Abstract
Recombination rates vary in intensity and location at the species, individual, sex and chromosome levels. Despite the fundamental biological importance of this process, the selective forces that operate to shape recombination rate and patterns are unclear. Domestication offers a unique opportunity to study the interplay between recombination and selection. In domesticates, intense selection for particular traits is imposed on small populations over many generations, resulting in organisms that differ, sometimes dramatically, in morphology and physiology from their wild ancestor. Although earlier studies suggested increased recombination rate in domesticates, a formal comparison of recombination rates between domestic mammals and their wild congeners was missing. In order to determine broad-scale recombination rate, we used immunolabeling detection of MLH1 foci as crossover markers in spermatocytes in three pairs of closely related wild and domestic species (dog and wolf, goat and ibex, and sheep and mouflon). In the three pairs, and contrary to previous suggestions, our data show that contemporary recombination rate is higher in the wild species. Subsequently, we inferred recombination breakpoints in sequence data for 16 genomic regions in dogs and wolves, each containing a locus associated with a dog phenotype potentially under selection during domestication. No difference in the number and distribution of recombination breakpoints was found between dogs and wolves. We conclude that our data indicate that strong directional selection did not result in changes in recombination in domestic mammals, and that both upper and lower bounds for crossover rates may be tightly regulated.
Keywords: Canis, Capra, immunolocalization, MLH1, Ovis, genomics, spermatocytes
Introduction
In the last few years, significant progress has been made in the understanding of recombination. This process of fundamental biological and evolutionary importance contributes to the proper disjunction of homologous chromosomes during the first meiotic division in many eukaryotes, and influences genomic architecture through allele shuffling and genome rearrangements. At the molecular level, many of the proteins involved have been identified and analyzed (Page and Hawley 2003; Tease and Hultén 2004; Baudat et al. 2013), and considerable variation has been found in recombination location and rate within and across individuals. However, the selective forces that might be important in shaping recombination rate and patterns are still unclear. Although the physiological and mechanistic constraints that operate at the molecular level to ensure the proper disjunction of chromosomes condition recombination, observations related to the intraspecific and interspecific heterogeneity in recombination rate and patterns are key to understand the selective pressures that may affect recombination over different genomic scales. Selection may operate to ensure the proper disjunction of chromosomes and thus reduce the rate of aneuploidy, to maintain genome integrity by lowering recombination rate locally where harmful effects such as changes in gene dosage and missense mutations may be the outcome, and by acting on recombination modifiers that may increase or decrease recombination rate and thus the degree of association between loci (Coop and Przeworski 2007).
Heterogeneity in recombination rate and patterns is observed at many levels. Recombination preferentially occurs, at least in certain organisms, in localized regions of the genome termed recombination hotspots (Arnheim et al. 2003), which seem to be ubiquitous in mammals (Kauppi et al. 2004). In humans, recombination mostly occurs in regions 1–2 kb long, 60–200 kb apart, where recombination rates can be 10–1,000 times higher than in surrounding areas (Kauppi et al. 2004; Coop and Przeworski 2007). Hotspot location is associated with a consensus sequence in humans (Myers et al. 2008) and with a different one in yeast (Steiner and Smith 2005), and recombination rate is correlated with nucleotide diversity and GC-content and increases from the centromere to the telomere in many organisms, including, for example, yeast, rodents, and humans (Kauppi et al. 2004; Coop and Przeworski 2007). There are also differences in recombination rate and location associated to the sexes (Lenormand and Dutheil 2005; Coop and Przeworski 2007); for example, in fish and most eutherian mammals studied, females have longer genetic maps, while in other instances the opposite is true or there are no differences between the sexes (Hansson et al. 2005; Calderón and Pigozzi 2006; Poissant et al. 2010; Samollow 2010; van Oers et al. 2014). Across species, recombination rates examined in 5 Mb orthologous regions of mouse, rat, and human were found to be weakly correlated (Jensen-Seaman et al. 2004), and humans and chimpanzees do not share hotspot locations, suggesting that their location evolved over timescales that are shorter than the separation of these two species (Winckler et al. 2005; Auton et al. 2012), about 5–6 Ma (Patterson et al. 2006), despite 99% identity at the sequence level. Several studies indicated that recombination rate is heritable (Charlesworth and Charlesworth 1985; Kong et al. 2002; Dumont et al. 2009; Tortereau et al. 2012) and recent research has shown that variation in the zinc-finger domain of a protein called PRDM9 plays a role in the localization of recombination hotspots in humans and mice (Baudat et al. 2010; Berg et al. 2010, 2011; Brick et al. 2012).
An extensive body of theoretical work has been dedicated to identify the conditions under which changes in recombination rate may be beneficial, and thus spread, in a population. The key idea is that recombination breaks up the association between loci, thus contributing to genetic diversity through the creation of new combinations of alleles that may result in novel phenotypes, or in new epistatic interactions, which will affect the organism's fitness and ability to respond to selection. Most successful and realistic explanatory models revolve around the idea of the presence of modifier loci that alter the frequency of recombination (Otto and Lenormand 2002). For example, a genetic modifier that increases recombination, even if it would decrease mean fitness in the short term, may be advantageous if it increases the variance in fitness, which would lead to an increased ability to respond to selection (Otto and Lenormand 2002; Butlin 2005). Higher recombination rate would be advantageous in small populations subject to strong selection due to Hill–Robertson interference, in a situation of weak negative epistasis between loci, or in spatially heterogeneous habitats when alleles are selected in the same direction in each population (either beneficial or deleterious in all populations), but with effects that covary negatively across populations (e.g., for loci A and B, selection is stronger in habitat 1 for locus A and in habitat 2 for locus B) (reviewed in Otto and Barton 2001; Otto and Lenormand 2002; Ross-Ibarra 2004; Butlin 2005; Coop and Przeworski 2007; Lenormand T, personal communication).
Domesticates offer a unique opportunity to study the interplay between recombination and selection. Domestication can be viewed as a long-term experiment in which animals and plants are subjected to intense selection for particular traits, in small populations and during thousands of generations, resulting in individuals that may differ, sometimes dramatically, in morphology and physiology from those in other populations and from their wild ancestors. It has been hypothesized that recombination played a key role in this process (reviewed in e.g., Ross-Ibarra 2004; Butlin 2005). According to Ross-Ibarra (2004), Rees and Dale (1974) proposed an increase in recombination rate in domestic species as a result of the selective forces imposed, whereas Gornall (1983) also expected a higher recombination frequency in domestic species, but hypothesized that high recombination rate would predate domestication, as higher recombination rate would increase response to selection and thus contribute to the success in domestication. Recombination would be particularly beneficial when genetic variability is limited by linkage disequilibrium (LD) subsequent to extensive population bottlenecks and drift (reviewed in Ross-Ibarra 2004). In small populations subject to strong selection, such as is the case for domestic species, simulations showed that high recombination would be beneficial (Otto and Barton 2001) and, in laboratory experiments, recombination increased in small animal populations subjected to strong selection for an unrelated trait (reviewed in Otto and Barton 2001; Otto and Lenormand 2002; Bell 2008). Increased recombination would be advantageous in breaking up the random association of alleles generated by drift, thus reducing Hill–Robertson interference (Otto and Lenormand 2002). However, a different hypothesis proposed a reduced recombination in domesticates in order to protect from maladaptive gene flow from wild relatives, such as between loci that were positively selected in domestic species (Lenormand and Otto 2000).
The wealth of theoretical studies dealing with the conditions for the evolution of recombination rates in small populations and domesticates contrasts with the few empirical studies. Ross-Ibarra (2004) used the number of chiasmata as a proxy for recombination rate and compared chiasma frequencies for 196 plants (including domesticated species and their wild progenitors and congeners); no support was found for the preadaptation hypothesis (comparing wild progenitors and congeners), and only a modest increase in recombination rate in domesticates as compared with wild congeners. The study concluded that “recombination rate is likely of little importance” in relation to plant domestication (Ross-Ibarra 2004). The study of Burt and Bell (1987), in which chiasmata counts for domestic mammals are reported, is often cited as evidence that domestic animals have higher recombination rate than their wild counterparts (see Schmidt-Hempel and Jokela 2002; Dumont and Payseur 2008; Groenen et al. 2009; Backström et al. 2010; Poissant et al. 2010; Smukowski and Noor 2011). However, domestic species were not compared with wild relatives in this study. A recent study has demonstrated a strong phylogenetic effect in recombination rate (Dumont and Payseur 2008), and a comparison of changes in the rates of recombination should take this effect into account. Additionally, in a study comparing linkage maps, the assumption of higher recombination rate in domesticates was found not to be supported in insects (Wilfert et al. 2007). Therefore, the evidence for an increase in recombination rate in domestic animals is inconclusive.
