Abstract
Rod monochromacy is a rare condition in vertebrates characterized by the absence of cone photoreceptor cells. The resulting phenotype is colourblindness and low acuity vision in dim-light and blindness in bright-light conditions. Early reports of xenarthrans (armadillos, sloths and anteaters) suggest that they are rod monochromats, but this has not been tested with genomic data. We searched the genomes of Dasypus novemcinctus (nine-banded armadillo), Choloepus hoffmanni (Hoffmann's two-toed sloth) and Mylodon darwinii (extinct ground sloth) for retinal photoreceptor genes and examined them for inactivating mutations. We performed PCR and Sanger sequencing on cone phototransduction genes of 10 additional xenarthrans to test for shared inactivating mutations and estimated the timing of inactivation for photoreceptor pseudogenes. We concluded that a stem xenarthran became an long-wavelength sensitive-cone monochromat following a missense mutation at a critical residue in SWS1, and a stem cingulate (armadillos, glyptodonts and pampatheres) and stem pilosan (sloths and anteaters) independently acquired rod monochromacy early in their evolutionary history following the inactivation of LWS and PDE6C, respectively. We hypothesize that rod monochromacy in armadillos and pilosans evolved as an adaptation to a subterranean habitat in the early history of Xenarthra. The presence of rod monochromacy has major implications for understanding xenarthran behavioural ecology and evolution.
Keywords: Xenarthra, opsins, rod monochromacy, subterranean mammals, pseudogenes
1. Introduction
Electrophysiological, molecular and genetic techniques have greatly increased our knowledge of the retinal basis for vision in mammals [1–4]. Cone photoreceptors—responsible for high acuity, colour vision in bright light—typically possess one of four spectral classes of photopigment called opsins. The presence of multiple cone opsins allows for the comparison of different wavelengths of light, whereas the dim-light sensitive rod photoreceptors possess a single type of opsin, precluding hue discrimination. The common ancestor of therian mammals probably possessed dichromatic colour vision (two cone classes: short-wavelength sensitive opsin 1 (SWS1) and long-wavelength sensitive opsin (LWS)) following the loss of two of four vertebrate cone types during a hypothesized ‘nocturnal bottleneck’ in the Mesozoic [5]. The loss of additional cone classes is relatively common and has evolved independently in assorted nocturnal, aquatic and subterranean mammals [3,6,7]. These losses presumably are a consequence of inhabiting dim-light niches in which colour discrimination is limited and provide well-documented cases of convergent, regressive evolution [2,3,6,7].
Xenarthrans (armadillos (Cingulata), sloths (Folivora) and anteaters (Vermilingua)) have been overlooked in vision research, despite being an ancient and evolutionarily distinct lineage of mammals [8–11]. Most xenarthran species do not occupy dim-light niches [12], but all three groups of xenarthrans are reported in behavioural [13–18] and anatomical studies [19–22] to have vision consistent with rod monochromacy wherein the retina lacks cones entirely. Rod monochromacy is characterized by low acuity and a complete lack of colour discrimination in dim-light, and blindness during the day (hemeralopia), as rod cells become saturated in bright light. Though pure-rod retinae have long been described in mammals [20], these reports have typically been refuted by the results of molecular and genetic studies (e.g. contrast [20], p. 216 with [7,23–25]). Only recently have genomic studies confirmed rod monochromacy in mammals [6,7], suggesting that this is a plausible phenotype for xenarthrans.
Using genomic and phylogenetic methods, we tested the hypotheses that xenarthrans are rod monochromats and that this condition was inherited from a common ancestor. Our results suggest that xenarthrans have a long history of rod monochromacy and that the most recent common ancestor of Xenarthra was at most an LWS-cone monochromat. These findings indicate that xenarthrans inhabited an extreme dim-light niche early in their evolution, which we suggest was a subterranean habit given fossorial adaptations in fossil and many living xenarthrans.
