein sequences, together with advances in bioinformatics and molecular biology methodologies, allow important issues in molecular evolution to be experimentally addressed on the basis of laboratory resurrections of ancestral proteins (Pauling and Zuckerkandl 1963; Benner et al. 2007). Recent examples include the adaptation of proteins to changing planetary conditions (Gaucher et al. 2008; Perez-Jimenez et al. 2011; Akanuma et al. 2013; Risso et al. 2013; Risso, Gavira, Sanchez-Ruiz 2014; Risso, Gavira, Gaucher, et al. 2014), the origin and evolution of thermophily (Hobbs et al. 2012), the origin of complexity in biomolecular machines (Finnigan et al. 2012), the role of epistasis in the emergence of new protein functions (Ortlund et al. 2007), the mechanisms of evolutionary innovation through gene duplication (Voordeckers et al. 2012), the degree of conservation of protein structure over planetary timescales (Ingles-Prieto et al. 2013), the evolutionary origin of detoxifying enzymes (Bar-Rogovsky et al. 2013), and the characterization of the evolutionary events leading to gene silencing (Kratzer et al. 2014). Here, we take advantage of the recent availability of phenotypically supported laboratory resurrections of Precambrian proteins to experimentally address the evolution of amino acid preferences on a timescale of ∼4 billion years (i.e., the time span of life on Earth). We used nearly 200 diverse extant thioredoxin sequences comprising the three domains of life to construct a highly articulated phylogenetic tree (Perez-Jimenez et al. 2011). In addition, we used a set of 75 chromosomal sequences of extant class A β-lactamases to construct a phylogenetic tree encompassing Gram-positive and Gram-negative bacteria (Risso et al. 2013). In both cases, the trees were sufficiently close to accepted organismal phylogenies to allow us to target well-defined Precambrian phylogenetic nodes (fig. 1) for which reliable age estimates are available (Hedges and Kumar 2009). Bayesian ancestral sequence reconstruction was used to obtain probabilistic estimates of the sequences at all the nodes of the phylogenetic trees. The genes encoded by the reconstructed sequences at the targeted nodes were synthesized and then expressed by Escherichia coli in the laboratory (or to use the parlance, “resurrected”) and exhaustively characterized in terms of structure, function, and stability (Perez-Jimenez et al. 2011; Ingles-Prieto et al. 2013; Risso et al. 2013; Risso, Gavira, Sanchez-Ruiz 2014; Risso, Gavira, Gaucher, et al. 2014). They were found to adopt the canonical fold of their modern counterparts despite a large number of mutational differences (close to 50% of the sequence in some cases) and their properties led to plausible evolutionary narratives that supported that proteins encoded by the reconstructed thioredoxin and β-lactamase sequences are credible phenotypic representations of the proteins that existed billions of years ago.
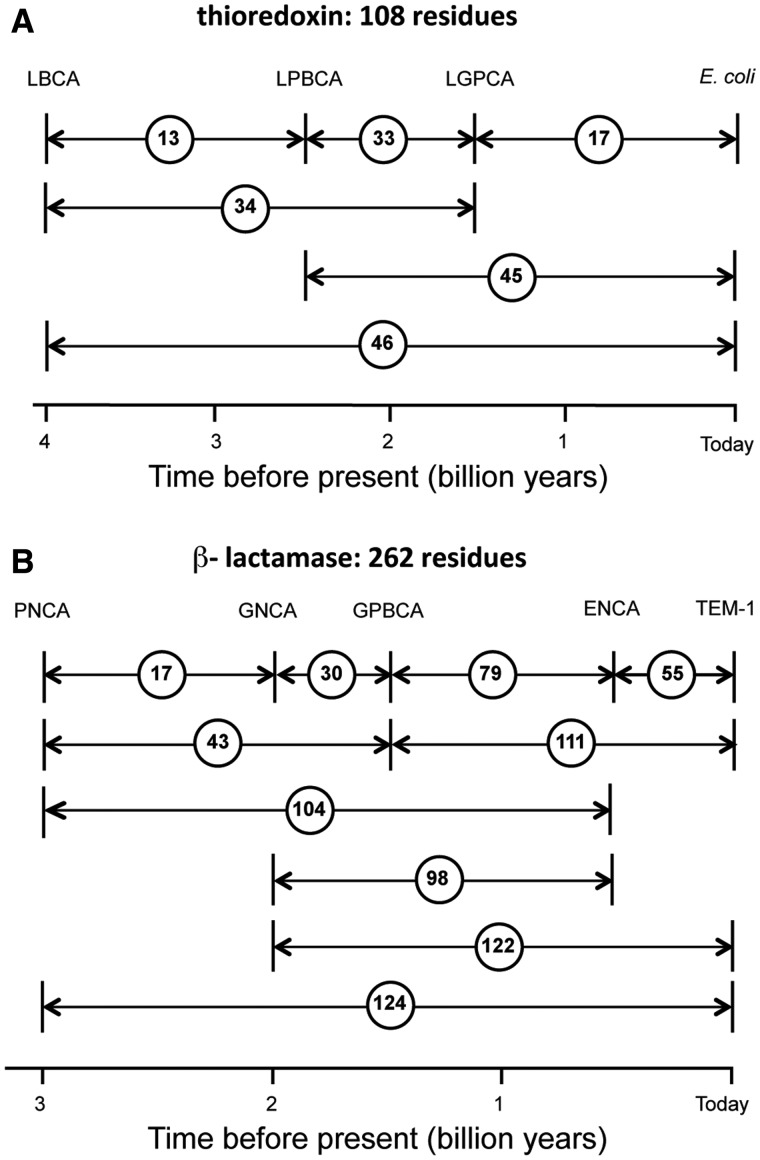
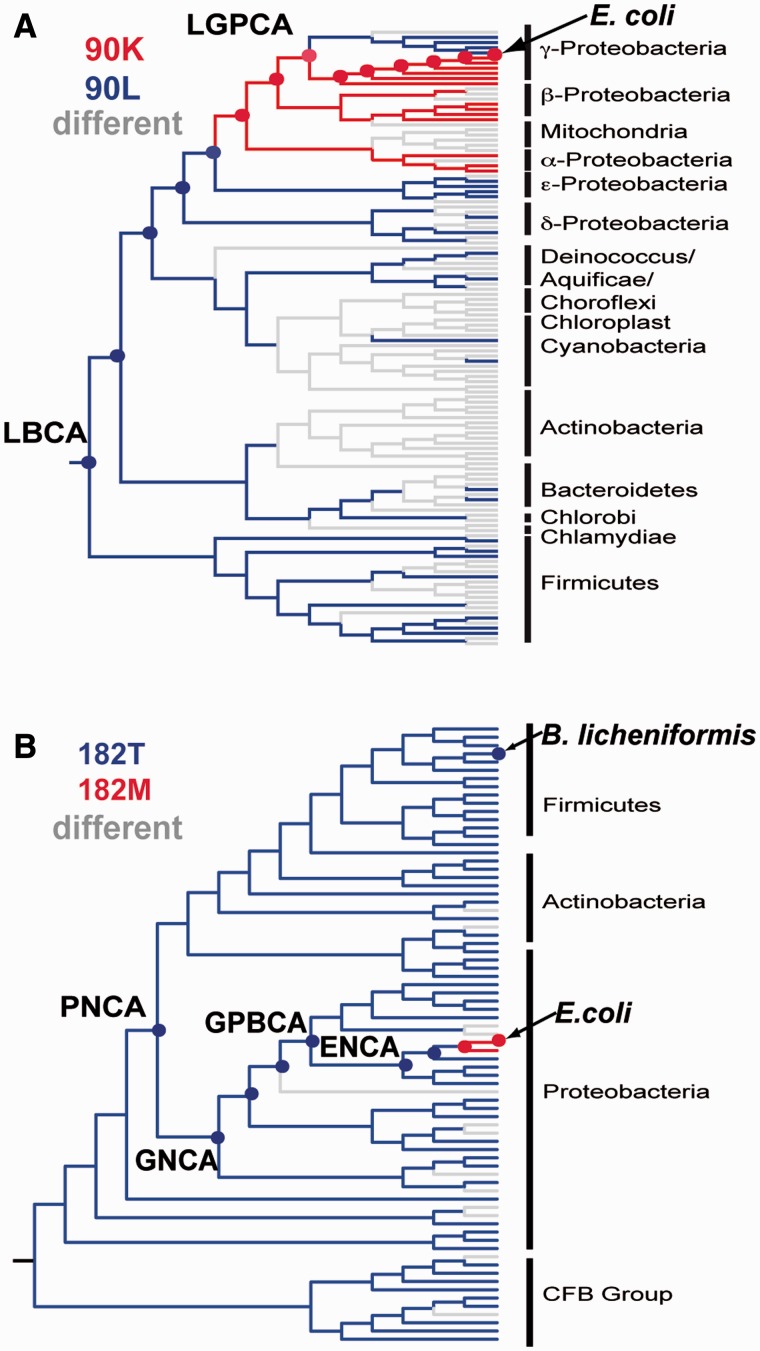
Fig. 1.
Amino acid differences between extant sequences and the reconstructed sequences for the Precambrian nodes targeted in this work. Age estimates are derived from the Timetree of Life (Hedges and Kumar 2009). (A) Thioredoxins. The ancestral nodes targeted are the following: LGPCA, last common ancestor of γ-proteobacteria; LPBCA, last common ancestor of the cyanobacterial, deinococcus, and thermus groups. Also shown is Escherichia coli thioredoxin. (B) β-lactamases. The ancestral nodes targeted are the following: PNCA, last common ancestor of various Gram-positive and Gram-negative bacteria; GNCA, last common ancestor of various Gram-negative bacteria; GPBCA, last common ancestor of γ-proteobacteria; ENCA, last common ancestor of enterobacteria. TEM-1 refers to E. coli TEM-1 β-lactamase.
The availability of phenotypically supported laboratory resurrections of Precambrian proteins allows us to address the evolution of amino acid preferences in a straightforward manner because: (1) Ancestral sequence reconstruction analyses lead to plausible estimates of the “age” of each given amino acid in a modern protein (i.e., the first appearance of the amino acid along the line of descent from the ancestor to the extant protein under study) and, therefore, to estimates of the geologic time available for energetic adjustment and (2) measurements of mutational effects on the stability of modern proteins can be compared with experimentally determined effects of the same mutations on the stability of the credible representations of their ancestors. Molecular clock age estimates are available for many Precambrian and Cambrian nodes for the tree of life (Hedges and Kumar 2009). We use these estimates as proxies to better understand the geologic timescales associated with site-specific amino acid preference. Given the controversial nature of molecular clocks, however, we also present our results in the context of sequence divergence (fig. 1).