Here, we measured recombination rate in domestic mammals and their wild counterparts using cytogenetic approaches. We counted the number of MLH1 foci, a marker of crossover sites (e.g., Lynn et al. 2004), along the synaptonemal complex in spermatocyte spreads of domestic mammals and wild relatives: dogs (Canis familiaris) and gray wolves (Canis lupus), goats (Capra hircus) and ibexes (Capra pyrenaica), and sheep (Ovis aries) and mouflons (Ovis musimon). MLH1 is a mismatch repair protein that is recruited to crossover sites during the pachytene stage of prophase I.
Counting MLH1 foci provides an estimate of broad-scale contemporary recombination rate, but it is not informative of recombination rate throughout the population history or the fine-scale location of recombination breakpoints. Statistical advances in coalescent modeling of genome-wide polymorphism data allow the estimation of fine-scale recombination rates averaged over many generations (Stumpf and McVean 2003; McVean et al. 2004; Auton and McVean 2007). Artificial selection may have favored individuals with increased recombination around genes associated with distinct phenotypes, so that these genes would be decoupled from surrounding regions, making artificial selection more efficient.
The association of recombination with genes associated with distinct phenotypes can be investigated in dogs. The dog and the gray wolf represent an interesting and valuable system to study the evolution of recombination at the fine scale. The dog has been subjected to intense artificial selection and it is the most phenotypically diverse mammal. The morphological (Wayne 1986a,b), physiological, and behavioral (Coppinger and Coppinger 2001) variation present among dogs is greater than across the entire family Canidae, which includes 36 species such as raccoon dog, foxes, wolves, jackals, and coyotes, which have evolved over about 15 My. A wealth of genetic resources exist for the dog and, in terms of recombination, Burt and Bell (1987) reported that the dog had the highest number of chiasmata among all the mammals included in their study. Moreover, canids are the only known eutherian mammals to carry a Prdm9 which has acquired disruptive mutations (Muñoz-Fuentes et al. 2011; Ponting 2011; Axelsson et al. 2012) (a marsupial, the opossum Monodelphis domestica, has a Prdm9 which lacks zinc fingers; Ponting 2011). In order to investigate this hypothesis, that is, whether artificial selection favored individuals with increased recombination around genes associated with distinct phenotypes, we selected 16 genomic regions associated with phenotypic characters that are candidates to have been selected during dog domestication (e.g., body size, coat type, color; table 1), potentially also by early breeders, as archaeological remains suggest for skeletal and size differences (Clutton-Brock 1999). We then used sequence data to test the hypothesis that increased recombination had been favored in these regions in dogs as compared with wolves. Therefore, in addition to the cytological techniques mentioned above to study contemporary patterns of broad-scale recombination rate in three domestic mammals and their wild congeners, we also investigated patterns of fine-scale recombination and the distribution of recombination breakpoints in both dogs and wolves around loci underlying potentially selected phenotypes in dogs.
Table 1.
Chromosomal Regions Studied, Each Containing Centrally a Locus Associated to a Distinct Phenotypic Character (morphological or behavioral) in Dogs.
| Chr | Target Gene | Trait | Start | End |
|---|---|---|---|---|
| 1 | MC2R, C18orf1 | Herding | 27,318,228 | 27,521,819 |
| 9 | STAT3 | Neck ratio | 23,799,353 | 24,030,794 |
| 10 | SILV = PMEL | Merle coat | 3,181,426 | 3,381,426 |
| 12 | Runx2 | Dorsoventral nose bend and midface length | 16,637,470 | 16,946,083 |
| 13 | RSPO2 | Furnishings | 11,484,766 | 11,784,766 |
| 15 | IGF-1 | Size | 44,115,824 | 44,381,171 |
| 16a | LMBR1 | “Dewclaw” or hind-limb-specificpreaxial polydactyly | 22,154,874 | 22,467,109 |
| 16b | K locus | Black coat | 61,752,782 | 62,052,782 |
| 17 | FOXI3 | Lack of hair | 40,996,789 | 41,197,329 |
| 18a | fgf4 | Short legs | 23,331,125 | 23,531,212 |
| 18b | FGF3 FGF4 FGF19 | Hair ridge | 51,298,518 | 51,631,941 |
| 20 | M promoter of MITF | White spotting | 24,747,309 | 25,049,039 |
| 22 | PCDH9 | Boldness | 25,055,041 | 25,259,603 |
| 25 | MLPH | Dilute coat | 51,044,488 | 51,244,488 |
| 27 | KRT71 | Curly coat | 5,442,806 | 5,642,806 |
| 32 | FGF5 | Coat length | 7,373,337 | 7,573,337 |
Note.—Start and end refer to the CanFam2 assembly coordinates.
Results
Cytological Estimates of Contemporary Recombination Rate
We estimated the number of genome-wide crossover events by counting the number of MLH1 foci along synaptonemal complexes in spermatocytes. We collected testes, and obtained good quality cell preparations for 6 dogs, 2 wolves, 6 goats, 6 ibexes, 6 sheep, and 5 mouflons (supplementary table S1, Supplementary Material online). We used fluorescently labeled antibodies to mark MLH1 and chromosome axes (table 2 and fig. 1). Experiments were also carried out on pig (Sus domesticus) and wild boar (Sus. scrofa) samples, but MLH1 labeling failed, which suggests that the antibodies (three different ones were tested) did not recognize pig and wild boar MLH1 proteins. Although MLH1 is well conserved across mammals, some key differences exist, and it is possible that the antibodies we used to detect MLH1 recognize an immunogenic peptide (or several) that is absent or is different in pig and boar.
Table 2.
Mean Number of MLH1 Foci per Cell and Standard Deviation (SD) Calculated over the Individual Means and the Estimated Number of Crossovers (COs) per Chromosome Pair.
| Species | N | n | Mean Number of MLH1 Foci/Cell | SD | Chromosome Pairs | COs/Chromosome Pair | Chromosome Type | COs/Chromosome Arm |
|---|---|---|---|---|---|---|---|---|
| Dog | 6 | 184 | 38.89 | 0.87 | 39 | 1.00 | Acrocentrica | 1.00 |
| Wolf | 2 | 45 | 40.94 | 1.61 | 39 | 1.05 | Acrocentrica | 1.05 |
| Goat | 6 | 109 | 61.24 | 4.03 | 30 | 2.04 | Metacentric | 1.02 |
| Ibex | 6 | 119 | 64.74 | 1.08 | 30 | 2.16 | Metacentric | 1.08 |
| Sheep | 6 | 125 | 63.47 | 3.42 | 27 | 2.35 | Metacentric | 1.18 |
| Mouflon | 5 | 113 | 69.03 | 2.49 | 27 | 2.56 | Metacentric | 1.28 |
Note.—N, number of individuals; n, total number of spermatocytes for which MLH1 was counted. Means calculated over all measurements were almost identical (not shown).
aAll autosomes are acrocentric.
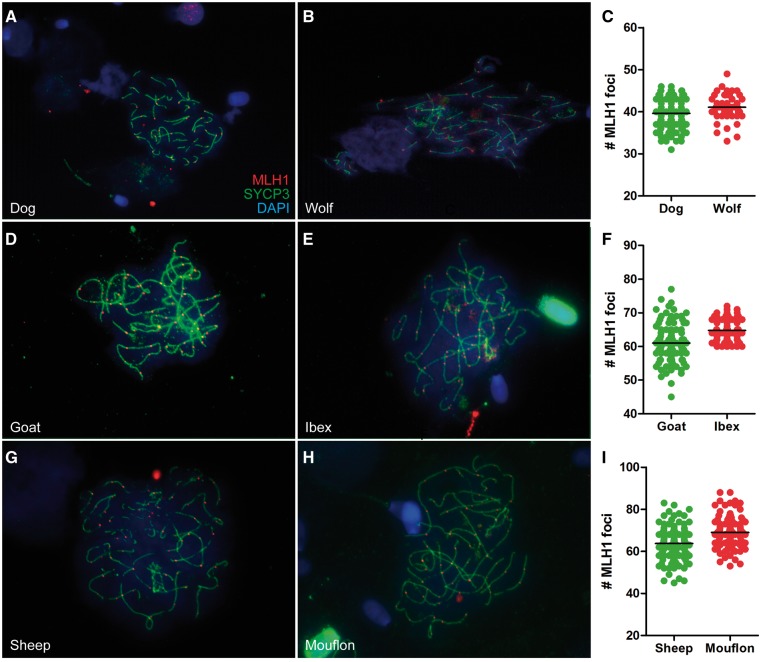
Fig. 1.
