Abstract
RNA editing is a posttranscriptional modification that can lead to a change in the encoded protein sequence of a gene. Although a few cases of mammalian coding RNA editing are known to be functionally important, the vast majority of over 2,000 A-to-I editing sites that have been identified from the coding regions of the human genome are likely nonadaptive, representing tolerable promiscuous targeting of editing enzymes. Finding the potentially tiny fraction of beneficial editing sites from the sea of mostly nearly neutral editing is a difficult but important task. Here, we propose and provide evidence that evolutionarily conserved or “hardwired” residues that experience high-level nonsynonymous RNA editing in a species are enriched with beneficial editing. This simple approach allows the prediction of sites where RNA editing is functionally important. We suggest that priority be given to these candidates in future characterizations of the functional and fitness consequences of RNA editing.
Keywords: editing level, evolution, human, nonsynonymous
RNA editing posttranscriptionally alters RNA sequences through insertion, deletion, or modification of nucleotides, with the exception of some common forms of RNA processing such as splicing, 5′-capping, and 3′-polyadenylation, which are not considered editing (Nishikura 2006; Farajollahi and Maas 2010). RNA editing evolved multiple times in various evolutionary lineages (Gray 2012). When occurring in protein-coding regions, RNA editing may lead to changes in protein sequence, structure, and function. Indeed, the functional importance of RNA editing has been demonstrated in a few cases (Nishikura 2010; Maas 2012). In the last few years, thanks to the genomic revolution, hundreds of thousands of RNA editing sites have been detected from the human genome, including over 2,000 sites in coding regions (Li et al. 2009; Bahn et al. 2012; Kleinman et al. 2012; Park et al. 2012; Peng et al. 2012; Ramaswami et al. 2012, 2013; Chen 2013; Bazak et al. 2014; Sakurai et al. 2014). However, our recent comparison of frequencies and levels of human coding RNA editing among various functional groups of sites suggested that most observed coding RNA editing is nonadaptive, representing tolerable promiscuous targeting of editing enzymes (Xu and Zhang 2014). If advantageous coding RNA editing is rare but present in more than the few known cases, how do we identify the other potentially beneficial cases from the sea of mostly nearly neutral editing? We focus on coding RNA editing because it is better studied and is likely more important than noncoding RNA editing. Here, we propose a simple method based on evolutionary principles and demonstrate its validity.
The rationale of our method is as follows. If RNA editing at a site is functionally important and beneficial, its editing level (i.e., fraction of RNA molecules edited) should be relatively high, because a higher level of editing at the site likely confers a higher fitness. Furthermore, beneficial editing is expected to be enriched at functionally constrained sites, compared with neutral editing, where a high editing level is selectively permitted but not advantageous. Because functional constraint implies evolutionary conservation (Kimura 1983), we expect beneficial editing sites to be evolutionarily relatively conserved. Two types of conservation are possible here. The first type is the among-species conservation of the pre-edited version, which would ensure editing. We refer to this type of editing sites as “conserved” sites. If only the postedited version is fully functional, it is possible that a genome directly encodes the postedited version. In other words, when the genomes of multiple species are examined, we will observe that some have the pre-edited version whereas others have the postedited version. Such sites are said to belong to the “hardwired” type. In short, we hypothesize that conserved or hardwired sites with high levels of RNA editing tend to be beneficial.
To test the above hypothesis, we should compare a set of beneficial editing sites with other editing sites. But, because only a few beneficial editing sites have been experimentally confirmed, we resort to a recently published list of shared RNA editing between genome-wide catalogs of editing sites found in human and mouse (Pinto et al. 2014). Under the presumption that RNA editing shared between human and mouse is likely to be functionally important and beneficial (see below), we can test our hypothesis by comparing the properties of these shared editing sites with those of other editing sites.