We first report a comparative experimental analysis on the effect of 21 mutations on the stability of both E. coli thioredoxin and on the laboratory resurrection corresponding to the thioredoxin of the last bacterial common ancestor (LBCA). The time span of this comparison is billions of years and all the mutations selected involve highly similar amino acids and very minor structural alterations. If our results demonstrate that amino acid preference is conserved across long evolutionary timescales, it may be reasonable to infer that this is a general phenomenon that holds for shorter timescales and, more importantly, for dissimilar amino acids. Despite the plausibility of this inference, we deemed it convenient to specifically test preference conservation in instances involving the exchange between highly dissimilar amino acids. We thus report the effect of the lysine/leucine exchange at position 90 on the stability of E. coli thioredoxin and several laboratory-resurrected Precambrian thioredoxins, so that the evolutionary history of the K versus L preference can be followed across large geologic timescales. Interestingly, the preference conservation found in this case (L is always energetically preferred over K at position 90, even for thioredoxins in which there is a K at that position) is linked to an unanticipated mechanism involving a local structural switch upon mutation. Finally, we consider the effect of the methionine/threonine exchange on the stability of two modern β-lactamases (E. coli TEM-1 and Bacillus licheniformis) and three laboratory-resurrected Precambrian β-lactamases dating up to about 3 billion years ago. This is a particularly interesting case because the M182T mutation in the TEM-1 β-lactamase gene is a global suppressor (Huang and Palzkill 1997; Wang et al. 2002; Bloom et al. 2005; Salverda et al. 2010) that appears linked to many clinical cases of emergence of resistance toward new antibiotics. In fact, our results suggest that a relationship between conservation of amino acid preference and the existence of global suppressor mutations exists for this protein family.
Results and Discussion
Comparative Analysis of the Effect of 21 Chemically Conservative Mutations on the Stability of the Thioredoxins from E. coli and the LBCA
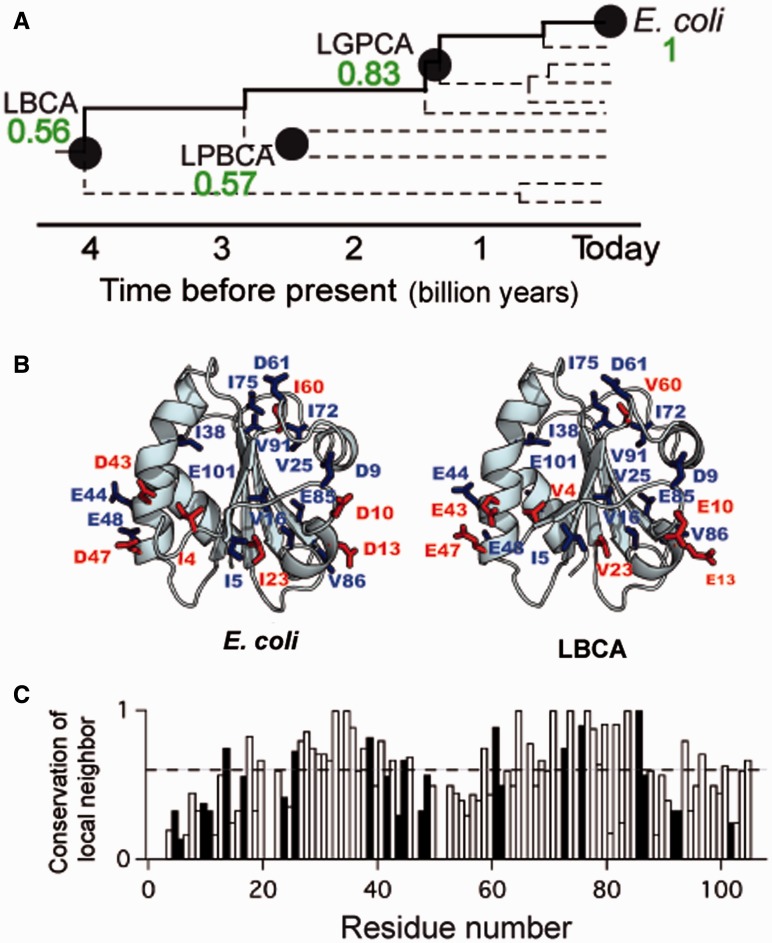
We previously reported the effects of a large number of mutations on the stability of the extant thioredoxin from E. coli (Godoy-Ruiz et al. 2004, 2005, 2006). All the mutations studied belong to the E→D, D→E, I→V, and V→I types and introduce, therefore, very small molecular changes: The presence or absence of a –CH3 in the case of an I↔V replacement versus the presence or absence of a –CH2- (and likely a small difference in the spatial position of the negative charge) in the case of an E↔D replacement. Here, we determine the effects of these mutations on the stability of the laboratory resurrection of the thioredoxin in the LBCA (fig. 2A). The mutations performed on the ancestral protein background can be classified into two groups. Fourteen mutations are identical, in terms of the residues involved and the direction of the mutation, with those we previously introduced in the E. coli background; for instance, there is a valine at position 16 in both E. coli and LBCA thioredoxins and therefore the effect of the V16I mutation can be studied in both backgrounds. On the other hand, seven mutations must be studied in opposite directions for the extant and ancestral backgrounds. For instance, there is an isoleucine at position 23 in E. coli thioredoxin, while a valine is present at the same position in LBCA thioredoxin. The I23V mutation is performed on the extant background and V23I is performed on the ancestral background. For comparison purposes, the stability effect of the V23I mutation is changed in sign to obtain the corresponding value in the “E. coli direction” (i.e., I23V). The positions corresponding to these two kinds of mutations are respectively labeled in blue and red in figure 2B (the same color code is used in supplementary table S1, Supplementary Material online, where the mutations are described in detail). It must be noted that, as expected from the sequence identity, the extant and ancestral proteins substantially differ in the residues present in the molecular neighborhoods of the positions targeted for mutation (nearly half the residues, on average, within a sphere of radius 6 Å around each position; see fig. 2C).
Fig. 2.
Ancestral and extant thioredoxins. (A) Schematic phylogenetic tree showing thioredoxin nodes relevant to this study (see fig. 1 for definitions). Numbers alongside the nodes stand for the values of the sequence identity with the extant protein. (B) Structures of E. coli thioredoxin and LBCA thioredoxin (PDB codes 2H6X and 4BA7). Positions targeted for mutation are shown (E. coli thioredoxin numbering) with a color that refers to the relation between the mutations actually performed in the two backgrounds. Blue, same direction in both backgrounds; red, opposite directions (see main text and supplementary table S1, Supplementary Material online, for details). (C) Fraction of amino acids present in the neighborhood (6 Å) of each LBCA thioredoxin residue that are identical with those in the neighborhood of the same position in E. coli thioredoxin. Positions targeted for mutation are shown with black bars. The horizontal dashed line corresponds to the sequence identity (0.55) between the two proteins.
The 21 variants of LBCA thioredoxin (supplementary table S1, Supplementary Material online) required to calculate the stability impact of the targeted mutations were prepared and their thermal denaturation was exhaustively characterized by differential scanning calorimetry (DSC). For all variants, experiments at different protein concentrations (supplementary table S2 and fig. S1, Supplementary Material online) were performed to rule out the possibility of association equilibria. Additional experiments were performed to assess the reversibility of the denaturation process and scan-rate effect on the denaturation process (supplementary table S3 and fig. S1, Supplementary Material online). These studies and the subsequent data analyses (see supplementary data, Supplementary Material online) support that the thermal denaturation of the LBCA thioredoxin variants conforms to a two-state equilibrium unfolding with some kinetic distortions at temperatures higher than the measured Tm. Such distortion precludes the determination of reliable values for the unfolding heat capacity change (ΔCP) and, consequently, prevents us from calculating mutational effects on unfolding free energy (ΔΔG values) on the basis of the integrated Gibbs–Helmholtz equation. Nevertheless, since mutational effects on denaturation temperature (ΔTm values) are small for the I/V and E/D exchanges, we could calculate ΔΔG’s from ΔTm’s using the approximate equation proposed by Schellman (1987) that does not require a ΔCP value. Note, however, that the same conclusions are reached using Tm as an empirical metric of stability and describing the mutational effects on stability by the ΔTm values; for reference, both ΔΔG and ΔTm values are shown in figure 3. In any case, the evolutionary stability threshold for thioredoxins is likely linked to kinetic stability and both ΔΔG and ΔTm can be viewed as metrics of the mutational effects on kinetic stability (see Godoy-Ruiz et al. 2006 for details).
Fig. 3.