Spermatocytes from wild mammals have more crossover markers than cells from domestic mammals. Spermatocytes from dog (A), wolf (B), goat (D), ibex (E), sheep (G), and mouflon (H) immunostained against the crossover marker MLH1 (red) and the synaptonemal complex protein SYCP3 (green), which allows to visualize chromosome axes. DNA is counter-stained with DAPI (blue). Scatterplots display the total number of MLH1 foci found in each spermatocyte, and black lines represent the average number of foci found in each species (C, F, and I). Domestic species, green; wild species, red.
Dog spermatocytes contained on average 38.89 ± 0.87 MLH1 foci per cell (mean ± SD calculated over the individual means, table 2) (fig. 1A–C), whereas wolf spermatocytes contained on average 40.94 ± 1.61 MLH1 foci per cell (fig. 1B and C). We used generalized linear-mixed models to account for mixed effects (species as a fixed factor and individual as a random factor) on the number of MLH1 foci, and found that species was a significant factor explaining the variation in the data (P = 0.024). Similarly, analyses of goats and ibexes showed that the average number of foci per cell was higher in the wild species (61.24 ± 4.03 for goat, 64.74 ± 1.08 for ibex; fig. 1D–F), and again species was significant at explaining the variation (P = 0.037). Likewise, sheep and mouflons yielded an average number of MLH1 foci higher in the wild species (63.47 ± 3.42 for sheep, 69.03 ± 2.49 for mouflon; fig. 1G–I), and again species significantly explained the variation present in the data (P = 0.002). In both ungulate pairs, the interindividual variation was larger across the domestic species (supplementary fig. S1B and C, Supplementary Material online). Thus, contrary to previous proposals based on the study of chiasma numbers, our results indicate that wild species had higher numbers of crossover markers than their domestic counterparts.
Given that recombination correlates with the number of chromosomes and it is proportional to the number of chromosome arms, at least in mammals (Pardo-Manuel de Villena and Sapienza 2001), we calculated the mean number of MLH1 foci (inferred crossovers) expected per chromosome arm. The haploid number of chromosomes for dog and wolf is 39, and all autosomes are acrocentric, thus we calculated an average of 1.00 and 1.05 crossovers per chromosome arm in dogs and wolves, respectively (table 2). The haploid number of chromosomes for the two Capra and Ovis species is 30 and 27, respectively (all metacentric), thus we inferred an average of 1.02 and 1.08 crossovers per arm in goat and ibex and 1.18 and 1.28 in sheep and mouflon, respectively.
Population-Genetic Estimates of Historical Breakpoints and Recombination Rate
We investigated whether artificial selection would have favored increased recombination in dogs, as compared with wolves, around loci associated with traits potentially subjected to intense selection in dogs. We sequenced 16 genomic regions, each containing a locus associated to a distinct dog phenotype (totaling ∼200–300 kb each; table 1), and we inferred recombination breakpoints from sequence data in both dogs and wolves. The number of segregating sites per region ranged between 46 (chr10) and 1,445 (chr27) (supplementary table S2, Supplementary Material online), and was similar for a given orthologous region across species or populations (wolf samples were grouped into populations with sample size similar to the sample size in dogs). In general, the number of segregating sites was lowest in dogs, and was followed in increasing order by the wolves from Spain + Italy, Sweden + Finland, and North America. We identified haplotype blocks using LDheatmaps for data previously adjusted for the same number of markers (single nucleotide polymorphisms [SNPs]). These maps showed more LD in dogs than in wolves and, in general, a linkage block observed in wolves could also be observed in dogs, but not the reverse (supplementary fig. S2, Supplementary Material online).
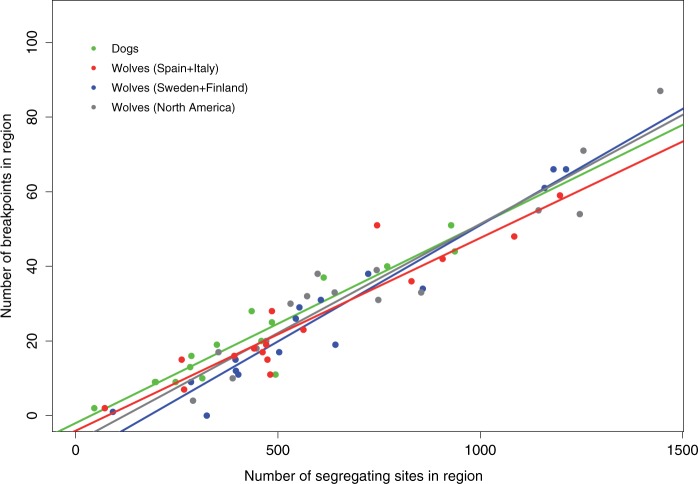
We estimated the number of historical recombination events using RDP3. These ranged from 2 (chr10) to 87 (chr27) (supplementary table S2, Supplementary Material online), and significantly correlated with the number of segregating sites present in that fragment (P < 0.0001) (fig. 2). This is expected, as a recombination event between two identical sequences is undetectable. An analysis of covariance indicated nonsignificant differences between the slopes (fig. 2, P = 0.4), suggesting that there were no differences in the rate of recombination in these regions across dogs and wolves.
Fig. 2.
Number of segregating sites and recombination breakpoints in each of the 16 genomic regions studied in dogs and wolves. Each dot represents the number of historical recombination breakpoints inferred in a particular genomic region and the number of segregating sites found in that region.
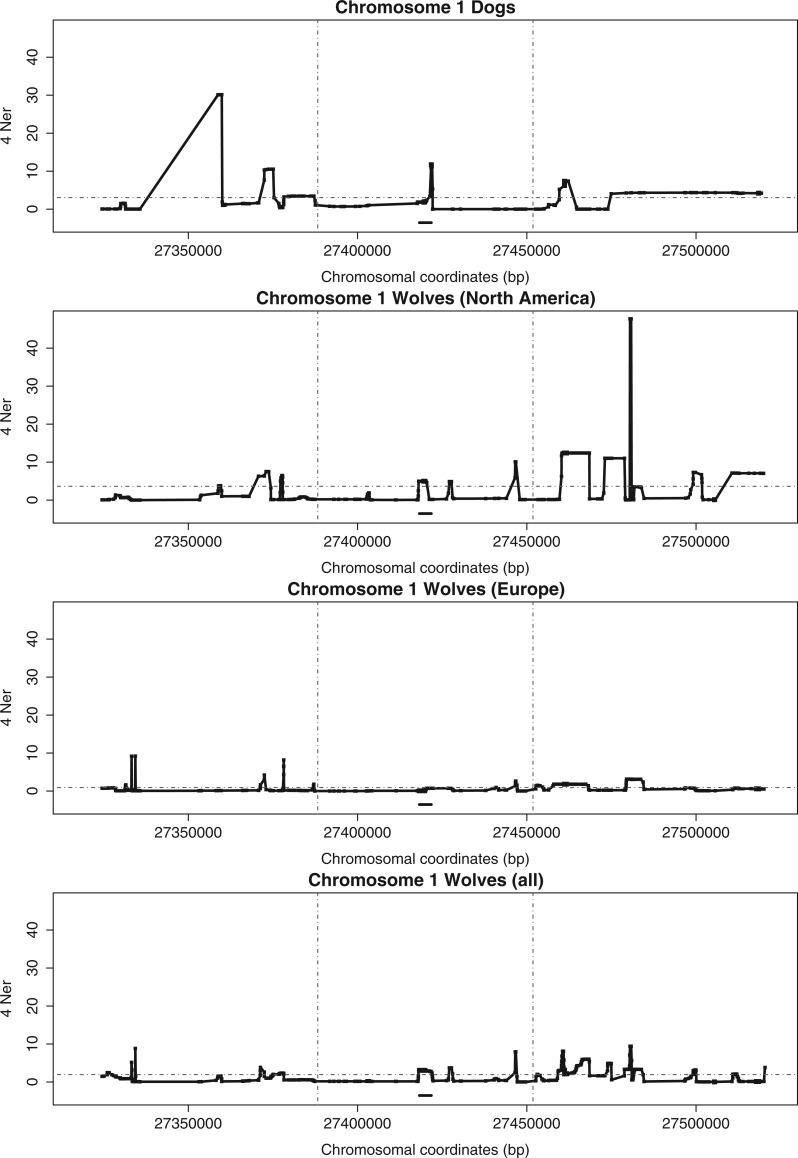
We used LDhat to obtain estimates of mean population recombination rate, ρ, between adjacent SNP pairs for each of the 16 targeted genomic loci (fig. 3 and supplementary fig. S3, Supplementary Material online). We counted the number of higher-than-average recombination rate peaks (HTAR peaks) in two flanking windows of 70 kb each and a central window containing the locus potentially under selection in dogs, wolves from Europe and wolves from North America (windows are designated by vertical dashed lines, fig. 3). We then compared the ratio between HTAR peaks around the locus (central window) and those in the remaining sequence (flanking windows) for dogs and each group of wolves. Our results showed no significant differences for the distribution of peaks in these regions between dogs and each group of wolves for any of the 16 genomic regions studied (Fisher’s exact probability test, P > 0.05 for 48 comparisons). When the 16 genomic regions were considered together, the differences were again nonsignificant (for dogs and North American wolves, P = 0.822; for dogs and European wolves, P = 0.831; and for dogs and all wolves, P = 1.000). Although this lack of differences could be partly due to limited detection power due to the small number of genomes and/or recombination breakpoints, we noted that the proportion of regions with high recombination in the central portion of the sequence was higher in dogs than in wolves for close to half of the comparisons (higher for dogs in 10 out of 16 comparisons when comparing with American wolves, 9 out of 16 when comparing with European wolves or all wolves), as would be expected if the recombination events were randomly distributed. Therefore, our results failed to support the notion of an increased recombination rate in dogs relative to wolves in regions potentially associated with selected phenotypes.