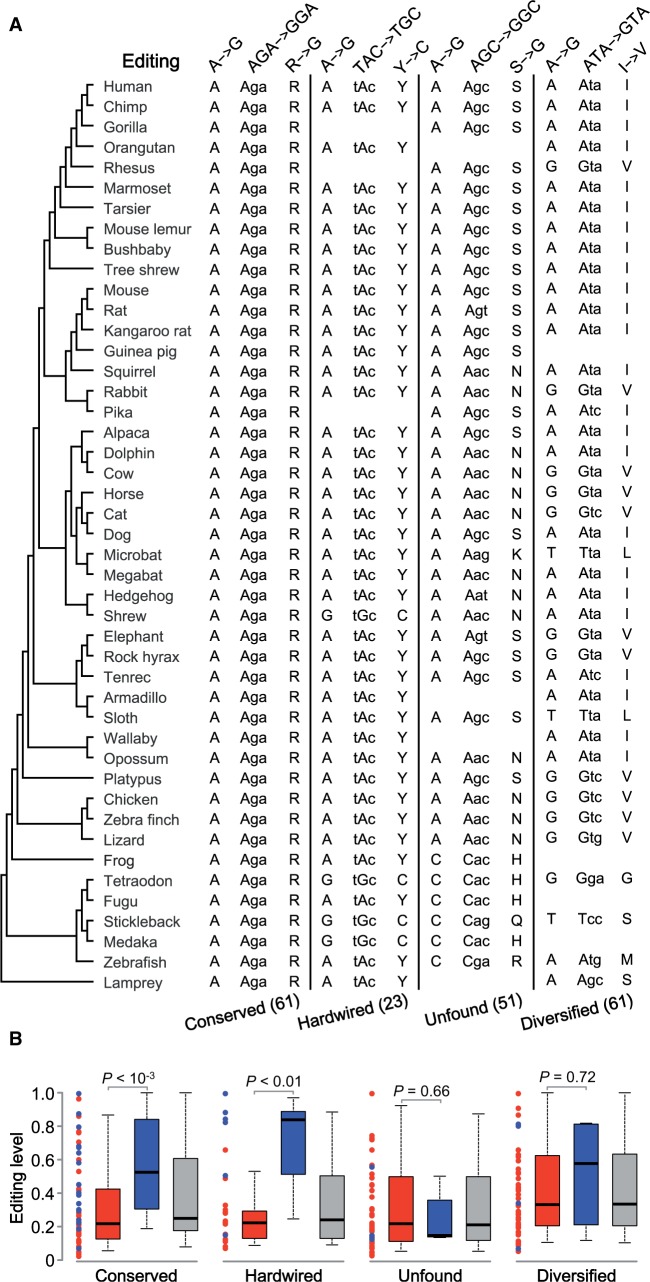
In animals, the predominant type of RNA editing is the hydrolytic deamination of adenosine (A) to inosine (I), catalyzed by adenosine deaminases acting on RNAs (Nishikura 2006). Because I is recognized as guanosine (G) by the translation machinery, this editing is also known as A-to-G editing (Nishikura 2006). We focus on A-to-G editing in this study, because only this type has sufficiently large data for statistically meaningful analysis. By comparing 1,432,743 human and 10,210 mouse A-to-G editing sites, most of which are located in lineage-specific, inverted repeats, Pinto et al. (2014) identified 58 human–mouse shared editing sites, 34 of which are in coding regions and will lead to nonsynonymous changes when edited. To compare these shared editing sites with other editing sites, we took advantage of a recently assembled list of 1,783 coding A-to-G editing sites identified from the human genome (Xu and Zhang 2014). We augmented this list with three additional data sets (see Materials and Methods), resulting in the total number of coding A-to-G editing sites being 2,042, including 679 synonymous and 1,363 nonsynonymous editing sites (supplementary data set S1, Supplementary Material online). Following Xu and Zhang (2014), for each nonsynonymous editing site, we retrieved the orthologous nucleotides and corresponding codons from the genome sequences of 44 nonhuman vertebrate species. We also compiled editing level information from human for these sites. If a site appeared in multiple data sets or tissues, the highest editing level reported was used. A total of 196 editing sites have editing level information and sufficient phylogenetic coverage (supplementary data set S2, Supplementary Material online). They were classified into 61 “conserved,” 23 “hardwired,” 51 “unfound,” and 61 “diversified” sites, according to the phylogenetic variations of the encoded amino acids (fig. 1A). As mentioned, at each conserved site, only the human genome-encoded amino acid is present in any species examined. At each hardwired site, either the human genome-encoded amino acid or the human edited amino acid is observed in each species. At each unfound site, the human edited amino acid is not found in the genome of any species, but the human pre-edited amino acid and at least another amino acid are found. At each diversified site, the human pre-edited, edited, and at least another amino acid are found in nonhuman species.