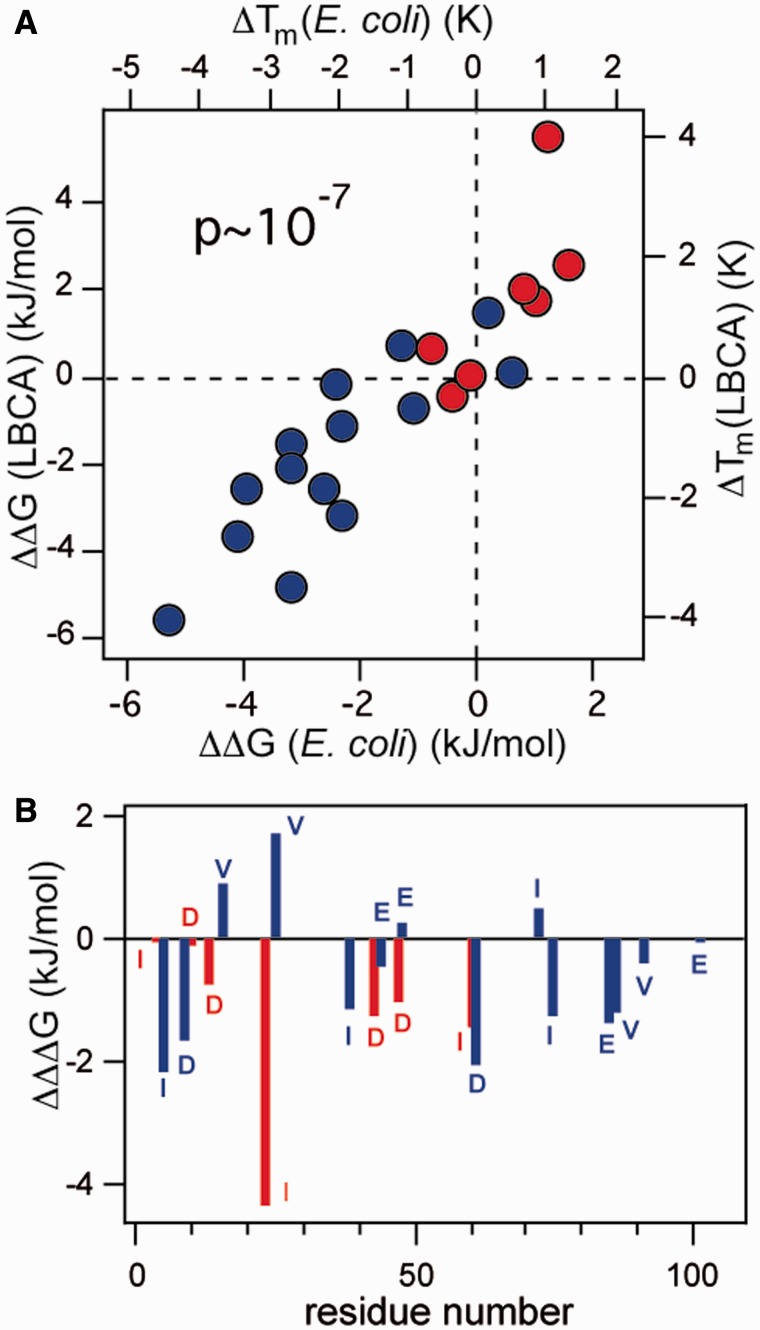
Comparison of mutation effects in E. coli thioredoxin versus LBCA thioredoxin. (A) Stability effects of mutations on LBCA thioredoxin versus the corresponding effects on E. coli thioredoxin. Mutational effects on unfolding free energies (ΔΔG values) and denaturation temperatures (ΔTm values) are calculated in the E. coli direction. The color code (the same as that used in fig. 2B and supplementary table S1, Supplementary Material online) refers to the relation between the mutations actually performed in the two backgrounds. Typical experimental scatter is smaller than the size of the points. (B) Mutational effects on E. coli thioredoxin stability minus the corresponding effects on LBCA thioredoxin stability (ΔΔΔG and ΔΔTm values).
A plot of mutation effects on the stability of the ancestral LBCA thioredoxin versus the corresponding effects on the stability of the extant E. coli thioredoxin shows a strong correlation (fig. 3A) with a Pearson correlation coefficient of 0.89, a slope close to unity (1.03 ± 0.21) and a value for the probability that the correlation occurs by random chance of P = 9 × ·10−8. Furthermore, the correlation holds for the positions in which the extant and ancestral amino acids differ (red data points in fig. 3A).
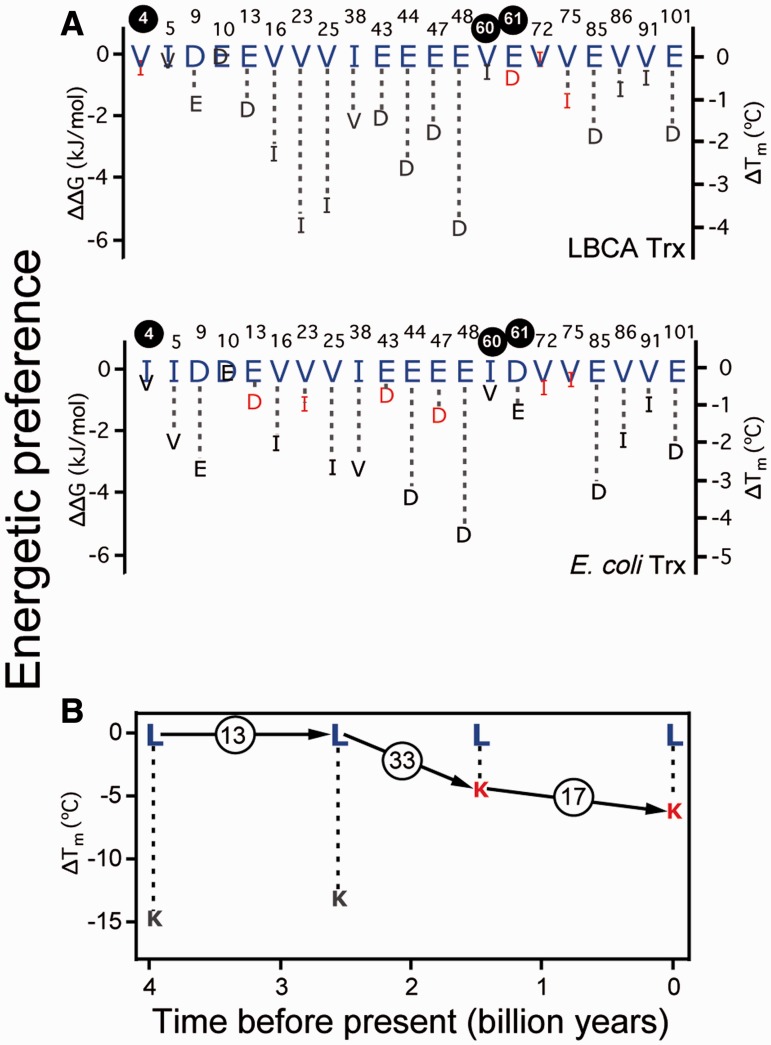
The excellent ancestral/extant correlation found, however, should not be taken to imply that the mutational energetics have not changed at all over the course of billions of years. In fact, when calculated in the E. coli direction, most of the mutations studied are more destabilizing on the modern E. coli thioredoxin as compared with the ancestral background (fig. 3B). This result appears consistent with some degree of evolutionary adjustment to the amino acid residues present in the extant protein, that is, with the evolutionary Stokes shift (Pollock et al. 2012). However, the extent of evolutionary adjustment is insufficient to erase the ancestral pattern of energetic preferences. To make this point visually clear, we have prepared plots of energetic preference versus position for the E. coli and LBCA thioredoxins (fig. 4A). The energetic preference scale is constructed in the following ways: (1) A value of zero is assigned to the energetically more preferred amino acid (i.e., if the X→Y mutation is destabilizing, X is the energetically more preferred amino acid; if the X→Y mutation is stabilizing, Y is the energetically more preferred amino acid) and (2) the less preferred amino acid is assigned a preference value equal to the mutational change in unfolding free energy (or the mutational change in denaturation temperature) associated to the replacement of the more preferred amino acid with the less preferred one (i.e., a negative value in all cases). There is a good agreement between the sets of more preferred amino acids for E. coli and LBCA thioredoxins: Only 3 discrepancies out of 21 instances were observed (positions 4, 60, and 61; see fig. 4A) and these corresponded to cases in which preference differences are quite minor. In contrast, there are 7 sequence differences between the extant and ancestral proteins at the 21 position studied. As such, the energetic amino acid preferences are more conserved than the residues themselves over evolutionary time.
Fig. 4.
Amino acid energetic preferences in modern and laboratory-resurrected Precambrian thioredoxins. For each position, two amino acids are considered. The more preferred amino acid is assigned a preference value of zero. The less preferred amino acid is assigned a (negative) preference value equal to minus the change in unfolding free energy or denaturation temperature (ΔΔG and ΔTm values, respectively) associated to its replacement with the more preferred amino acid. In all cases, the more preferred amino acid is shown in large blue letters. Red color is used for the less preferred amino acid whenever it is the amino acid present in the non-mutated (wt) protein. (A) Amino acid preferences for several positions in E. coli and LBCA thioredoxins. Positions at which more preferred amino acids differ between E. coli and LBCA thioredoxins (Chou and Fasman 1974; Mitraki et al. 1991; Baroni et al. 2004) are labeled with white numbers inside black circles. Position 10 is not labeled because there is no significant difference in preference. (B) Amino acid preference at position 90 of thioredoxins as a function of geologic time. Total numbers of amino acid differences between consecutive nodes are shown within circles.
Leucine Versus Lysine Preferences at Position 90 in Thioredoxins
The results summarized in the preceding section support the notion that preferences among biochemically similar amino acids may be conserved even over long evolutionary timescales. One obvious implication is that conservation of amino acid preference is likely widespread and that it can be expected to hold over shorter timescales and also for dissimilar amino acids. A particularly illustrative instance of the latter case is described below.
A lysine residue is present at position 90 in the modern E. coli thioredoxin, while leucine is the ancestral residue at this position along the line of descent from the ancestor to the extant E. coli protein (see phylogenetic tree annotated with amino acids at position 90 in fig. 5A). Specifically, the leucine residue present at position 90 is inferred for thioredoxins of the LBCA (last bacterial common ancestor, about 4 billion years before present) and the last common ancestor of the cyanobacterial, deinococcus, and thermus groups (LPBCA, about 2.5 billion years before present). Thus, we prepared variants of these proteins with the L90K mutation. On the other hand, a lysine residue is present at position 90 in the extant thioredoxin from E. coli and inferred for the laboratory resurrection corresponding to the thioredoxin of the last common ancestor of γ-proteobacteria (LGPCA, about 1.5 billion years before present); therefore, we prepared variants of these proteins with the K90L mutation. We determined the stability of the “wild-type” (wt) proteins and the corresponding mutant variants using DSC. Some of the determined mutational effects on denaturation temperature were very large (up to about 15°) and, therefore, the use of Schellman equation (Schellman 1987) to calculate mutational effects of unfolding free energy (ΔΔG values) was not advisable in this case. As an alternative, we elected to use denaturation temperature values as a metric for stability and to construct the amino acid preference scale on the basis of the mutation ΔTm’s. The results are summarized by plotting amino acid preference against a geologic timescale (fig. 4B). In all cases, mutations were found to be stabilizing in the K→L direction (i.e., K90L was found to be stabilizing in the E. coli and LGPCA background and L90K was found to be destabilizing in the LPBCA and LBCA backgrounds; see supplementary fig. S3, Supplementary Material online). Therefore, the energetic preference of L over K is conserved over ∼4 billions of years, despite the fact that, according to the ancestral reconstruction, the lysine at position 90 appeared about 2 billion years ago in the line of descent leading to the extant E. coli protein (figs. 4B and 5A). There is certainly evidence of adjustment to the “new” lysine residue, as the effect of mutation in the K→L direction is more stabilizing for the oldest thioredoxins in which leucine is the residue present at position 90 (fig. 4B). This is consistent with either preadjusting or permissive changes (previous mutations in the spatial neighborhood of position 90 “permitted” the introduction of a lysine residue at position 90) or the evolutionary Stokes shift (Pollock et al. 2012) (energetic adaptation to the new residue after it has been introduced). The adjustment, however, does not change the ranking of amino acid preferences at this site and replacement with the ancestral amino acid (i.e., the mutation K90L) does stabilize E. coli thioredoxin (see fig. 4 and supplementary fig. S3, Supplementary Material online).