Fig. 3.
Genetic maps for dogs and wolves. Genomic region 1 (table 1) is shown here (see supplementary fig. S3, Supplementary Material online, for all regions). Each dot indicates the mean ρ = 4Ner estimates between each pair of SNPs (penalty 5). The horizontal thick line above the x-axis shows the location of the locus associated with the phenotypic character (see table 1). The dashed horizontal line indicates the average recombination rate for the region. The vertical dashed lines designate three windows, two of 70 kb in size at the two ends of the region, and a central window of size between 60 and 160 kb (see Results and Methods for details).
Discussion
Reduced Recombination Rate in Domestic Mammals As Compared with Their Wild Relatives
Analysis of the number of MLH1 foci as markers for crossover events showed that, for the three domestic-wild species pairs examined here, dog versus wolf, goat versus ibex, and sheep versus mouflon, the wild species had higher number of crossovers per cell than the domestic counterpart. Our data accounted for an average of crossovers per bivalent that ranged between 1 and 2.56 (the autosomes of dogs and wolves are acrocentric, see table 2), and between 1.00 and 1.28 per chromosome arm, in agreement with the requirement of one crossover per arm (except short arms in acrocentric chromosomes) for the correct segregation of chromosomes during meiosis (Hultén 1974; Pardo-Manuel de Villena and Sapienza 2001).
The Number of MLH1 Foci As an Estimate of Broad-Scale Recombination Rate
Our estimates for broad-scale recombination rate based on the number of MLH1 foci for the dog are similar to those previously reported in cytological studies in this species (Wada and Imai 1995; Basheva et al. 2008) and, for goat and sheep they are similar or slightly higher than the mean chiasma counts for both spermatocytes and oocytes previously obtained from diplotene, diakinesis, and metaphase I stage cells (Datta 1970; Jagiello et al. 1974; Logue 1977; Long 1978). The goat and sheep data are in agreement with the chiasma counts reported in Burt and Bell (1987), which were mostly based on the same studies (Burt A, personal communication). However, the dog estimate reported by Burt and Bell (1987) differs by 2-fold from estimates in this study and the other studies mentioned above, and was based on chiasma observations carried out on three male dogs corresponding to three breeds (Ahmed 1941; Burt A, personal communication). It is well possible that a technical problem resulted in an overestimation in that study.
Concern has been expressed as to whether chiasma numbers are good indicators of genetic length (e.g., Hultén 1974; Wada and Imai 1995), and mapping crossovers using MLH1 foci is now established as a more accurate procedure than using chiasma counts. True chiasmata can be identified with confidence only after the complete dissolution of the synaptonemal complex and the condensation of chromosomes upon entry into meiotic metaphase. Unfortunately, the condensed nature of the chromosomes at this stage makes it difficult to accurately identify chiasmata from cell preparations alone in the absence of molecular markers. In addition, the presence of pseudochiasmata due to residual synapsis between homologous chromosome axes and the twisting of the bivalents in diplotene stage cells during prophase I of meiosis may lead to an overestimation of chiasma numbers (Hultén 1974; Wada and Imai 1995). Indeed, the numbers of perceived “chiasmata” decrease through diplotene, diakinesis, and metaphase I (Datta 1970), although the numbers of crossovers are expected to remain the same. In mammals, most crossovers are formed through an MLH1-dependent pathway, whereas a marginal fraction depends on MUS81 activity (Holloway et al. 2008). There is now good evidence that MLH1 foci recognize the sites of meiotic exchange and provide an estimate of recombination rate that avoids the ambiguities associated with chiasma counts (Baker et al. 1996; Barlow and Hultén 1998; Anderson et al. 1999; Lynn et al. 2004; Cohen and Holloway 2010). Therefore, to count the number of MLH1 foci on chromosome axes in meiocytes, as we have done in this study, is a more precise way of estimating broad-scale recombination rate than chiasma counts.
Linkage Maps for Dog, Goat, and Sheep and MLH1 Counts Provide Similar Estimates of the Number of Crossovers
Availability of male linkage maps for dog, goat, and sheep allowed us to compare estimates of crossovers based on map length with the average number of crossovers estimated from the number of MLH1 foci. The number of crossovers based on the count of MLH1 foci, we obtained for dogs suggests that each dog chromosome pair usually contains just one crossover, which agrees with estimates based on the male dog linkage map, in which 1,910 cM (Wong et al. 2010) would account for 0.98 crossover per chromosome pair (about one crossover for 50 cM over 39 chromosome pairs). For the two domestic Capra and Ovis species, 2.0 and 2.4 crossovers per chromosome pair were estimated from the number of MLH1 foci, respectively, which are similar to the number of crossovers estimated from the male linkage map (goat, 2,737 cM, Schibler et al. 1998; sheep, 3,876 cM, Maddox and Cockett 2007) of 1.8 and 2.9 crossovers per chromosome pair for goat and sheep (30 and 27 chromosome pairs). Therefore, the average numbers of crossovers per chromosome pair as inferred from spermatocyte MLH1 counts in this study were in agreement with those estimated from male linkage maps.
In this study, we have only attempted to estimate recombination rate in males. It could be claimed that the recombination rate could still be larger in domestic mammals considering only recombination in females. However, linkage maps for the dog and the sheep have also been obtained for females (2,388 cM, Wong et al. 2001; 3,278 cM, Maddox and Cockett 2007), which account for 1.2 crossovers per chromosome arm. Considering the agreement between MLH1 estimates and recombination rate estimated from linkage maps, it seems unlikely that recombination rate is greatly increased in the females of domestic mammals.
Although linkage maps provide both female and male broad-scale recombination rate estimates, there are instances in which the number of MLH1 foci (reflecting only recombination in males) might be the only way to obtain such estimates, even when access to spermatocytes might be complicated. The construction of genetic linkage maps requires access to both an extensive number of markers that provide a good coverage of the genome and large-known pedigrees. Although the former is becoming less challenging with current developments in genomic technology, large pedigrees of wild species are rare. In addition, estimates of broad-scale recombination rates based on the number of MLH1 foci might be preferred to estimates based on linkage maps. The resolution of maps is compromised by marker coverage, in particular for the telomeres, which may lead to overestimate the sex-differences in recombination rate (Coop and Przeworski 2007); for example, in humans and other placental mammals, males recombine more toward the telomeres, whereas females have higher recombination rates near the centromeres. In addition, linkage mapping is based on transmitted chromosomes, and thus provide no information about half the crossovers that occur in meiosis or about gametes that may be selected against (Vallente et al. 2006).