Fig. 1.
Human nonsynonymous A-to-G editing sites with conserved or hardwired phylogenetic patterns and ≥30% editing levels are enriched with human–mouse shared editing. (A) Editing sites are classified into four groups based on the evolutionary variations among human and 44 other vertebrate species. The observed nucleotides and corresponding codons and amino acids at each site are shown for one example of each group, with the number of identified cases in each group provided in the parentheses. The four listed examples are GRIA4 (edited position chr11.105804694), GRIK2 (chr6.102337702), AZIN1 (chr8.103841636), and MYH1 (chr17.10400445). The tree topology follows http://hgdownload.cse.ucsc.edu/goldenpath/hg19/phyloP46way/, last accessed November 15, 2014, and the branches are not drawn to scale. Our analysis does not depend on the accuracy of the tree. (B) Comparison in human editing level between human–mouse shared editing sites and other editing sites. Each dot represents a human editing site, with human–mouse shared editing in blue and others in red. The boxes show the human editing level distributions for shared editing (blue), other editing (red), and all editing sites (gray). The values of upper quartile, median, and lower quartile are indicated in each box, whereas the bars outside the box show the 5th and 95th percentiles. The P values are from two-tailed Mann–Whitney U test.
All 34 human–mouse shared nonsynonymous editing sites are on our list of 1,363 human nonsynonymous editing sites and within the 196 sites that have sufficient phylogenetic coverage. Apparently, the criterion of having sufficient phylogenetic coverage already enriches human–mouse shared editing, because many human editing sites are so unimportant that they either have no identifiable orthologous sites in many other species or their orthologous sites in many other species are noncoding.
Among the 196 human editing sites with sufficient phylogenetic coverage, 32.8% (20/61) of conserved sites, 26.1% (6/23) of hardwired sites, 7.8% (4/51) of unfound sites, and 6.6% (4/61) of diversified sites show shared editing between human and mouse (table 1). Consistent with our hypothesis, the fraction of sites with shared editing is significantly greater among the conserved and hardwired sites (31%) than among the unfound and diversified sites (7.1%) (P = 1.7 × 10−5, Fisher’s exact test). Also consistent with our hypothesis, the editing levels of the human–mouse shared editing sites are significantly higher than those of unshared editing sites in the conserved (P = 7.4 × 10−4, Mann–Whitney U test) and hardwired (P = 3.3 × 10−3) groups, but not in the unfound (P = 0.66) and diversified (P = 0.72) groups (fig. 1B).
Table 1.
Fractions of Sites with Shared Nonsynonymous RNA Editing between Human and Mouse.
| Conserved Type |
Hardwired Type |
Unfound Type |
Diversified Type |
|||||
|---|---|---|---|---|---|---|---|---|
| Editing level (%) | >0 | ≥30 | >0 | ≥30 | >0 | ≥30 | >0 | ≥30 |
| Number of editing sites | 61 | 27 | 23 | 9 | 51 | 20 | 61 | 33 |
| Number of sites with shared editing | 20 | 16 | 6 | 5 | 4 | 1 | 4 | 3 |
| Fraction of sites with shared editing | 0.328 | 0.593 | 0.261 | 0.556 | 0.078 | 0.050 | 0.066 | 0.091 |
Given the small fraction of coding RNA editing that is functionally important, the precision of our prediction is quite high. Among conserved and hardwired editing sites with ≥30% editing levels, 58.3% (21/36) are human–mouse shared editing (table 1). This is a 3.4-fold enrichment of shared editing (P < 10−6; Fisher’s exact test) compared with the 196 sites analyzed (34/196 = 17.3%), and a 23.4-fold enrichment (P < 10−21) compared with the 1,363 human nonsynonymous editing sites (34/1,363 = 2.5%).