2. Material and methods
(a). Data collection
We used BLASTN to search the publically available genomes of Dasypus novemcinctus (nine-banded armadillo) and Choloepus hoffmanni (Hoffmann's two-toed sloth) for DNA sequences of cone and rod phototransduction genes, other cone- and rod-specific genes and genes expressed in both rods and cones (electronic supplementary material, dataset S1). We used mRNA transcripts from GenBank for reference sequences. We also mined NCBI's Sequence Read Archive (SRA) for sequences from the genome of an extinct ground sloth, Mylodon darwinii. The SRA sequences were converted into FASTA format and imported into Geneious v. 7.0.5 [26]. In Geneious, we gathered sequences with BLASTN using exons and at least 60 bp of flanking intron/untranslated region (UTR) sequence on each side for reference (Ensembl). Results were assembled into contigs with the de novo assembly tool in Geneious. For comparison, we searched for cone phototransduction genes in the genomes of two known rod monochromats (Physeter macrocephalus (giant sperm whale) and Balaenoptera acutorostrata (minke whale)), a xenarthran analogue (Manis pentadactyla (Chinese pangolin)) and an LWS-cone monochromat (Tursiops truncatus (bottlenose dolphin)) (electronic supplementary material, dataset S1). Individual exons and splice acceptor/donor sites were manually aligned with Se-Al v. 2.0a11 [27] and inspected for inactivating mutations.
We used PCR and Sanger sequencing to confirm shared inactivating mutations in SWS1 and PDE6C in six armadillos, three sloths and three anteaters (electronic supplementary material, dataset S1). After aligning exon sequences for D. novemcinctus and C. hoffmanni, we designed primers based on the flanking introns/UTRs (electronic supplementary material, table S1). We performed PCR with Ramp-Taq DNA polymerase (Denville Scientific Inc.) in 50 µl reactions using the following thermal cycling parameters: template denaturation at 95°C for 7 min, followed by 45 cycles of 1 min at 95°C (denaturation), 1 min at 50°C (annealing) and 2 min at 72°C (extension), followed by an extension at 72°C for 10 min. Genomic DNA (500–750 ng) was used as the template for the initial PCR reaction, and 1–1.5 µl of the PCR product was used in the nested PCR reactions. PCR products were assayed on 1% agarose gels, excised with razor blades and cleaned with a Bioneer AccuPrep Gel Purification kit. Cleaned PCR products were sequenced in both directions using an automated DNA sequencer (ABI 3730xl) at the UCR Core Instrumentation Facility. Contig assembly was performed in Geneious using the Muscle alignment tool [28].
(b). Inactivating mutations and pseudogene dating analyses
Three general types of inactivating mutations were searched for manually in Se-Al: splice donor/acceptor mutations, premature stop codons and frameshift indels. Owing to the relatively high frequency of GC as an alternative splice donor in mammals [29], this variant was not considered an inactivating mutation. Sequences with splice site mutations alone were not considered pseudogenes owing to the possibility of functional splice variants. All putative mutations were compared to outgroups to determine whether they were uniquely derived. Inactivation times of pseudogenes were estimated using previously described methods [7,30]. The alignments used for the analyses can be found in the electronic supplementary material, dataset S2. We assumed phylogenetic relationships and divergence time (global means) from [31] for these calculations.