Recombination Around Genes Associated with Phenotypic Characters in Dogs
Direct methods, such as counting MLH1 foci or sperm-typing studies, provide a contemporary measure of recombination rate, but may not be fully informative about historical recombination at the population level. In humans, significant discrepancies have been found between sperm crossover frequencies and historical recombination rates at specific sites, which have been attributed to the rapid evolution of hotspots and their transient activity (Jeffreys and Neumann 2005, 2009). Although our results above indicate that recombination rate may not have changed at the genome-wide level during domestication, it is possible that artificial selection may have favored individuals with increased recombination around loci associated with selected phenotypes, so that these genes would be decoupled from surrounding regions, making artificial selection more efficient. Our results showed, for the 16 genomic regions studied, no difference in the overall number of recombination events across dogs and wolves (equality of the slopes, fig. 2). Likewise, we found no differences in the proportion of peaks with HTAR in central versus flanking windows in dogs as compared with wolves.
LDhat analyses provide population recombination rate estimates. We did not attempt to identify hotspots or compare the intensity of recombination between wolves and dogs, and thus we did not rescale the population recombination rate, ρ = 4Ner, to per-generation recombination rate, r (measured in cM/Mb) using the effective population size, Ne, of dogs or wolves. Indeed, great uncertainty surrounds Ne estimates for these species. Axelsson et al. (2012) and Auton et al. (2013) obtained estimates of Ne for dogs that differed 4-fold, and Freedman et al. (2014) detected very large changes in effective population size since the time of domestication and in different wolf lineages. Although demography and selection are confounders of recombination rate (see Pritchard and Przeworski [2001], Clark et al. [2010], and Chan et al. [2012] for excellent reviews), here we assess the distribution of recombination breakpoints along the regions for each species or population, and thus differences in demography and selection across species or populations should not bias our results. Even if the power to detect recombination events were not the same in all groups of samples, the recombination events detected did not tend to accumulate in the center of the chromosomal region under study in dogs more than in wolves. Consequently, our results do not support an increase recombination in these regions in dogs.
Chan et al. (2012) indicated that LDhat may spuriously detect hotspots in the presence of a selective sweep. In this respect, our results are conservative, because the bias in LDhat should lead to a higher number of hotspots toward the center of the chromosomal regions under study in dogs, where the loci under selection are located, and we observed no differences between the two species.
Domestication and Changes in the Recombination Rate
Phylogenetic relationships (Dumont and Payseur 2008) as well as the number of chromosome arms (Pardo-Manuel de Villena and Sapienza 2001) have been shown to have an effect on recombination rates. In this study, we included species pairs separated by small phylogenetic distance; the dog was compared with its direct wild ancestor, the gray wolf (Vilà et al. 1997) and, in the case of the sheep and the goat, for which the wild ancestors are less clear (Bruford and Townsend 2006; Luikart et al. 2006) and the candidate species are in most cases vulnerable or threatened, we chose to work with closely related wild congeners. In addition, the two species being compared had equal chromosome number and organization (meta- or acrocentric).
Ross-Ibarra (2004) distinguished between wild progenitors and congeners to test both the Rees and Dale (1974) hypothesis for an increase in recombination rate in domestic species, as well as the Gornall (1983) hypothesis of preadaptation, in which higher recombination rate would predate, and contribute, to success in domestication. Our results do not support the hypothesis that domestic animals have higher recombination rate than their wild counterparts. Although only three pairs of species were compared, they represent early domesticates, with large diversity and world-wide distribution, and all of them showed higher recombination rate in the wild species. In addition, given that the domestic species we investigated had an average number of crossover markers close to the minimum expected for the correct segregation of chromosomes, we find that the preadaptation hypothesis is not likely either. Ross-Ibarra (2004) did not find support for the preadaptation hypothesis in plants, and concluded that increased recombination rate was of little importance in the process of domestication. Our results show that domestication may have not been associated with an increase in the recombination rate in mammals, even though the Burt and Bell (1987) study is often cited as an example (see Introduction). This study did not include wild progenitors of the domestic species and, therefore, it may not have been conclusive in assessing whether domestication resulted in an increased recombination rate (Ross-Ibarra 2004; Coop and Przeworski 2007).
Even if broad-scale recombination rates had not changed, it was possible that strong artificial selection may have contributed to increased recombination around loci associated to distinct phenotypes, so that these genes would be decoupled from surrounding regions, making artificial selection more efficient. Our results in dogs and gray wolves showed no evidence for differences in the number and distribution of recombination breakpoints in 16 genomic regions (200–300 kb each) around loci potentially associated to phenotypes subject to strong selection in dogs. In these species, even for purebred dogs, LD decay is rapid over less than 50 kb, and is very limited over 100 kb (Lindblad-Toh et al. 2005; Axelsson et al. 2012). Thus, the length of the regions studied seems adequate to detect changes in recombination rate. Although these results may reflect genome-wide patterns, due to the small number of regions studied here, a more conclusive confirmation of the results may require individual recombination maps (see below) on a larger number of samples.
An alternative prediction proposed that selection for reduced recombination in domesticated species may protect from maladaptive gene flow from wild relatives (Lenormand and Otto 2000). For the three pairs of species studied here, the overall recombination rate is lower for the domestic counterpart, apparently supporting this hypothesis. Nevertheless, it is not clear if this process should affect all domestic mammals, because many of them spread far beyond the distribution range of the ancestor species soon after domestication (sheep and goat, e.g., for other species, the wild ancestor became extinct, as is the case of the horse), thus decreasing the chances for intercrossing and the possible selection for lower recombination rate.
Based on the results presented in this article, we find no support for the idea that strong directional selection resulted in the evolution of increased recombination rate in domestic mammals, or that increased recombination associated to selected loci during domestication facilitated a response to selection. It has been proposed that rates of recombination may evolve neutrally, with selection pushing them back to the neutral range if they drift toward low or high recombination rates (Dumont and Payseur 2008).
Current advances in genome sequencing allow massive parallel whole-genome amplification of single sperm cells followed by high-throughput genotyping to construct an individual’s recombination map (Lu et al. 2012; Wang et al. 2012; Kirkness et al. 2013). Phased SNPs or haplotypes are obtained, which enable recombination events and possibly also gene conversion events to be directly identified, and individual high-resolution maps to be built, irrespective of a preselection of candidate genes or loci as we have done in this study. Although sperm cells might be obtained in large numbers from adult males, via ejaculation or from dead animals (e.g., Muñoz-Fuentes et al. 2014), applying this technique to oocytes remains challenging, mainly due to the temporal aspects of mammalian oogenesis. Thus, these studies, like previous ones, may be potentially limited to the more readily available male samples. Applying this technique to dogs and wolves will be particularly interesting as, like other canids analyzed (Muñoz-Fuentes et al. 2011; Axelsson et al. 2012), they seem to lack a functional PRDM9. So far, it seems that PRDM9 in Ovis sp and Capra sp has not been investigated.
Materials and Methods
Cytology and Immunofluorescence Assays
Samples
Testes from dogs, wolves, pigs, wild boar, sheep, mouflons, goats, and ibexes were opportunistically collected and tissue samples snap frozen in liquid nitrogen. All samples used in immunofluorescence analyses were obtained in Spain and were available for reasons other than this study. We contacted veterinary clinics for the dog samples (derived from castration), zoos for the wolf samples (from dead wolves), slaughterhouses for pig, sheep, and goat samples, and attended hunting events to collect wild boar, mouflon, and ibex samples.
We obtained high quality cell preparations for spermatocyte spreads against MLH1 from 6 dogs, 2 wolves, 6 goats, 6 ibexes, 6 sheep, and 5 mouflons (supplementary table S1, Supplementary Material online). In addition, these experiments were also carried out for pig (S. domesticus) and wild boar (S. scrofa) spermatocytes (seven and ten individuals, respectively), but we were unable to visualize MLH1 proteins. Because these samples were collected as the others and were preserved and processed in the same way, we attribute the lack of success to the antibodies not recognizing pig and wild boar MLH1 proteins.