Previous functional studies demonstrated the importance of human coding RNA editing at a small number of sites (Maas 2012), including chr12.5021742 in KCNA1; chr4.158281294 and chr4.158257875 in GRIA2; chrX.122598962 in GRIA3; chr11.105804694 in GRIA4; chr21.30953750 in GRIK1; chrX.151358319 in GABRA3; chr6.102337689, chr6.102337702, and chr6.102372589 in GRIK2; and chrX.114082682, chrX.114082684, chrX.114082688, chrX.114082689, and chrX.114082694 in HTR2C. Among these 15 sites, only 2 (chrX.114082688 and chrX.114082689) in HTR2C could not pass our criteria, because chrX.114082688 has a less than 30% editing level and chrX.114082689 belongs to the unfound group. The remaining 13 sites have editing levels higher than 50% and belong to either the conserved or the hardwired group.
One criterion of our method is that the edited site should be conserved or hardwired, based on the phylogenetic variation of the genome-encoded amino acid in 45 vertebrate species. Previous authors classify an editing site as conserved or hardwired based on whether it has A or G across species rather than based on the encoded amino acid (Nishikura 2010). If we had used this definition, among conserved and hardwired sites with ≥30% editing in human, only 34.8% (16/46) would show human–mouse shared editing, significantly lower than what our method achieves (58.3%, P = 0.04, Fisher’s exact test). This comparison demonstrates that it is more informative to classify an editing site based on the phylogenetic variation of the encoded amino acid rather than nucleotide. This is hardly surprising, because the function of a coding site is more directly determined by its amino acid rather than nucleotide. We also examined the receiver operating characteristic curve of our method (supplementary fig. S1, Supplementary Material online) and found that the use of 30% editing level as the cutoff ensures a sufficiently high true positive rate without producing many false positives.
One potential weakness of our method is that the classification of an editing site to one of the four groups depends on the number and specific genome sequences available for phylogenetic survey. Obviously, when the number of genome sequences surveyed increases, the probability that a site belongs to the conserved group will decrease, whereas the probability that it belongs to the hardwired group may increase or decrease. To examine how sensitive the classification is to the number of genomes surveyed, we randomly removed 10 of the 44 nonhuman genomes used in figure 1 and repeated this experiment 10,000 times. We found that the total number of conserved and hardwired sites increased slightly but not significantly upon the removal of ten genomes (supplementary fig. S2, Supplementary Material online), suggesting that our method is relatively robust to a moderate (10/44 = 23%) change in the number of genomes surveyed. In the future, it will be important to develop a quantitative phylogenetic score rather than the qualitative classification to assist the prediction of beneficial RNA editing.
We found that 38.2% (13/34) of human–mouse shared nonsynonymous editing sites reside in essential genes (essentiality based on mouse orthologs at http://ogeedb.embl.de, last accessed November 15, 2014), significantly greater than the fraction (140/1,329 = 10.5%) of unshared human nonsynonymous editing sites that reside in essential genes (P = 3.1 × 10−5; Fisher’s exact test). Thus, localization of an editing site in an essential gene can also be used to improve the prediction of beneficial editing. However, we decide not to adopt this criterion, because genome-wide gene essentiality information is available only for a few species and gene essentiality is not always transferable between species (Liao and Zhang 2008); relying on this criterion would thus limit the utility of our method.