3. Results and discussion
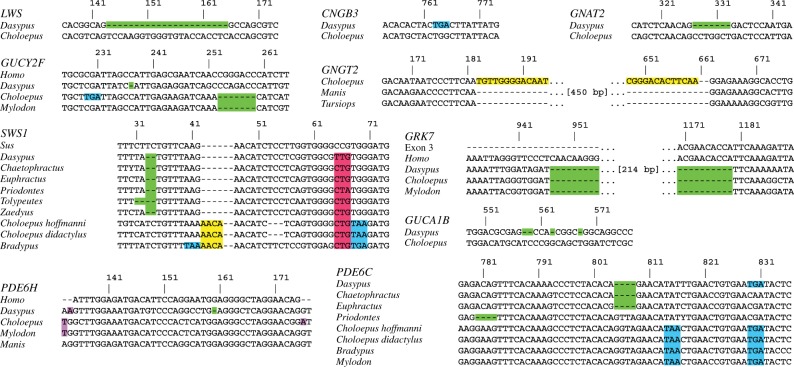
Dasypus novemcinctus has seven inactivated cone-specific genes (SWS1, LWS, GNAT2 (cone transducin alpha subunit), PDE6C (cone phosphodiesterase 6C), PDE6H (cone phosphodiesterase 6H), CNGB3 (cone cyclic nucleotide-gated channel beta subunit), GRK7 (cone G-protein-coupled receptor)) and two pseudogenic rod and cone genes (GUCA1B (guanylate cyclase activator 1B) and GUCY2F (guanylate cyclase 2F)) (figures 1 and 2). By contrast, all rod-specific genes are intact (electronic supplementary material, table S1). In theory, inactivating mutations in both cone opsins (SWS1 and LWS) and/or any of the subsequent genes in the cone phototransduction cascade (CNGA3 (cone cyclic nucleotide-gated channel alpha subunit; [33–35]), CNGB3 [35–37], GNAT2 [38,39], GNGT2 (cone transducin gamma subunit; [40]), PDE6C [41–43]) should result in non-functional or absent cones. The exceptions are PDE6H [44] and GNB3 (cone transducin beta subunit; [45]), which lead to partial rod monochromacy and reduced light-sensitivity in their respective absence. The inactivation of both cone opsins (SWS1 and LWS), GNAT2, PDE6C and CNGB3 all indicate that D. novemcinctus is a rod monochromat. For C. hoffmanni, SWS1, PDE6C, PDE6H, GNGT2, GRK7 and GUCY2F are pseudogenic (figures 1 and 2; electronic supplementary material, table S2). A rod phototransduction gene, PDE6B (rod phosphodiesterase 6B), has a 2 bp deletion in exon 21, but this deletion is near the 3′-end of this long gene so it may not result in inactivation. The retention of all other rod-specific genes in C. hoffmanni suggests this gene is probably still functional. Mylodon darwinii's low coverage genome was examined only for genes that are pseudogenic in C. hoffmanni, and we found inactivating mutations in SWS1, PDE6C, GRK7 and GUCY2F, several of which are shared with C. hoffmanni (electronic supplementary material, table S3). A splice acceptor mutation in PDE6H shared between Mylodon and C. hoffmanni suggests this gene may be inactivated in Mylodon as well (figure 1). The inactivation of PDE6C in C. hoffmanni and M. darwinii, as well as GNGT2 in the former, confirm rod monochromacy in both taxa.
Figure 1.
Examples of inactivating mutations for all retinal genes found to be inactivated in one or more xenarthrans. Green, frameshift deletion; yellow, frameshift insertion; blue, premature stop codon; purple, splice site mutation; red, P23L missense mutation. The numbering of the nucleotide positions corresponds to those in the electronic supplementary material, dataset S1, and includes artificial gaps after frameshift insertions to maintain the original reading frame. Choloepus, Choloepus hoffmanni, unless otherwise noted.