Immunofluorescent Localization of MLH1 Protein on Spermatocytes Synaptonemal Complexes
Spreading and immunostaining of spermatocytes was performed as in Roig et al. (2004). Briefly, a piece of frozen testis was minced in Phosphate Buffer Saline (PBS) to obtain a cell suspension. Cell membrane was disrupted with the addition of 1% Lipsol (DH Scientific) (diluted in water) and incubating at 4 °C for approximately 14 min. Cells were fixed on slides for 2 h with 1% paraformaldehyde, 0.15% Triton X-100, and 1× protease inhibitor cocktail (Roche) in water. Slides were rinsed in 1% Photo-Flo solution (Kodak) and blocked with PBS with 0.05% Tween-20, 0.2% Bovine Serum Albumin (BSA), and 0.2% Gelatin (PTBG, PBS with 0.1% Tween, 0.2% BSA & 0.1% Gelatin). Incubation of rabbit polyclonal antibody against SYCP3 (dilution 1:200, Abcam) to mark chromosomes and mouse monoclonal antibody against MLH1 (dilution 1:50, Pharmingen) was performed at 4 °C overnight. After four washes with PTBG, CY3-conjugated antibody against rabbit and a Fluorescein Isothiocyanate (FICT)-conjugated antibody against mouse antibodies (dilution 1:100, Jackson Immunoresearch) were incubated 1 h at 37 °C. Slides were then washed four times in PTBG and DNA counterstained with DAPI dissolved in Vectashield mounting medium (Vector Lab).
Slides were analyzed using a Zeiss Axioskop fluorescence microscope. Only well spread cells displaying bright foci were captured and processed by Progress Capture software (Jenoptik). Images were further enhanced using Adobe Photoshop version CS2 to match the fluorescent intensity seen in the microscope. To avoid biases, for a subset of samples, MLH1 foci were counted by at least two investigators. In all these cases, similar results were obtained by the different researchers (data not shown). Furthermore, the same person counted the foci in each domestic and wild species pair. Foci were counted in 14–75 spermatocytes per individual.
To investigate the variation in the number of MLH1 foci, we used the generalized linear mixed model function in R ver. 3.0.3 (R Core Team 2014) “lmer()” in the package “lme4” (Bates et al. 2014). We set the number of foci per cell as the dependent variable, species as fixed factor (wild or domestic), and individual as random factor. Assuming a normal error distribution, the model fitted the data well, with the residuals following a straight line in a normal probability plot (QQ plot). To compute P values, we used the function “cftest()” in the package “multcomp” (Hothorn et al. 2008).
Population Genomic Inferences of Recombination
Sample Collection and Sequencing
Mouth swab samples were collected from five mongrel dogs (dogs of unknown ancestry, except one pure breed German shepherd) and blood or tissue samples from 22 dead wolves from widespread geographic locations (BC, Canada, n = 2; Finland, n = 3; Italy, n = 3; North Western Territories, Canada, n = 3; Spain, n = 4; Sweden, n = 4; United States, n = 2) (supplementary table S1, Supplementary Material online). Mongrel dogs were preferentially analyzed for two reasons; first, to increase the number of polymorphisms (SNPs) per individual, which would increase the power to detect recombination, and, second, to avoid biases that could be associated with certain breeds, as the rate of recombination is a heritable trait and inbreeding could lead to interbreed differences. Except for the wolves from the United States, which were captive and from which blood samples were obtained, all other wolves were wild and died for reasons unrelated to this study.
We extracted DNA using the QIAgen DNeasy kit and prepared Illumina paired-end libraries for each sample using the Agilent protocol for indexed paired-end Illumina libraries and Agilent SureSelect capture system. Briefly, DNA was sheared with a Covaris S2 device (Covaris, Inc. Woburn, MA, USA), end-repaired, A-tailed, ligated with Illumina’s indexing-specific paired-end adaptors and polymerase chain reaction (PCR) amplified for five cycles.
We enriched for 16 chromosomal regions, each containing a locus associated with a distinct phenotypic character (morphological or behavioral) in dogs (table 1) in a central position, and 100–150 kb upstream and downstream (totaling ∼200–300 kb in length). We enriched with a custom Agilent SureSelect RNA oligo kit. The oligo-targeted regions added up to 2.48 Mb (repetitive regions excluded) and encompassed approximately 3.96 Mb of the dog genome. Libraries were then Illumina indexed/barcoded in a PCR of 13 cycles. A Nanodrop spectrophotometer and a Bioanalyzer instrument were used to assess both quality and quantity of the samples at various steps during the laboratory procedures. Libraries were validated using real-time quantitative PCR, pooled and then 90- or 100-bp paired-end sequenced on four lanes of an Illumina GAIIx machine, yielding 84,682,789 of paired-end reads. All read data were submitted to the European Nucleotide Archive (ENA) and are accessible under the Short Read Archive (SRA) accession number PRJEB7877.
Alignment of Reads and SNP Calling
We followed the Broad Institute Best Practice Variant Detection guide (http://www.broadinstitute.org/gatk/guide/topic?name=best-practices, last accessed November 25, 2014) for data processing and analysis. Briefly, we aligned raw reads using BWA 0.6.1-r104 (Li and Durbin 2009) at four edit distance to the CanFam2 reference assembly downloaded from the UCSC (University of California Santa Cruz) Genome Browser. PICARD TOOLS 1.66 (http://broadinstitute.github.io/picard/, last accessed November 25, 2014) and SAMTOOLS 0.1.18 (Li et al. 2009) were used to remove PCR duplicates and multimapping reads, respectively, and at various stages during the mapping and SNP calling procedures to manage files. We used the Genome Analysis Toolkit (GATK) 2.1.9 (McKenna et al. 2010) to realign around indels, perform base quality recalibration, call SNPs using UnifiedGenotyper, and then filter using VariantFiltration to avoid false-positive SNP calls (DePristo et al. 2011). We excluded indels and filtered variants following Auton et al. (2012) with some modifications. We used BEDTOOLS (Quinlan and Hall 2010) to extract information at various stages during the bioinformatic procedures. On average, we mapped 97% of the reads per sample, which was reduced to 79% after removing PCR duplicates and multimapping reads. The proportion of reads on target was 40–60%. Our filtered SNP set consisted of 22,614 SNPs, of which 17,390 were typed in all individuals. Data were phased using BEAGLE (Browning and Browning 2007).
Population-Genetic Inferences of Recombination
As a measure of LD, or population-level nonrandom association of alleles at two loci, we used the r2 statistic. It ranges from 0 to 1, and it equals 0 when the two alleles are in equilibrium, that is, the loci are independent of one another. We calculated r2 on the phased data using VCFTOOLS 0.1.10 (Danecek et al. 2011) and constructed LD maps, in which pairwise LD measures are plotted between each pair of SNPs, using the R function “LDheatmap()” (Shin et al. 2006). Wolf genotype data were previously thinned to match the dog data in the number and location of SNPs, by selecting the SNP with the same or the closest coordinates to each dog SNP.
Representations such as LD maps based on r2 allow the identification of haplotype blocks, but they do not allow us to directly associate differences in patterns between a pair of SNPs with differences in the underlying recombination rate. In order to characterize the nonrandom association of alleles in the population due to recombination, methods have been developed to statistically determine recombination breakpoints or to estimate the likelihood of the observed sample data under population models that assume different sets of population genetic parameters (e.g., recombination rate, mutation rate) and that attempt to include all the information present in the data through the underlying genealogy (Posada et al. 2002; Stumpf and McVean 2003). The latter is generally computationally intractable for large data sets using full-likelihood methods and, in this respect, an important contribution has been the development of approximate-likelihood methods to infer the population recombination rate, ρ = 4Ner, from a large number of markers (McVean et al. 2004; Auton and McVean 2007).
We used RDP, GENECONV, BOOTSCAN, MAXCHI, CHIMAERA, SISCAN, and 3SEQ, as implemented in RDP3 (Martin et al. 2010), to simultaneously estimate the number of recombination breakpoints. We also used LDhat 2.2 (McVean et al. 2004; Auton and McVean 2007), which implements a coalescent-based model to infer population recombination rates between adjacent SNPs. Because the wolves came from different populations, we performed the analyses for the wolves in separate groups according to continent of origin (North America, n = 7; Europe (Spain, Italy, Sweden and Finland), n = 14; and all together, n = 21). Due to computation limitations in RDP3, we further divided the wolves from Europe in two groups (Italy and Spain, and Finland and Sweden; n = 7 in each case) for those analyses.