Fig. 5.
Evolutionary histories of the amino acid present at position 90 in thioredoxins (A) and position 182 in β-lactamases (B). Phylogenetic trees used for ancestral sequence reconstruction of thioredoxins (Perez-Jimenez et al. 2011) and β-lactamases (Risso et al. 2013) are color coded according to the residue present: Red (extant), blue (ancestral), and grey (other). Small circles are used to highlight the evolutionary trajectory from the oldest ancestor to the extant E. coli protein.
Methionine Versus Threonine Preferences at Position 182 in β-Lactamases
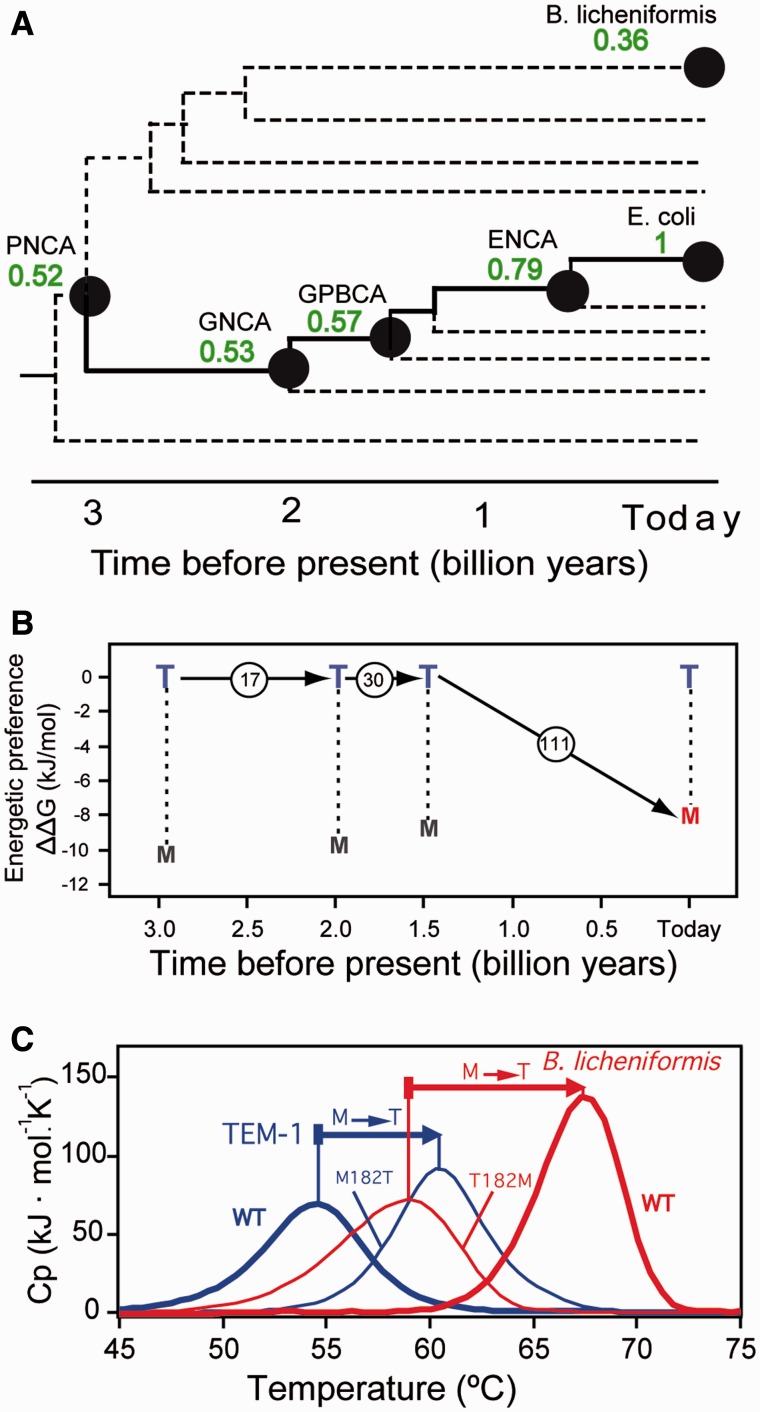
The global suppressor M182T mutation appears often in TEM-1 β-lactamases linked to clinical cases of emergence of antibiotic resistance. The mutation is known to be stabilizing in the extant TEM-1 background and this stabilizing effect has been proposed to permit the acquisition of destabilizing mutations that enhance catalytic efficiency toward a new antibiotic (Huang and Palzkill 1997; Wang et al. 2002; Bloom et al. 2005; Salverda et al. 2010). Reconstruction of ancestral lactamase sequences (Risso, Gavira, Gaucher, et al. 2014) supports that the methionine residue at position 182 in TEM-1 β-lactamase appeared comparatively recently in the line of descent leading to the extant TEM-1 protein (figs. 5B and 6 and supplementary fig. S4, Supplementary Material online), while a threonine is present in sequences of many modern β-lactamases and also in the reconstructed sequences corresponding to the last common ancestors of γ-proteobacteria (GPBCA, about 1.5 billion years before present), various Gram-negative bacteria (GNCA, about 2 billion years before present), and various Gram-positive and Gram-negative bacteria (PNCA, about 3 billion years before present) (fig. 6A). We thus prepared the proteins encoded by these reconstructed sequences with and without the T182M mutation, while the extant TEM-1 β-lactamase was prepared with and without the original global suppressor mutation M182T (see supplementary data, Supplementary Material online, for details). DSC studies on the thermal denaturation of all these wt and variant forms showed that the energetic/structural preference of T over M is conserved over billions of years (fig. 6B). That is, the mutation T182M in the ancestral backgrounds is destabilizing and the mutation M182T in the extant TEM-1 background is stabilizing (supplementary fig. S4, Supplementary Material online). No clear evidence of adjustment to the new residue is apparent in this case. This is perhaps due to the inference that a methionine at position 182 appeared only recently in the evolutionary trajectory leading to the TEM-1 β-lactamase (figs. 5B and 6). We have also studied the effect of the M/T exchange on the stability of the extant β-lactamase from B. licheniformis (figs. 5B and 6A). In this case, a threonine residue is present in the wt protein and, therefore, we prepared the β-lactamase from B. licheniformis with and without the T182M mutation. The corresponding scanning calorimetry profiles are compared with those for the TEM-1 lactamase in figure 6C. M182T is stabilizing in the TEM-1 background, while T182M is destabilizing in the B. licheniformis background. That is, in both instances the mutation is stabilizing in the M→T direction, further supporting the conservation of energetic preference. It is to be noted that TEM-1 β-lactamase and the β-lactamase from B. licheniformis share a common ancestor on the order of 3 billion years ago (figs. 5B and 6A). These extant proteins, therefore, may be viewed as being separated by ∼6 billion years of evolution and, in fact, they show only 37% sequence identity.
Fig. 6.
Amino acid energetic preferences at position 182 for modern and laboratory-resurrected Precambrian β-lactamases (see main text and legend to fig. 4 for a description of the preference scale). (A) Schematic phylogenetic tree showing β-lactamase nodes relevant to this work (see fig. 1 for definitions). Numbers alongside the nodes stand for the values of the sequence identity with the extant protein. (B) Amino acid preferences at position 182 in β-lactamases as a function of geologic time. The data point at “today” corresponds to the E. coli TEM-1 lactamase. Total numbers of amino acid differences between consecutive nodes are shown within circles. (C) Effect of the M/T exchange at position 182 on the stability of two extant β-lactamases as followed by DSC. A methionine is present at position 182 in E. coli TEM-1 lactamase, while threonine is the residue at that position in the lactamase from Bacillus licheniformis. M182T is stabilizing in the TEM-1 background and T182M is destabilizing in the B. licheniformis background. In both cases, the mutation is stabilizing in the M→T direction, indicating preference conservation.
A Plausible Evolutionary Explanation for the Occurrence and Persistence of Less Preferred Amino Acids
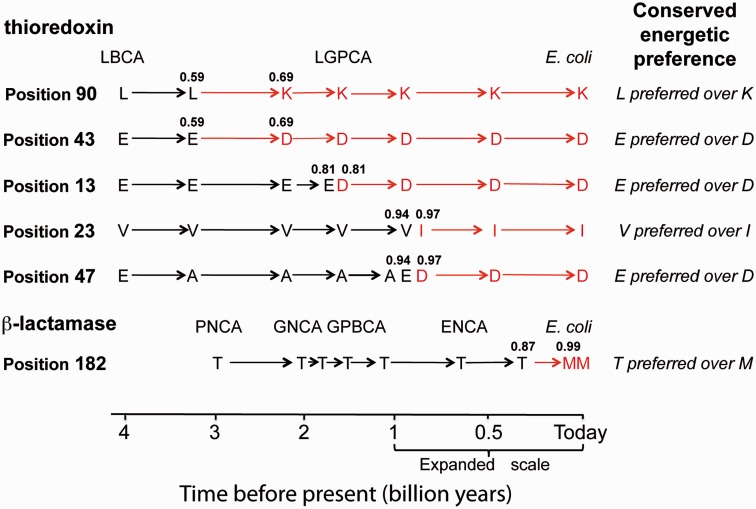
A rather noteworthy result reported in this work is the observation that billion-year-old conservation of energetic preference exists at positions in which the amino acid present has changed in the line of descent leading from the oldest ancestor to the extant protein. We found six examples of this scenario. They are summarized in figure 7 where the common pattern is apparent. The residue present in the extant protein (the “extant” amino acid) differs from the residue in the oldest ancestral protein (the “ancestral” amino acid) but the energetic preference for the ancestral amino acid over the extant one is conserved. Ancestral sequence reconstruction provides estimates of the geologic time at which the extant amino acid first appeared in the line of descent leading to the extant protein (see fig. 7 and annotated phylogenetic trees in fig. 5 and supplementary figs. S2 and S4, Supplementary Material online). Such times range from several hundred million years (for the methionine residue at position 182 in β-lactamase) to about 2 billion years (for the aspartate residue at position 43 and the lysine residue at position 90 in thioredoxins) and correspond to differences in sequence identity that range between 0.59 and 0.69.