We assumed that human–mouse shared nonsynonymous RNA editing is beneficial. Indeed, there are nshared = 34 shared A-to-G nonsynonymous editing sites but only sshared = 3 shared A-to-G synonymous editing sites (Pinto et al. 2014). There are Nshared = 947,261 A sites shared between human and mouse genomes that would have nonsynonymous changes if edited to G, and Sshared = 258,138 shared A sites that would have synonymous changes if edited to G. Hence, the frequency of human–mouse shared nonsynonymous editing is nshared/Nshared = 3.6 × 10−5, whereas that of shared synonymous editing is sshared/Sshared = 1.2 × 10−5; the former is significantly greater than the latter (P = 0.029, Fisher’s exact test). That human–mouse shared nonsynonymous editing is significantly more frequent than shared synonymous editing is in sharp contrast to the previous finding that human nonsynonymous editing in general is significantly less frequent than synonymous editing and supports our assumption that human–mouse shared nonsynonymous editing is likely beneficial. Our observation also suggests that human–mouse shared nonsynonymous editing is unlikely an inevitable consequence of sequence conservation due to processes unrelated to editing (e.g., RNA folding and transcription factor binding), because these processes should not preferentially constrain nonsynonymous editing sites.
By assuming functional importance of nonsynonymous editing shared between human and mouse, we demonstrated an over 20-fold enrichment of beneficial editing among human nonsynonymous editing sites that have both high editing levels ( ≥30%) and conserved or hardwired phylogenetic patterns. Our method makes it possible to predict beneficial RNA editing in the lack of suitable comparative data of RNA editing. For instance, RNA editing has been extensively surveyed in the model organism Drosophila melanogaster (Stapleton et al. 2006; Graveley et al. 2011; Rodriguez et al. 2012; Ramaswami et al. 2013; St Laurent et al. 2013) but not in other arthropods. Nevertheless, one can predict beneficial editing in D. melanogaster using the two criteria established above, because of the availability of multiple arthropod genome sequences. Even in the present case of mammalian RNA editing, not many tissues have been surveyed in nonhuman species, which has likely resulted in some false negatives in the list of human–mouse shared editing sites. Consequently, some of the human editing sites that pass the two criteria but are not on the current list of shared editing sites may turn out to be shared and beneficial. In other words, our method is also useful even when comparative data are available but are limited in tissue coverage. Because generating RNA editing data from many tissues in multiple species is expensive and time-consuming whereas genome sequences of many species are already available, our method has advantages over the use of shared editing sites in predicting beneficial editing.
We demonstrated that a human nonsynonymous editing site passing the two established criteria has a reasonably high probability (58.3%) to exhibit shared editing with mouse. As aforementioned, 58.3% is likely an underestimate due to the false negatives in the current list of shared editing sites. Although applying the two criteria also filtered out 38% (13/34) of shared editing sites, this is less of a concern, because at this stage the main task is to identify functionally important RNA editing sites rather than identify them comprehensively. Thus, we propose that our method be used to predict beneficial editing sites for functional verification (Li and Church 2013), which has become feasible in a variety of species at a relatively large scale, thanks to the rapid progress in genome engineering technologies (Hsu et al. 2014). The accumulation of functionally validated beneficial editing cases will allow a mechanistic understanding of why RNA editing is advantageous in these cases and how it originated in evolution.