We made input alignments files for RDP3 with a custom script and accepted breakpoints that were detected by at least two methods (Posada et al. 2002; Martin et al. 2010). We then compared the number of recombination breakpoints across regions for dogs and wolves.
In order to run the program interval as implemented in the LDhat package, we downloaded a lookup table for n = 50 sequences and a population mutation rate θ = 0.001 per site from http://ldhat.sourceforge.net/instructions.shtml (last accessed November 25, 2014). We then generated adequate lookup tables for the number of sequences in our data set using the program lkgen from the LDhat package. Recombination rates were estimated with a block penalty of 5 and 10 million MCMC iterations, and we sampled from the chain every 5,000 iterations and discarded the first 100,000 as burn-in, following recommendations in the manual. Because no reliable estimates of effective population size were available for dogs and wolves (Axelsson et al. 2012; Auton et al. 2013), we report only the estimates of the population recombination rate parameter as obtained with this method (see Discussion).
We then identified the number of HTAR peaks (number of regions with recombination rate above the average as inferred by LDhat, indicated by a horizontal dashed line in fig. 3 and supplementary fig. S3, Supplementary Material online) along each of the 16 genomic regions in three windows of 70 kb in size at the two ends of the region, and a central window of size between 60 and 160 kb (represented by vertical dashed lines in fig. 3). The central window length varied according to the length of the chromosomal region captured and was longer in the cases in which the locus associated with the dog trait was larger (haplotype instead of a point mutation) (table 1). However, because the size of the fragments in dogs and wolves were equal in size, no bias was introduced in this respect in the comparisons. We used a Fisher’s exact probability test to compare the ratios of HTAR peaks in central to flanking windows in dogs and North America wolves, dogs and European wolves, and dogs and all (North American and European) wolves (16 × 3 = 48 comparisons performed).
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are extremely thankful for help in sample collection to C. Asa and K. Bauman (Saint Louis Zoo, USA), E. Martínez Nevado (Madrid Zoo, Spain), M. Amundin and B. Röken (Kolmårdens Djurpark, Sweden), J. Vicente, O. Rodríguez, D. González Barrio, J. Á. Armenteros Santos, and J. Cabello Stom (IREC-CSIC, Spain), D. Cluff (Environment and Natural Resources, Canada), E. Randi (ISPRA, Italy), J. Aspi (University of Oulu, Finland), I. Paredes, I. Sánchez, and M. Camacho (EBD-CSIC, Spain), B. Herrera Díaz (Cuevas Minadas, Spain), P. Prieto, J. Monje Samblás, H. Peralta Cózar, Á. Lara, E. García, M. Franco, F. J. Pérez,and J. C. Soria (Parque Natural de Cazorla, Segura y Las Villas, Spain), K. Ocejo, M. Pariente, R. Franco, Cárnicas Coviher, L. Urquijo, and the veterinarian clinics Los Corrales, Sevilla Este, Santa Eufemia, Alcosa, and Dirus (in Seville, Spain). The authors are grateful to M. Przeworski for fruitful discussions during the early phases of this project, A. Muñoz-Pomer, M. Fernández, M. Melé, and T. Marques-Bonet for advice during the bioinformatic analyses, M. P. Höppner for writing a script for us that converted the phased data from BEAGLE into an RDP3 alignment, I. Sánchez and B. O'Hara for guidance with mixed models, and A. Auton, T. Lenormand and two anonymous reviewers for a critical reading of our manuscript, which helped to improve it. We also wish to thank Jennifer A. Leonard for logistic support. This work was supported by the JAE fellowship program of the Spanish Research Council, CSIC (grant JAEDOC-2009.031), the EU Marie Curie Individual Fellowship Programme (MEIF-CT-2006-041193) and a mobility grant awarded by the Spanish Ministry of Science and Innovation (grant PA1003151) to V.M.-F., the Spanish FPI fellowship programme to M.M.-O. (grant BES-2011-045381), the Deutsche Gesellschaft für Forschung (DGF) to K.D. (grants DGF: TO 421/3-2 and 4-2) and to A.T. (grant DFG:TO 421/5-1), National Institutes of Health (grant GM83098), and the Programa de Captación del Conocimiento para Andalucía (C2A) to C.V.
References
- Ahmed IA. IX.—Cytological analysis of chromosome behaviour in three breeds of dogs. Proc R Soc Edinb Biol. 1941;61:107–118. [Google Scholar]
- Anderson LK, Reeves A, Webb LM, Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, Calabrese P, Nordborg M. Hot and cold spots of recombination in the human genome: the reason we should find them and how this can be achieved. Am J Hum Genet. 2003;73:5–16. doi: 10.1086/376419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Fledel-Alon A, Pfeifer S, Venn O, Ségurel L, Street T, Leffler EM, Bowden R, Aneas I, Broxholme J, et al. A fine-scale chimpanzee genetic map from population sequencing. Science. 2012;336:193–198. doi: 10.1126/science.1216872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, Li YR, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 2013;9:e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, McVean G. Recombination rate estimation in the presence of hotspots. Genome Res. 2007;17:1219–1227. doi: 10.1101/gr.6386707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 2012;22:51–63. doi: 10.1101/gr.124123.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Torbjörn Ö, Schneider M, Kempenaers B, et al. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010;20:485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- Barlow AL, Hultén MA. Crossing over analysis at pachytene in man. Eur J Hum Genet. 1998;6:350–358. doi: 10.1038/sj.ejhg.5200200. [DOI] [PubMed] [Google Scholar]
- Basheva EA, Bidau CJ, Borodin PM. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res. 2008;16:709–719. doi: 10.1007/s10577-008-1221-y. [DOI] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2014. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1-5 [cited 2014 Nov 25]. Available from: http://CRAN.R-project.org/package=lme4, last accessed 25 November 2014.
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, De Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Bell G. Selection. 2nd edn. New York: Oxford University Press; 2008. [Google Scholar]
- Berg IL, Neumann R, Lam KWG, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet. 2010;42:859–863. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg IL, Neumann R, Sarbajna S, Odenthal-Hesse L, Butler NJ, Jeffreys AJ. Variants of the protein PRDM9 differentially regulate a set of human meiotic recombination hotspots highly active in African populations. Proc Nat Acad Sci U S A. 2011;108:12378–12383. doi: 10.1073/pnas.1109531108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;484:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruford MW, Townsend SJ. Mitochondrial DNA diversity in modern sheep. In: Zeder MA, Bradley DG, Emshwiller E, Smith BD, editors. Documenting domestication. New genetic and archaeological paradigms. Berkeley: University of California Press; 2006. pp. 306–316. [Google Scholar]
- Burt A, Bell G. Mammalian chiasma frequencies as a test of two theories of recombination. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- Butlin R. Recombination and speciation. Mol Ecol. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Calderón PL, Pigozzi MI. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosome Res. 2006;14:605–612. doi: 10.1007/s10577-006-1059-0. [DOI] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 2012;8:e1003090. doi: 10.1371/journal.pgen.1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Genetic variation in recombination in Drosophila. I. Responses to selection and preliminary genetic analysis. Heredity. 1985;54:71–83. [Google Scholar]
- Clark AG, Wang X, Matise T. Contrasting methods of quantifying fine structure of human recombination. Annu Rev Genomics Hum Genet. 2010;11:45–64. doi: 10.1146/annurev-genom-082908-150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutton-Brock J. A natural history of domesticated mammals. 2nd edn. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Cohen PE, Holloway JK. Predicting gene networks in human oocyte meiosis. Biol Reprod. 2010;82:469–472. doi: 10.1095/biolreprod.109.083014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Przeworski M. An evolutionary view of human recombination. Nat Rev Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- Coppinger R, Coppinger L. Dogs. Chicago: The University of Chicago Press; 2001. [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker R, Lunter G, Marth G, Sherry ST, et al. The Variant Call Format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta M. Reinvestigation of meioiss in the male goat, Capra hircus, with special reference to chiasma formation in the sex and autosomal bivalents. Cytologia. 1970;35:344–353. doi: 10.1508/cytologia.35.344. [DOI] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Broman KW, Payseur BA. Variation in genomic recombination rates among heterogeneous stock mice. Genetics. 2009;182:1345–1349. doi: 10.1534/genetics.109.105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont BL, Payseur BA. Evolution of the genomic rate of recombination in mammals. Evolution. 2008;62:276–294. doi: 10.1111/j.1558-5646.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B, et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornall RJ. Recombination systems and plant domestication. Biol J Linn Soc Lond. 1983;20:375–383. [Google Scholar]