Fig. 7.
Evolutionary history of the amino acid occupancy at several positions in thioredoxins and β-lactamases as inferred from ancestral sequence reconstruction. In all the positions shown, the amino acid present in the extant protein (the “extant amino acid”) is not the energetically more preferred. Extant amino acids are shown in red color, in such a way that the time in which they appeared is visually apparent. Two consecutive one-letter amino acid codes are used when different amino acids are present at two nodes of very similar age (if the same amino acid is present at two nodes of similar age, a single letter is shown). Values of sequence identity with the extant protein are shown for the sequences involved in the ancestral to extant amino acid transition. (see supplementary fig. S2, Supplementary Material online for further details).
Overall, the six cases collected in figure 7 provide clear evidence that energetic preferences may be conserved over planetary timescales even when the amino acid residues themselves change during evolution. However, they also pose some obvious evolutionary questions that need to be addressed. It follows from the preference conservation that the mutation to the extant (less preferred) amino acid was destabilizing when it occurred at a particular time before present (on the order of billions of years in most cases). As previously pointed out, however, the stability of natural proteins is marginal (Taverna and Goldstein 2002; Bershtein et al. 2006; Godoy-Ruiz et al. 2006; Bloom et al. 2007; Tokuriki et al. 2007; Sikosek and Chan 2014), that is, just slightly above an evolutionary stability threshold, and, thus, even a moderately destabilizing mutation could potentially impair proper folding or facilitate degradation with a subsequent deleterious impact on organismal fitness. According to a stability threshold selection scenario, destabilizing mutations are still often accepted (otherwise, protein stability would not be marginal). Acceptance of a mutation with a destabilizing effect may require that a previous stabilizing mutation (i.e., a compensating or permissive mutation) pave the way for the acceptance of the destabilizing one (Weinreich et al. 2006; Bloom et al. 2007; Ortlund et al. 2007; Wyganowski et al. 2013). It is important to note in this context that protein stability thresholds are unlikely to remain constant during evolution. Actually, several threshold-relaxing events may have plausibly occurred and facilitated the acceptance of destabilizing mutations: (1) The development of efficient chaperone systems may lower stability thresholds, as suggested by the fact that chaperonin overexpression can promote enzyme evolution by allowing the folding of variants with functionally useful but destabilizing mutations (Tokuriki and Tawfik 2009; Wyganowski et al. 2013); (2) according to one proposal, the temperature of the oceans has decreased over billions of years (Knauth and Lowe 2003; Gaucher et al. 2008), thus providing ample opportunities to relax the stability threshold for ancient life living in ancient oceans; (3) organismal migration from a high-temperature local environment (hydrothermal systems for instance; Lane and Martin 2013) to more temperate environment could also lower the stability threshold for the proteins of the migrating organisms; and (4) comparatively short periods of sharp decreases in planetary temperature are known to have occurred (global glaciations, usually referred to as Snowball Earths; Hoffman et al. 1998; Kirschvink et al. 2005) and may have possibly facilitated the occurrence of some highly destabilizing mutations.
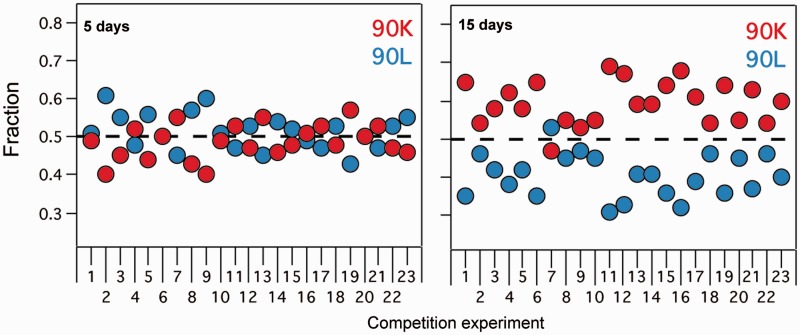
Overall, it is clear that several plausible scenarios may explain why proteins accumulate less energetically preferred amino acids. However, some specific explanation is required for the fact that these “energetically sub-optimal” amino acids persist over significant evolutionary periods, since estimates of neutral mutation rates (Ochman et al. 1999) obviously predict that mutational changes will occur over the billion years timescale of figure 7. The simplest explanation for the evolutionary persistence of less preferred amino acids is that they allow protein functional properties that lead to enhanced organismal fitness. In order to obtain some experimental insights into this scenario, we have considered the functional impact of the K/L exchange in thioredoxins—proteins that regulate many cellular processes (Holmgren 1985) and that proteomic analyses (Kumar et al. 2004) have identified as having a large number of protein binding partners in vivo. It is clear, therefore, that activity assays in vitro are of limited usefulness in this context, as they cannot provide information about the effect of the K/L exchange on the multitude of biomolecular processes/interactions in which thioredoxin participates in vivo. Consequently, we elected to directly measure the effect of the K/L exchange at position 90 of thioredoxins on organismal fitness. It is obviously difficult to perform fitness studies on Precambrian micro-organisms, but the effects of these proteins on modern organisms can be determined. We thus complemented a thioredoxin-deficient E. coli strain with plasmids containing either the wt thioredoxin gene or the gene carrying the K90L mutation. We performed competition experiments in batch culture for long periods of time (about 2 weeks) without addition of nutrients. The rationale behind this approach is that conditions in long-term stationary-phase cultures have been proposed to mimic conditions found in natural environments (Finkel 2006). Briefly, we set up pair competition experiments of a single clone of the strain complemented with wt thioredoxin versus a single clone of the strain complemented with the K90L variant. The proportions of the two variants in each population were determined at 5 and 15 days after the start of the competition using Sanger sequencing and the QSVanalyzer program (Carr et al. 2009), a methodology that does not require the use of markers (which could potentially have an effect on fitness). In order to rule out the possibility that the “winner” of the competition is determined by fitness differences between clones that are not related to the K90L mutation in thioredoxin, we performed 23 independent competition experiments (i.e., with 23 independent pairs of clones). After 5 days of competition, wt/K90L population ratios for the 23 experiments showed some dispersion although the average value was close to unity, indicating no systematic bias (fig. 8). On the other hand, after 15 days 22 (out of 23) competition experiments displayed a wt/K90L population ratio higher than unity, indicating a clear preference of the “wt strain” over the “K90L” strain (fig. 8). The interpretation of these organismal fitness experiments is complex. One possibility is that the genetic alterations leading to the growth advantage in the stationary phase phenotype (Finkel 2006) are more probable to occur in the wt strain, thus amplifying an originally small difference in fitness. This notwithstanding, the fitness experiments summarized in figure 8 are consistent with an evolutionary narrative that involves acceptance of a destabilizing mutation linked to relaxation of the stability threshold and persistence of the less preferred amino acid because of functional advantages that translate into enhanced organism fitness.
Fig. 8.
Effect of the K90L mutation in thioredoxin on E. coli fitness. The results of wt/K90L competition experiments in batch cultures for long periods of time are shown. Twenty-three independent experiments were set up and the fractions of cells containing wt (red) and K90L (blue) thioredoxins were determined after 5 and 15 days, respectively. Note that in 22 (out of 23) experiments, cells containing wt thioredoxin “win” the competition after 15 days.
On the Molecular Basis of Amino Acid Preference Conservation
The experimental results reported in this work show that amino acid preferences in proteins can be conserved over diverse geological and evolutionary timescales. This result may seem surprising when considering the large changes in sequence and, therefore, in residue–residue interactions, which proteins experience over billions of years. However, reasonable and convincing molecular explanations can be synthesized for many instances. These explanations are described below and categorized in terms of the molecular effect invoked.
Secondary Structure-Forming Tendencies
We start by considering the correlation (fig. 3) found between the effect of E/D exchanges on the stability of the thioredoxins from E. coli and the LBCA (about 4 billion years before present). In some cases, the correlation may be simply linked to fold conservation through secondary structure-forming tendencies. For instance, six of the studied E↔D exchanges are in α-helix positions (one in a β-strand while three are in loops; see fig. 1 in Ingles-Prieto et al. 2013) and glutamate is considered to be a better helix former than aspartate (Pace and Scholtz 1988). Not unexpectedly, mutations at those six positions are stabilizing in the D→E direction (see supplementary table S4, Supplementary Material online).
Hydrophobic Packing
The kind of interpretation provided in the preceding section does not apply to the studied I↔V exchanges analyzed in this study: Seven of them were introduced at β-strand positions (two at α-helices and four at loops) and there appears to be little difference between isoleucine and valine in terms of β–strand-forming tendency (see fig. 3 in Kim and Berg 1993). In fact, the stability impacts of the mutations at the buried positions more likely reflect hydrophobic packing effects (Shortle and Lin 1985; Lim and Sauer 1989; Wilson et al. 1992; Gromiha et al. 2013). Consider, for instance, an isoleucine residue located at a given position in a well-packed hydrophobic core. Replacement to valine will remove a methyl group and this is known to cause strain, distortion, or elimination of stabilizing interactions, with the consequent protein destabilization (Wilson et al. 1992). Local compensation of this destabilization would plausibly require that a methyl group be reintroduced at a compensatory location without disturbing local packing, a result that can hardly be achieved through a single second-site mutation. Indeed, it has been known for many years that local stability-compensating mutations within a protein core are highly uncommon (Shortle and Lin 1985; Lim and Sauer 1989; Wilson et al. 1992). We could expect then that the energetic preference for isoleucine over valine at the position under consideration be conserved over evolutionary time (even if the residue at the position changes). Of course, the same argument holds if, at the different position, packing and interactions are optimized for valine in the ancestral protein. In such a case, the energetic preference for valine over isoleucine would be conserved.