Materials and Methods
We first collected RNA editing data from three recently published studies (Chen 2013; Bazak et al. 2014; Sakurai et al. 2014) to augment our list of human coding A-to-G editing sites (Xu and Zhang 2014). Note that editing sites identified from cancer cell lines (Chen 2013) were excluded. For data sets with no annotation of synonymous or nonsynonymous editing (Chen 2013; Bazak et al. 2014), we used ANNOVAR (Wang et al. 2010) to annotate each site. As defined previously (Xu and Zhang 2014), a synonymous (or nonsynonymous) editing site is a site where A-to-G editing causes a synonymous (or nonsynonymous) change. We then retrieved for each nonsynonymous editing site the codon in which an editing site resides and the homologous codons in 44 other vertebrate species from pairwise genomic alignments in the Ensembl database (version 75) (Flicek et al. 2013) using Ensembl Perl API. For a human gene that has multiple homologous genes in another species, the pairwise alignment was generated using the one with the highest similarity to the human gene. Only sites with enough phylogenetic variation information and human editing level information were retained for further analyses. According to our definition, as long as the homologous codons encode for at least one more type of amino acid besides the human pre-edited and edited amino acids, we classify this site as diversified. For the other three groups, we require representatives from at least two different orders in addition to primates. Editing level information was collected from various data sets of previous studies (Li et al. 2009; Kleinman et al. 2012; Park et al. 2012; Peng et al. 2012; Ramaswami et al. 2012, 2013; Chen 2013; Bazak et al. 2014; Sakurai et al. 2014). If the same site appeared in multiple data sets or various tissues, the highest reported editing level was used in subsequent analysis. A human gene is considered essential if its one-to-one mouse ortholog is essential, and the list of essential genes was acquired from a recent study (Xu and Zhang 2014). To count the number (Nshared) of human–mouse shared A sites that would be nonsynonymous if edited to G and the number (Sshared) that would be synonymous if edited to G, we obtained ENSEMBL gene IDs for all 15,182 human–mouse one-to-one orthologous genes through Ensembl BioMart. We used the principal transcript of each gene defined by the APPRIS system (http://appris.bioinfo.cnio.es, last accessed November 15, 2014) to generate a pairwise protein sequence alignment, based on which the coding DNA sequence alignment was created. Using these alignments, we obtained Nshared and Sshared by custom R scripts.
Supplementary Material
Supplementary data sets S1 and S2 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Chuan Li, Jian-Rong Yang, and an anonymous reviewer for valuable comments. This work was supported in part by research grants from the US National Institutes of Health (R01GM103232) to J.Z. and National Natural Science Foundation of China (31100170) to G.X.
References
- Bahn JH, Lee JH, Li G, Greer C, Peng GD, Xiao XS. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 2012;22:142–150. doi: 10.1101/gr.124107.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Characterization and comparison of human nuclear and cytosolic editomes. Proc Natl Acad Sci U S A. 2013;110:E2741–E2747. doi: 10.1073/pnas.1218884110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. Evolutionary origin of RNA editing. Biochemistry. 2012;51:5235–5242. doi: 10.1021/bi300419r. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kleinman CL, Adoue V, Majewski J. RNA editing of protein sequences: a rare event in human transcriptomes. RNA. 2012;18:1586–1596. doi: 10.1261/rna.033233.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 2013;16:1518–1522. doi: 10.1038/nn.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. Posttranscriptional recoding by RNA editing. Adv Protein Chem Struct Biol. 2012;86:193–224. doi: 10.1016/B978-0-12-386497-0.00006-2. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Williams B, Wold BJ, Mortazavi A. RNA editing in the human ENCODE RNA-seq data. Genome Res. 2012;22:1626–1633. doi: 10.1101/gr.134957.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Pinto Y, Cohen HY, Levanon EY. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014;15:R5. doi: 10.1186/gb-2014-15-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O’Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Menet JS, Rosbash M. Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Mol Cell. 2012;47:27–37. doi: 10.1016/j.molcel.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai M, Ueda H, Yano T, Okada S, Terajima H, Mitsuyama T, Toyoda A, Fujiyama A, Kawabata H, Suzuki T. A biochemical landscape of A-to-I RNA editing in the human brain transcriptome. Genome Res. 2014;24:522–534. doi: 10.1101/gr.162537.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Tackett MR, Nechkin S, Shtokalo D, Antonets D, Savva YA, Maloney R, Kapranov P, Lawrence CE, Reenan RA. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 2013;20:1333–1339. doi: 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
- Stapleton M, Carlson JW, Celniker SE. RNA editing in Drosophila melanogaster: new targets and functional consequences. RNA. 2006;12:1922–1932. doi: 10.1261/rna.254306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li MY, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Zhang J. Human coding RNA editing is generally nonadaptive. Proc Natl Acad Sci U S A. 2014;111:3769–3774. doi: 10.1073/pnas.1321745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.