- Groenen MAM, Wahlberg P, Foglio M, Cheng HH, Megens H-J, Crooijmans RPMA, Besnier F, Lathrop M, Muir WM, Wong GK-S, et al. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B, Åkesson M, Slate J, Pemberton JM. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc R Soc Lond B Biol Sci. 2005;272:2289–2298. doi: 10.1098/rspb.2005.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3–independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Hultén MAJ. Chiasma distribution at diakinesis in the normal human male. Hereditas. 1974;76:55–78. doi: 10.1111/j.1601-5223.1974.tb01177.x. [DOI] [PubMed] [Google Scholar]
- Jagiello GM, Miller WA, Ducayen MB, Lin JS. Chiasma frequency and disjunctional behavior of ewe and cow oocytes matured in vitro. Biol Reprod. 1974;10:354–363. doi: 10.1095/biolreprod10.3.354. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R. The rise and fall of a human recombination hot spot. Nat Genet. 2009;41:625–629. doi: 10.1038/ng.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Furey TS, Payseur BA, Lu Y, Roskin KM, Chen C-F, Thomas MA, Haussler D, Jacob HJ. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14:528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Grindberg RV, Yee-Greenbaum J, Marshall CR, Scherer SW, Lasken RS, Venter JC. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–32. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J. Recombination Difference between Sexes: A Role for Haploid Selection. PLoS Biol. 2005;3:e63. doi: 10.1371/journal.pbio.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Otto SP. The evolution of recombination in a heterogeneous environment. Genetics. 2000;156:423–438. doi: 10.1093/genetics/156.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ III, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Logue DN. Meiosis in the domestic ruminants with particular reference to Robertsonian translocations. Genet Sel Evol. 1977;9:1–15. doi: 10.1186/1297-9686-9-4-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. Chiasma counts and non-disjunction frequencies in normal ram and in rams carrying the Massey I (t1) translocation. J Reprod Fertil. 1978;53:353–356. doi: 10.1530/jrf.0.0530353. [DOI] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Fernández H, Mashkour M, England PR, Taberlet P. Origins and diffusion of domestic goats inferred from DNA markers. In: Zeder MA, Bradley DG, Emshwiller E, Smith BD, editors. Documenting domestication. New genetic and archaeological paradigms. Berkeley: University of California Press; 2006. pp. 294–305. [Google Scholar]
- Lynn A, Ashley T, Hassold T. Varitation in human meiotic recombination. Annu Rev Genomics Hum Genet. 2004;5:317–349. doi: 10.1146/annurev.genom.4.070802.110217. [DOI] [PubMed] [Google Scholar]
- Maddox JF, Cockett NE. An update on sheep and goat linkage maps and other genomic resources. Small Rumin Res. 2007;70:4–20. [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Muñoz-Fuentes V, Di Rienzo A, Vilà C. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One. 2011;6:e25498. doi: 10.1371/journal.pone.0025498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fuentes V, Linde Forsberg C, Vilà C, Morrell JM. Single-layer centrifugation separates spermatozoa from diploid cells in epididymal samples from gray wolves, Canis lupus (L.) Theriogenology. 2014;82:773–776. doi: 10.1016/j.theriogenology.2014.04.029. [DOI] [PubMed] [Google Scholar]
- Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- Otto SP, Barton NH. Selection for recombination in small populations. Evolution. 2001;55:1921–1931. doi: 10.1111/j.0014-3820.2001.tb01310.x. [DOI] [PubMed] [Google Scholar]
- Otto SP, Lenormand T. Resolving the paradox of sex and recombination. Nat Rev Genet. 2002;3:252–261. doi: 10.1038/nrg761. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Recombination is proportional to the number of chromosome arms in mammals. Mamm Genome. 2001;12:318–322. doi: 10.1007/s003350020005. [DOI] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Poissant J, Hogg JT, Davis CS, Miller JM, Maddox JF, Coltman DW. Genetic linkage map of a wild genome: genomic structure, recombination and sexual dimorphism in bighorn sheep. BMC Genomics. 2010;11:524. doi: 10.1186/1471-2164-11-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. What are the genomic drivers of the rapid evolution of PRDM9? Trends Genet. 27. 2011:165–171. doi: 10.1016/j.tig.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Holmes EC. Recombination in evolutionary genomics. Annu Rev Genet. 2002;36:75–97. doi: 10.1146/annurev.genet.36.040202.111115. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Rees H, Dale PJ. Chiasmata and variability in Lolium and Festuca populations. Chromosoma. 1974;47:335. [Google Scholar]
- Roig I, Liebe B, Egozcue J, Cabero L, Garcia M, Scherthan H. Female-specific features of recombinational double-stranded DNA repair in relation to synapsis and telomere dynamics in human oocytes. Chromosoma. 2004;113:22–33. doi: 10.1007/s00412-004-0290-8. [DOI] [PubMed] [Google Scholar]
- Ross-Ibarra J. The evolution of recombination under domestication: a test of two hypotheses. Am Nat. 2004;163:105–112. doi: 10.1086/380606. [DOI] [PubMed] [Google Scholar]
- Samollow P. Marsupial linkage maps. In: Deakin JE, Waters PD, Marshall Graves JA, editors. Marsupial genetics and genomics. The Netherlands: Springer; 2010. pp. 75–99. [Google Scholar]
- Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP. Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res. 1998;8:901–915. doi: 10.1101/gr.8.9.901. [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P, Jokela J. Socially structured populations and evolution of recombination under antagonistic coevolution. Am Nat. 2002;160:403–408. doi: 10.1086/341517. [DOI] [PubMed] [Google Scholar]
- Shin J-H, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Soft. 2006;16 Code Snippet 3. [Google Scholar]
- Smukowski CS, Noor MAF. Recombination rate variation in closely related species. Heredity. 2011;107:496–508. doi: 10.1038/hdy.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MPH, McVean GAT. Estimating recombination rates from population-genetic data. Nat Rev Genet. 2003;4:959–968. doi: 10.1038/nrg1227. [DOI] [PubMed] [Google Scholar]
- Tease C, Hultén MA. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet Genome Res. 2004;107:208–215. doi: 10.1159/000080599. [DOI] [PubMed] [Google Scholar]
- Tortereau F, Servin B, Frantz L, Megens H-J, Milan D, Rohrer G, Wiedmann R, Beever J, Archibald A, Schook L, et al. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics. 2012;13:586. doi: 10.1186/1471-2164-13-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallente R, Cheng E, Hassold T. The synaptonemal complex and meiotic recombination in humans: new approaches to old questions. Chromosoma. 2006;115:241–249. doi: 10.1007/s00412-006-0058-4. [DOI] [PubMed] [Google Scholar]
- van Oers K, Santure AW, De Cauwer I, van Bers NE, Crooijmans RP, Sheldon BC, Visser ME, Slate J, Groenen MA. Replicated high-density genetic maps of two great tit populations reveal fine-scale genomic departures from sex-equal recombination rates. Heredity. 2014;112:307–316. doi: 10.1038/hdy.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Wada MY, Imai HT. Theoretical analyses of chiasmata using a novel chiasma graph method applied to Chinese hamsters, mice, and dog. Jpn J Genet. 1995;70:233–265. doi: 10.1266/jjg.70.233. [DOI] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake Stephen R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK. Cranial morphology of domestic and wild canids—the influence of development on morphological change. Evolution. 1986a;40:243–261. doi: 10.1111/j.1558-5646.1986.tb00467.x. [DOI] [PubMed] [Google Scholar]
- Wayne RK. Limb morphology of domestic and wild canids—the influence of development on morphological change. J Morphol. 1986b;187:301–319. doi: 10.1002/jmor.1051870304. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Gadau J, Schmid-Hempel P. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity. 2007;98:189–197. doi: 10.1038/sj.hdy.6800950. [DOI] [PubMed] [Google Scholar]
- Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, Bontrop RE, McVean GA, Gabriel SB, Reich D, Donnelly P, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- Wong AK, Ruhe AL, Dumont BL, Robertson KR, Guerrero G, Shull SM, Ziegle JS, Millon LV, Broman KW, Payseur BA, et al. A comprehensive linkage map of the dog genome. Genetics. 2010;184:595–605. doi: 10.1534/genetics.109.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.