Helix Capping
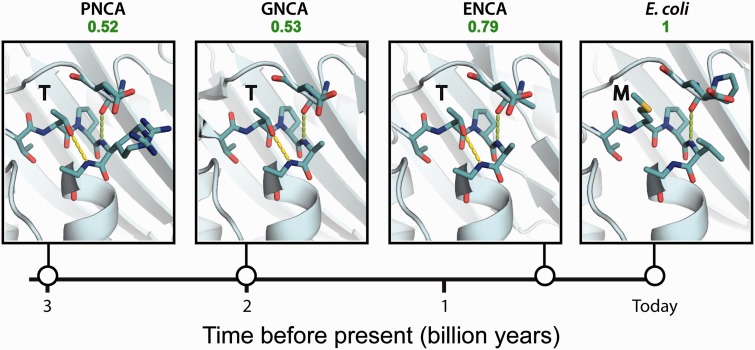
Energetic preference for threonine over methionine at position 182 in lactamases is very likely related to the fact that 182 occupies the amino-capping (Ncap) position for the 183–195 helix (Kather et al. 2008) and that threonine is an excellent Ncap residue (Harper and Rose 1993) while methionine is not. Indeed, threonine is the ancestral residue at position 182 in the laboratory-resurrected β-lactamases corresponding to the Precambrian ENCA, GNCA, and PNCA nodes and the three-dimensional (3D) structures of these proteins (Risso et al. 2013; Risso, Gavira, Gaucher, et al. 2014) show the expected hydrogen bonding between the Ncap and N3 residues in the capping motif (fig. 9), while this interaction is not possible with a methionine at 182, as shown by the structure of the extant TEM-1 β-lactamase (fig. 9).
Fig. 9.
Structural basis for preference conservation at position 182 in β-lactamases. The region of the 3D structure around position 182 is shown for the extant TEM-1 lactamase and for the lactamases corresponding to several Precambrian nodes: ENCA, GNCA, and PNCA (see figs. 1 and 6A). The N-terminal end of the 183–192 helix and the corresponding capping box are shown. Note the canonical hydrogen bond between the Ncap residue (threonine) and the N3 residue in the ancestral proteins. Such interaction is absent in the extant TEM-1 lactamase because methionine is present at the Ncap position.
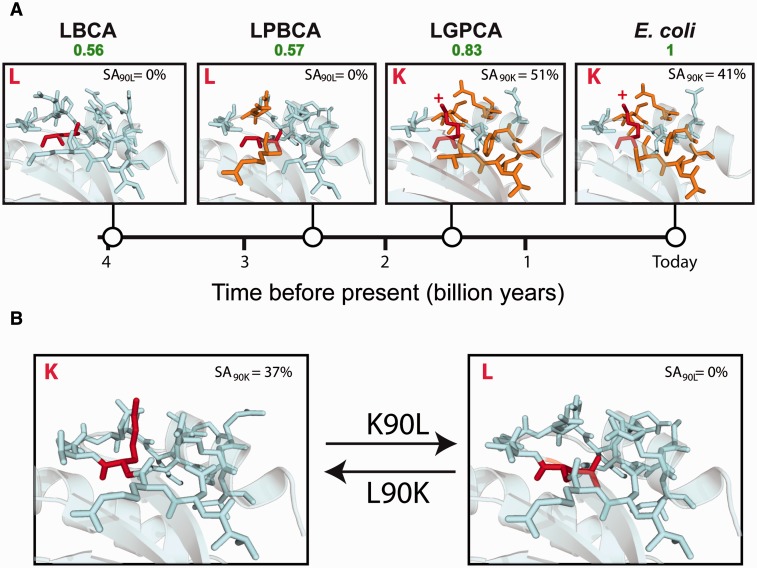
A Local Structural Switch
The kind of explanations adduced in the preceding paragraphs hardly apply to the conservation of the leucine over lysine preference at position 90 in thioredoxins because the energetically favored situations for hydrophobic and ionizable residues are quite different. Hydrophobic residues tend to be found at buried positions, while ionizable residues tend to be on the protein surface with the charged moiety exposed to the aqueous solvent. Indeed, examination of the previously determined 3D structures for extant and laboratory-resurrected Precambrian thioredoxins reveals a buried leucine residue in the “oldest” LBCA and LPBCA thioredoxins and an exposed lysine residue in the “younger” LGPCA and E. coli thioredoxins (fig. 10). Mutations involving the exchange between hydrophobic and ionizable residues are found to be experimentally destabilizing (in the wt to variant direction) in most cases (Isom et al. 2008; Pey et al. 2010). This is a reasonable result given that a single molecular context cannot be energetically favorable for two highly dissimilar amino acids. The only possible explanation for the stabilizing character of the K90L mutation in the E. coli thioredoxin background is, therefore, that, upon mutation, a local structural rearrangement takes place with concomitant burial of the new leucine residue. Likewise, the L90K mutation in the ancestral LBCA and LPBCA thioredoxins must be accompanied by a local rearrangement that allows the introduced lysine reside to be exposed to the solvent, although in this case the stabilization of the exposed lysine does not fully compensate the destabilizing effect of removing the buried leucine. In other words, the K/L exchange at position 90 in thioredoxins involves a local structural switch that allows the optimization of the molecular surroundings for the residue present, leucine or lysine, although such optimization does not reverse the overall preference of leucine over lysine at position 90. This interpretation assumes, of course, that the switch has been conserved over billions of years, despite the fact that some of the residues in the neighborhood of position 90 have changed over that period of time (fig. 10). Crystallographic structures of the extant E. coli thioredoxin and the laboratory-resurrected thioredoxins corresponding the LBCA, LPBCA, and LGPCA nodes (Ingles-Prieto et al. 2013) are consistent with the structural switch hypothesis, as a buried leucine is seen in the 3D structures of LBCA and LPBCA thioredoxins, while an exposed lysine appears, at a different orientation, in the structures of LGPCA and E. coli thioredoxins (fig. 10A). In order to directly observe the switch, we have determined the crystal structure of the resurrected LPBCA thioredoxin with the mutation L90K. The structural switch is clearly apparent on comparison with the structure of the nonmutated LPBCA thioredoxin (fig. 10B).
Fig. 10.
Structural switch linked to L over K preference conservation at position 90 in thioredoxins. (A) The region of the structure around position 90 is shown for the extant E. coli thioredoxin and laboratory resurrections of Precambrian thiroredoxins corresponding to the LGPCA, LPBCA, and LBCA nodes (see figs. 1 and 2A). Residues (other than that at position 90) that differ from those in LBCA thioredoxin are shown in orange. The residue at position 90 (K or L) is shown in red and the SA of its side chain is given. (B) Region of the structure around 90 for LPBCA thioredoxin with and without the L90K mutation. SA, solvent accessibility.
Site-Specific Preference Conservation Provides One Plausible Evolutionary Explanation for the Existence of Intragenic Global Suppressor Mutations
The M182T mutation in the TEM-1 β-lactamase gene has been found to independently occur in many cases of emergence of resistance against extended spectrum cephalosporins (Huang and Palzkill 1997; Wang et al. 2002; Bloom et al. 2005; Salverda et al. 2010). It has by itself little effect on catalysis and, in fact, it is always reported to occur coupled to other mutations that are actually the ones responsible for the increased rate of hydrolysis of the antibiotic. These catalysis-enhancing mutations have, however, a destabilizing effect that is compensated by the stabilizing M182T mutation. M182T is, therefore, a paradigmatic example of an intragenic global suppressor, that is, a mutation that can rescue mutations at several sites. M182T is often considered as an intriguing mutation (Salverda et al. 2010). However, the results and analyses reported here provide a credible and straightforward explanation for its evolutionary origin. Threonine is the ancestral and energetically preferred residue at position 182. Still, the destabilizing mutation to methionine did occur along the evolutionary trajectory leading to TEM-1 β-lactamase (fig. 7) likely linked to one of the several scenarios for acceptance of a less preferred amino acid we have previously discussed (see section “A Plausible Evolutionary Explanation for the Occurrence and Persistence of Less-Preferred Amino Acids”). The presence of methionine, however, does not change the energetic preference ranking at position 182, which favors threonine over methionine even in the extant TEM-1 β-lactamase (fig. 6). This preference conservation makes sense from a structural point of view given that, with methionine at position 182, the canonical Ncap–N3 hydrogen bond of the N-capping motif for the 183–195 helix cannot be formed (fig. 9) and no second-site mutation can re-establish this stabilizing interaction. We may reasonably assume that the destabilizing presence of methionine at position 182 brings about some fitness advantage under “normal” circumstances but, nevertheless, we may expect the reversion to the energetically preferred threonine to readily occur in those cases in which the concomitant stabilizing effect has an adaptive value. This is in fact the scenario created by the challenge of a new antibiotic, as the mutations that enhance the lactamase-catalyzed hydrolysis of the antibiotic are typically destabilizing.
Besides M182T, several other examples of global suppressors are known for the β-lactamase gene (Salverda et al. 2010). Furthermore, intragenic global suppressors have been identified for other protein systems, such as staphylococcal nuclease (Shortle and Lin 1985), the transcription factor p53 (Baroni et al. 2004), the bacteriophage P22 tailspike protein (Mitraki et al. 1991), and the phage lambda repressor (Hetch and Sauer 1985). Although different mechanisms of suppression are possible and have been discussed, global stabilization has been demonstrated or proposed in many instances (Hetch and Sauer 1985; Shortle and Lin 1985; Nikolova et al. 2000; Baroni et al. 2004; Salverda et al. 2010). In view of the discussion provided in the preceding paragraph, it appears plausible that many stability-linked global suppressor mutations may in fact be reversions to the ancestral, energetically preferred amino acid. The intriguing possibility thus arises such that global suppressor mutations can be predicted on the basis of ancestral sequence reconstruction.
Conclusions
An amino acid replacement is more likely to be accepted at a given site if it contributes stabilizing interactions with the surrounding residues in the protein structure. Accordingly, a ranking of site-specific amino acid preferences applies to each site in a protein. There have been recent discussions about this topic in the literature (Pollock et al. 2012; Ashenberg et al. 2013; Pollock and Goldstein 2014), specifically in regards to whether amino acid preferences remain approximately constant or substantially change during the course of evolution. This is a crucial issue that bears not only on methodologies used for phylogenetic analysis, but also on the general descriptions and models of molecular evolutionary processes. We have provided experimental evidence here that, while evolutionary adjustments to a new amino acid may certainly occur, the extent of such adjustments are insufficient to erase the primitive rankings for amino acid preferences. Needless to say, our studies do not rule out the possibility that in some cases an adjustment (the evolutionary Stokes shift) does reverse the original preferences and we suspect that those cases are of particular interest. Yet generally, our results support the model that site-specific selective constraints were conserved throughout evolution despite sequence divergence that generates chemical diversity in protein space. It is important to note that such evolutionary conservation of amino acid preferences (and the concomitant conservation of mutation effects on protein stability) does not imply that proteins evolve without epistasis. This point has been eloquently made by Bloom and coworkers (Ashenberg et al. 2013) and there is no need to repeat their arguments here.