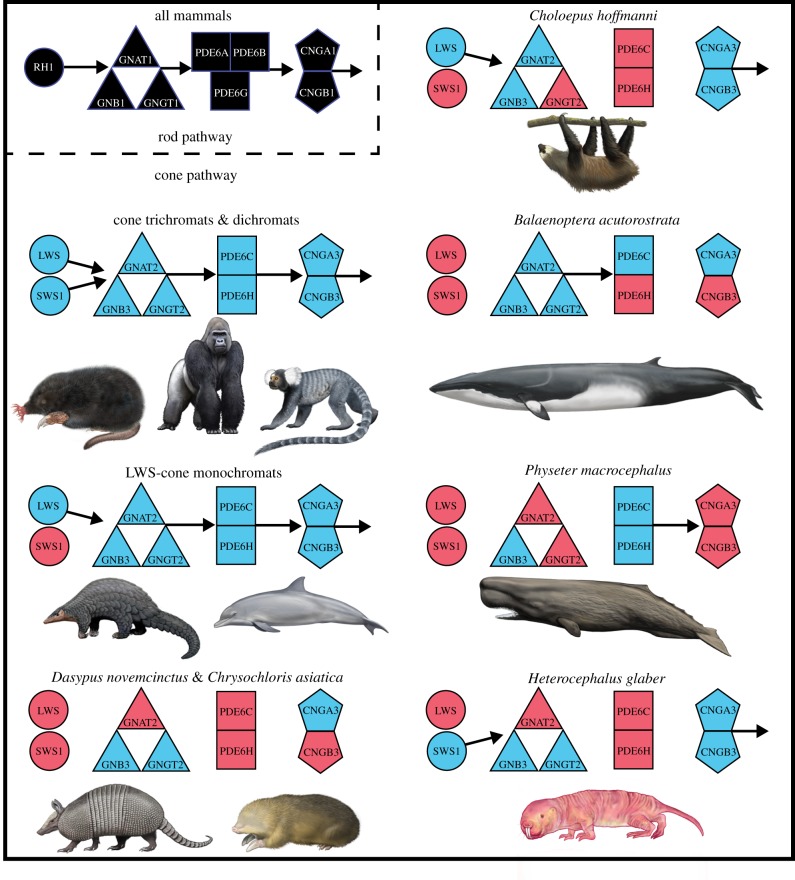
Figure 2.
Patterns of protein loss in the phototransduction cascades of various mammals. Black symbols correspond to rod phototransduction proteins. All mammals investigated so far retain the entire rod pathway (though see note about sloth PDE6B in §3). Blue and red symbols correspond to intact and inactivated cone phototransduction genes, respectively. Arrows indicate the directionality of the phototransduction cascade beginning with the absorption of light by the opsins (RH1, SWS1 and LWS), activation of transducin (GN proteins), activation of phosphodiesterase (PDE proteins) and hyperpolarization of the photoreceptor by cGMP-gated channels (CNG proteins). The absence of an arrow indicates the predicted disruption of that portion of the cascade. Species not reported on in this paper are from references [7,32]. All paintings by Carl Buell (copyright John Gatesy) except star-nosed mole and naked mole-rat (Michelle S. Fabros).
For the comparison groups, M. pentadactyla's SWS1 gene is inactivated, but all other cone phototransduction genes are functional, as is the case for T. truncatus (figure 2; electronic supplementary material, table S2). The rod monochromats B. acutorostrata and P. macrocephalus both have inactivated copies of CNGB3, B. acutorostrata has a PDE6H pseudogene and P. macrocephalus has inactivated copies of CNGA3, GNAT2 and GNGT2 (figure 2; electronic supplementary material, table S2). These results, combined with previous studies [7,32], confirm that rod monochromats are unique in having inactivated cone phototransduction genes, with the exception of SWS1 in LWS-cone monochromats (figure 2). Rod monochromats show a mosaic of pseudogenization with most of the phototransduction genes inactivated in multiple lineages, including SWS1, LWS, CNGB3, GNAT2, GNGT2, PDE6C and PDE6H. The pleiotropic GNB3 [46,47] is the only gene that has remained functional in all rod monochromats examined (figure 2).