Our study is based on extensive mutational analysis of proteins encoded by reconstructed ancestral sequences corresponding to Precambrian nodes in the evolution of thioredoxins and β-lactamases. Admittedly, the reconstruction of these ancestral sequences is based on simple models of evolution. However, two important points must be noted in this regard. First, the properties of the experimental representations of Precambrian thioredoxins and β-lactamases used in this work have been previously found to conform to convincing evolutionary narratives that support their plausibility as phenotypic representations of the proteins that actually existed billions of years ago (Perez-Jimenez et al. 2011; Ingles-Prieto et al. 2013; Risso et al. 2013; Risso, Gavira, Sanchez-Ruiz 2014; Risso, Gavira, Gaucher, et al. 2014). Second, it appears highly unlikely that the unavoidable simplifications used in ancestral sequence reconstruction procedures bias the results of the mutational analyses specifically toward the conservation of energetic preferences. This is further supported when considering that the number of sequence differences between the extant proteins and the corresponding ancestral reconstructions is very large, approaching about 50% of the sequence for the oldest nodes.
We have discussed energetic preferences from a molecular point of view and we have shown that several straightforward mechanisms can reasonably explain their conservation over billions of years. Our analyses support that many cases of preference conservation may be due to the unavailability of local “second-site” stability-compensating mutations, although fold conservation through secondary structure-forming tendencies may also play a role in some instances. Furthermore, we have provided experimental evidence that conservation of the preference for an ancestral amino acid may in some cases involve reorganization on mutation to the ancestral local structure around the mutation site. This unanticipated structural switch mechanism implies a kind of structural memory effect in proteins and may potentially be highly relevant for the understanding of molecular evolution. Work is currently under way to ascertain the scope and impact of this memory effect.
We have shown that the M182T global suppressor mutation in the TEM-1 β-lactamase gene (linked to many clinical cases of resistance against new antibiotics) can be viewed as a return to the ancestral energetically preferred state. Similar explanations may plausibly hold for other intragenic global suppressors and, therefore, the intriguing possibility arises that ancestral sequence reconstruction can be used to predict global suppressor mutations.
Materials and Methods
Purification of the different thioredoxin variants to be used in stability measurements was performed as previously described (Perez-Jimenez et al. 2011). Briefly, genes were cloned into a pQE80L vector and transformed in E. coli BL21(DE3) cells and the His-tagged proteins were purified by affinity chromatography (His GraviTrap, GE Healthcare). Thioredoxins for crystallization experiments were prepared without a His-tag following a procedure we have previously described in detail (Ingles-Prieto et al. 2013). Purification of the different β-lactamases was performed as previously described (Risso et al. 2013). Briefly, genes were cloned into a pET24 vector with kanamycin resistance and transformed in E. coli BL21(DE3) cells. The proteins were purified by osmotic shock and gel filtration. Oligonucleotides used for mutagenesis were obtained from Eurofins MWG Operon (Ebersberg, Germany). Mutations were introduced using the Quikchange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and were verified by DNA sequence analysis (see supplementary data, Supplementary Material online, for further details on protein preparation).
Thermal stabilities of all the protein variants studied in this work were determined in Hepes buffer pH 7 with a VP-Capillary DSC (Microcal, Malvern) following protocols well established in our laboratory (Godoy-Ruiz et al. 2004, 2005; Perez-Jimenez et al. 2011; Risso et al. 2013; Risso, Gavira, Gaucher, et al. 2014). A typical calorimetric run involved several buffer–buffer baselines to ensure proper equilibration of the calorimeter followed by runs with several protein variants with intervening buffer–buffer baselines. For most variants, were performed detailed DSC studies into the reversibility of the calorimetric transitions, their scan-rate dependence, and the effect of protein concentration on the denaturation temperature (see supplementary data, Supplementary Material online, for details). The subsequent exhaustive data analyses (described in detail in the supplementary data, Supplementary Material online) supported in many cases the applicability of a two-state equilibrium model to the calculation of mutation effects on thermodynamic stability from the corresponding mutation effects on denaturation temperature.
Crystallization (using the counter-diffusion technique) and X-ray structural determination for the L90K variant of LPBCA thioredoxin were carried out as previously described in detail for several resurrected Precambrian thioredoxins (Ingles-Prieto et al. 2013) with only minor changes [capillaries of 0.3-mm inner diameter were used in initial crystallization screenings; data collection was done at the European Synchrotron Radiation Facility using beam line BM30; co-ordinates from the LPBCA thioredoxin (PDB.ID 2yj7) were used as search model for molecular replacement]. Crystallization methodologies and conditions are summarized in supplementary table S5, Supplementary Material online. The co-ordinates and the experimental structure factors have been deposited in the Protein Data Bank (PDB.ID 4ulx).
To study the impact of the K90L mutation in thioredoxin on organism fitness, we used an E. coli strain deficient in thioredoxins 1 and 2 (DHB4 derivative strain FA41, a gift from Dr Jonathan Beckwith, Harvard University) and we complemented it with plasmids containing the genes for wt thioredoxin and the variant with the K90L mutation. We deemed convenient to use this complementation approach, rather than allelic replacement, to avoid fitness effects associated with regulatory changes in expression levels triggered by the stress conditions created during fitness experiments. The genes coding for E. coli wt and mutant K90L thioredoxins were introduced in pET30a(+) (Novagen) derivative plasmids, in which target gene expression is under the control of a T7 promoter. In order to express the desired gene using this system, the cell requires the presence of the RNA polymerase specific from the T7 phage. This gene was introduced in FA41 by lysogenization with λDE3 (λDE3 Lisogenizaton kit, Novagen), a lambda-derivative phage bearing the T7 RNA polymerase under an Isopropyl beta-D-1-thiogalactopyranoside (IPTG)-inducible promoter. This is a system often used in our laboratory because it allows IPTG-induced overexpression for protein preparation purposes, as well as meaningful fitness studies in the absence of IPTG induction. The reason for the latter possibility is that the system is leaky, that is, even under non-inducing conditions there is basal expression from the T7 promoter. Actually, when the thioredoxin plasmid is introduced in the Trx-deficient strain, this basal expression is sufficient to compensate for the deficiency in growth of the Trx minus strain. For competition assays, 23 independent clones of FA41λDE3 with plasmid pET30a(+)::trxA and the same number of the strain bearing pET30a(+)::trxA K90L were separately grown overnight at 37 °C in LB medium. Cultures were then diluted 1/1,000 in minimal medium supplemented with glucose and grown at 37 °C to an OD600 of 0.2. At that point, one culture of wt thioredoxin and one culture of the mutant variant were mixed in 1/1 proportion. Mixed cultures were incubated at 37 °C for 15 days. At time points 5 and 15 days, 5 ml of mixed cultures were taken for plasmid extraction and sequencing. The obtained electropherograms were analyzed using the quantitative sequence variant analyzer (Carr et al. 2009) for quantification of the relative proportions of DNA from the two variants. Further details are provided in the supplementary data, Supplementary Material online.
Supplementary Material
Supplementary figures S1–S5 and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grants BIO2012-34937, CSD2009-00088 (J.M.S.R), BIO2010-16800; “Factoría Española de Cristalización,” Consolider-Ingenio 2010 (J.A.G) from the Spanish Ministry of Economy and Competitiveness, P09-CVI-5073 (B.I.M.) from the “Junta de Andalucía,” FEDER Funds (J.M.S.R., B.I.M., and J.A.G.), DuPont Young Professor Award (E.A.G.), and grants NNX13AI08G and NNX13AI10G (E.A.G.) from NASA Exobiology. We would like to thank the staff at BM30, Ref.Mx1541 (ESRF, Grenoble, France), for support during data collection.
References
- Akanuma S, Nakajima Y, Yokobori S, Kimura M, Nemoto N, Mase M, Miyazono K, Tanokura M, Yamagishi A. Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci U S A. 2013;110:11067–11072. doi: 10.1073/pnas.1308215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O, Gong LI, Bloom JD. Mutational effects on stability are largely conserved during protein evolution. Proc Natl Acad Sci U S A. 2013;110(52):21071–21076. doi: 10.1073/pnas.1314781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroni TE, Wang T, Qian H, Dearth LR, Truong LN, Zeng J, Denes AE, Chen SW, Brachmann RK. A global suppressor motif for p53 cancer mutants. Proc Natl Acad Sci U S A. 2004;101(14):4930–4935. doi: 10.1073/pnas.0401162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Rogovsky H, Hugenmatter A, Tawfik DS. The evolutionary origins of detoxifying enzymes. The mammalian serum paraoxonases (PONs) relate to bacterial homoserine lactonases. J Biol Chem. 2013;288:23914–23927. doi: 10.1074/jbc.M112.427922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Sassi SO, Gaucher EA. Molecular paleoscience: systems biology from the past. Adv Enzymol Relat Areas Mol Biol. 2007;75:1–132. doi: 10.1002/9780471224464.ch1. [DOI] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature. 2006;444:929–932. doi: 10.1038/nature05385. [DOI] [PubMed] [Google Scholar]
- Bloom JD. An experimentally determined evolutionary model dramatically improves phylogenetic fit. Mol Biol Evol. 2014;31:1956–1978. doi: 10.1093/molbev/msu173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Arnold FH, Wilke CO. Breaking proteins with mutations: threads and thresholds in evolution. Mol Syst Biol. 2007;3:76. doi: 10.1038/msb4100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci U S A. 2005;102(3):606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr IM, Robinson JI, Dimitriou R, Markham AF, Morgan AW, Bonthron DT. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics. 2009;25:3244–3250. doi: 10.1093/bioinformatics/btp583. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman G. Conformational parameters of amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Evolution of increased complexity in a molecular machine. Nature. 2012;481:360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451:704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
- Godoy-Ruiz R, Ariza F, Rodriguez-Larrea D, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM. Natural selection for kinetic stability is a likely origin of correlations between mutational effects on protein energetics and frequencies of amino acid occurrences in sequence alignments. J Mol Biol. 2006;362:966–978. doi: 10.1016/j.jmb.2006.07.065. [DOI] [PubMed] [Google Scholar]
- Godoy-Ruiz R, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM. Relation between protein stability, evolution and structure, as probed by carboxylic acid mutations. J Mol Biol. 2004;336:313–318. doi: 10.1016/j.jmb.2003.12.048. [DOI] [PubMed] [Google Scholar]
- Godoy-Ruiz R, Perez-Jimenez R, Ibarra-Molero B, Sanchez-Ruiz JM. A stability pattern of protein hydrophobic mutations that reflects evolutionary structural optimization. Biophys J. 2005;89:3320–3331. doi: 10.1529/biophysj.105.067025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromiha MM, Pathak MC, Saraboji K, Ortlund EA, Gaucher EA. Hydrophobic environment is a key factor for the stability of thermophilic proteins. Proteins. 2013;81:715–721. doi: 10.1002/prot.24232. [DOI] [PubMed] [Google Scholar]
- Halpern AL, Bruno WJ. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol Biol Evol. 1998;15:910–917. doi: 10.1093/oxfordjournals.molbev.a025995. [DOI] [PubMed] [Google Scholar]
- Harper ET, Rose GD. Helix stop signals in proteins and peptides: the capping box. Biochemistry. 1993;32:7605–7669. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. The timetree of life. New York: Oxford University Press; 2009. [Google Scholar]
- Hetch MH, Sauer RT. Phage lambda repressor revertants. Amino acid mutations that restore activity to mutant proteins. J Mol Biol. 1985;186:53–63. doi: 10.1016/0022-2836(85)90256-6. [DOI] [PubMed] [Google Scholar]
- Hobbs JK, Shepherd C, Saul DJ, Demetras NJ, Haaning S, Monk CR, Daniel RM, Arcus VL. On the origin and evolution of thermophily: reconstruction of functional precambrian enzymes from ancestors of Bacillus. Mol Biol Evol. 2012;29:825–835. doi: 10.1093/molbev/msr253. [DOI] [PubMed] [Google Scholar]
- Hoffman PF, Kaufman AJ, Halverson GP, Schrag DP. A neoproteorozoic snowball Earth. Science. 1998;281:1342–1346. doi: 10.1126/science.281.5381.1342. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;254:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Huang W, Palzkill T. A natural polymorphism in beta-lactamase is a global suppressor. Proc Natl Acad Sci U S A. 1997;94(16):8801–8806. doi: 10.1073/pnas.94.16.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles-Prieto A, Ibarra-Molero B, Delgado-Delgado A, Perez-Jimenez R, Fernandez JM, Gaucher EA, Sanchez-Ruiz JM, Gavira A. Conservation of protein structure over four billion years. Structure. 2013;21:1690–1697. doi: 10.1016/j.str.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Cannon BR, Castañeda C, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2008;105(46):17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kather I, Jakob RP, Dobbek H, Schmid FX. Increased folding stability of TEM-1 β-lactamase by in vitro selection. J Mol Biol. 2008;383:238–251. doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- Kim CA, Berg JM. Thermodynamic β-sheet propensities measured using a zinc-finger host peptide. Nature. 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- Kirschvink JL, Gaidos EJ, Bertani LE, Beukes NJ, Gutzmer J, Maepa LN, Steinberger RE. Paleoproteorozoic snowball Earth: extreme climate and geochemical global change and its biological consequences. Proc Natl Acad Sci U S A. 2005;97(4):1400–1405. doi: 10.1073/pnas.97.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauth LP, Lowe DR. High Archean climatic temperature inferred from oxygen isotope geochemistry of cherts in the 3.5 Ga Swaziland Supergroup, South Africa. Geol Soc Am Bull. 2003;115:566–580. [Google Scholar]
- Kratzer JT, Lanaspa MA, Murphy MN, Graves CL, Tipton PA, Ortlund EA, Johnson RJ, Gaucher EA. Evolutionary history and metabolic insights of ancient mammalian uricases. Proc Natl Acad Sci U S A. 2014;111(10):3763–3768. doi: 10.1073/pnas.1320393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:3759–3764. doi: 10.1073/pnas.0308701101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Martin WF. The origin of membrane bioenergetics. Cell. 2013;151:1406–1416. doi: 10.1016/j.cell.2012.11.050. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Phil Trans R Soc Lond B. 2008;363:3965–3976. doi: 10.1098/rstb.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Sauer RT. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989;339:31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Mitraki A, Fane B, Haase-Penttingel C, Sturtevant J, King J. Global suppression of protein foding defects and inclusion body formation. Science. 1991;253:54–58. doi: 10.1126/science.1648264. [DOI] [PubMed] [Google Scholar]
- Myers JK, Pace CN, Scholtz JM. A direct comparison of helix propensity in peptides and proteins. Proc Natl Acad Sci U S A. 1997;94:2833–2837. doi: 10.1073/pnas.94.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova PV, Wong KB, DeDecker B, Henckel J, Fersht AR. Mechanism of rescue of common p53 cancer mutations by second-site suppressor mutations. EMBO J. 2000;19:370–378. doi: 10.1093/emboj/19.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak AM. Evolutionary dynamics: exploring the equations of life. Cambridge (MA): Harvard University Press; 2006. [Google Scholar]
- Ochman H, Elwyn S, Moran NA. Calibrating bacterial evolution. Proc Natl Acad Sci U S A. 1999;96:12638–12643. doi: 10.1073/pnas.96.22.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CM, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1988;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L, Zuckerkandl E. Chemical paleogenetics: molecular “restoration studies” of extinct forms of life. Acta Chem Scan. 1963;17:S9–S16. [Google Scholar]
- Perez-Jimenez R, Inglés-Prieto A, Zhao ZM, Sanchez-Romero I, Alegre-Cebollada J, Kosuri P, Garcia-Manyes S, Kappock TJ, Tanokura M, Holmgren A, et al. Single-molecule paleoenzymology probes the chemistry of resurrected enzymes. Nat Struct Mol Biol. 2011;18:592–596. doi: 10.1038/nsmb.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pey AL, Rodriguez-Larrea D, Gavira JA, Garcia-Moreno B, Sanchez-Ruiz JM. Modulation of buried ionizable groups in proteins with engineered surface charge. J Am Chem Soc. 2010;132:1219–1219. doi: 10.1021/ja909298v. [DOI] [PubMed] [Google Scholar]
- Pollock DD, Goldstein RA. Strong evidence for protein epistasis, weak evidence against it. Proc Natl Acad Sci U S A. 2014;111(15):E1450. doi: 10.1073/pnas.1401112111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock DD, Thiltgen G, Goldstein RA. Amino acid coevolution induces an evolutionary Stokes shift. Proc Natl Acad Sci U S A. 2012;109(21):E1352–E1359. doi: 10.1073/pnas.1120084109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of α helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Gaucher EA, Sanchez-Ruiz JM. Phenotypic comparisons of consensus variants versus laboratory resurrections of Precambrian proteins. Proteins. 2014;82:887–896. doi: 10.1002/prot.24575. [DOI] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian β-lactamases. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- Risso VA, Gavira JA, Sanchez-Ruiz JM. Thermostable and promiscuous Precambrian proteins. Environ Microbiol. 2014;16:1485–1489. doi: 10.1111/1462-2920.12319. [DOI] [PubMed] [Google Scholar]
- Rodrigue N, Philippe H, Lartillot N. Mutation-selection models of coding sequence evolution with site-heterogeneous amino acid fitness profiles. Proc Natl Acad Sci U S A. 2010;107(10):4629–4634. doi: 10.1073/pnas.0910915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salverda ML, De Visser JA, Barlow M. Natural evolution of TEM-1 β-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34:1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- Schellman JA. The thermodynamic stability of proteins. Annu Rev Biophys Biophys Chem. 1987;16:115–137. doi: 10.1146/annurev.bb.16.060187.000555. [DOI] [PubMed] [Google Scholar]
- Serrano L, Day AG, Fersht AR. Step-wise mutation of barnase to binase. A procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J Mol Biol. 1993;233:305–312. doi: 10.1006/jmbi.1993.1508. [DOI] [PubMed] [Google Scholar]
- Shortle D, Lin B. Genetic analysis of staphylococcal nuclease: identification of three intragenic “global” suppressors of nuclease-minus mutations. Genetics. 1985;110:539–555. doi: 10.1093/genetics/110.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikosek T, Chan HS. Biophysics of protein evolution and evolutionary protein biophysics. J R Soc Interface. 2014;11:20140419. doi: 10.1098/rsif.2014.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CK, Withka JM, Regan L. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- Smith JM. Natural selection and the concept of a protein space. Nature. 1970;225:563–564. doi: 10.1038/225563a0. [DOI] [PubMed] [Google Scholar]
- Tamuri AU, dos Reis M, Goldstein RA. Estimating the distribution of selection coefficients from phylogenetic data using sitewise mutation-selection models. Genetics. 2012;190:1101–1115. doi: 10.1534/genetics.111.136432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna DR, Goldstein RA. Why are proteins marginally stable? Proteins. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Stricher F, Schymkowitz J, Serrano L, Tawfik DS. The stability effects of protein mutations appear to be universally distributed. J Mol Biol. 2007;369:1318–1332. doi: 10.1016/j.jmb.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- Voordeckers K, Brown CA, Vanneste K, van der Zande E, Voet A, Maere S, Verstrepen KJ. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 2012;10:e1001446. doi: 10.1371/journal.pbio.1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HC, Li K, Susko E, Roger AJ. A class frequency mixture model that adjusts for site-specific amino acid frequencies and improves inference of protein phylogeny. BMC Evol Biol. 2008;8:331. doi: 10.1186/1471-2148-8-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- Wilson KP, Malcolm BA, Matthews BA. Structural and thermodynamic analysis of compensating mutations within the core of chicken egg white lysozyme. J Biol Chem. 1992;267:10842–10849. [PubMed] [Google Scholar]
- Wyganowski KT, Kallenback M, Tokuriki N. GroEL/ES buffering and compensatory mutations promote protein evolution by stabilizing folding intermediates. J Mol Biol. 2013;425:3403–3414. doi: 10.1016/j.jmb.2013.06.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.