Dasypus novemcinctus, C. hoffmanni and M. darwinii share a large deletion in the cone-specific GRK7 (figure 1; electronic supplementary material, table S3). We estimate that this was inactivated in a stem xenarthran approximately 95 Ma (figure 3; electronic supplementary material, table S4). Dasypus novemcinctus and C. hoffmanni also share a unique missense mutation in SWS1, possessing a leucine at residue 23 rather than a proline (bovine RH1 numbering; figure 1; electronic supplementary material, table S3). A proline is present in all opsins across vertebrates [49], with missense mutations in rod opsin (RH1) resulting in a non-functional pigment in vitro (P23H [50]), progressive photoreceptor degeneration in vivo (P23H [51,52]), reduction in chromophore yield owing to a decrease in cell-surface transportation (P23H, P23L [53]) and high amounts of misfolding, with P23L having the highest degree of misfolding among seven RH1 mutants [54]. Consistent with these data, Kogia breviceps (pygmy sperm whale [6]) and Megaderma lyra (greater false vampire bat (AWHB01305061–AWHB01305064)) both have SWS1 pseudogenes with L23. We confirmed that this mutation is present in two additional sloths (Bradypus tridactylus (pale-throated three-toed sloth), Choloepus didactylus (Linnaeus' two-toed sloth)) and five armadillos (Euphractus sexcinctus (six-banded armadillo), Chaetophractus villosus (big hairy armadillo), Tolypeutes matacus (southern three-banded armadillo), Priodontes maximus (giant armadillo), Zaedyus pichiy (pichi)) (figure 1). We were unable to amplify the exon containing this mutation in two anteaters (Myrmecophaga tridactyla (giant anteater) and Tamandua tetradactyla (southern tamandua)), but confirmed that SWS1 is pseudogenic in these species (electronic supplementary material, tables S2 and S3). Using a molecular phylogenetic method to date gene inactivations [27], we estimated that SWS1 was pseudogenized approximately 80 Ma in a stem xenarthran (figure 3; electronic supplementary material, table S4), rendering the earliest crown xenarthrans at most LWS-cone monochromats. This provides evidence of the earliest acquisition of LWS-cone monochromacy in mammals (electronic supplementary material, table S5). Though LWS-cone monochromacy is frequently associated with nocturnality [3], the nocturnal bottleneck hypothesis posits that placental mammals were nocturnal through the end of the Mesozoic [5,20,55,56] owing to competition and/or predation pressures from diurnal sauropsids. Yet xenarthrans represent the only extant lineage of mammals that appear to have disposed of SWS1 prior to the end of the Mesozoic approximately 65.5 Ma, (electronic supplementary material, table S5) suggesting that factors other than nocturnality may explain SWS1 and GRK7 inactivation in this lineage (see §4).
Figure 3.
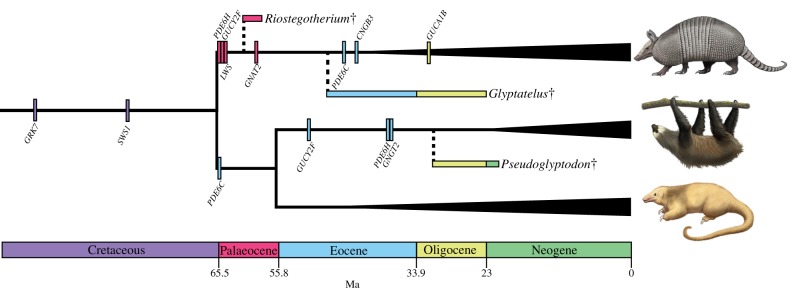
A timetree depicting the loss of photoreceptor genes in xenarthrans. The origins of crown armadillos, sloths and anteaters are indicated by the vertical widening of the branches leading to their representative taxa. Dates for crown Xenarthra, Pilosa (anteaters and sloths), Vermilingua (anteaters) and Folivora (sloths) are derived from [31]; date for crown armadillos is from [48]. Small vertical bars correspond to the averaged inactivation estimates for each gene (electronic supplementary material, table S4). Horizontal bars indicate geological ranges of the oldest xenarthran (Riostegotherium), glyptodont (Glyptatelus) and pilosan fossils (Pseudoglyptodon). Dashed branches arbitrarily connect to the earliest occurrence of extinct taxa and should not be interpreted as divergence time estimates. Colours of horizontal and vertical bars correspond to the colours of the geological strata at the bottom of the figure. Daggers indicate extinct taxa. Note: GUCA1B was demonstrated to be inactivated only in Dasypus novemcinctus, not other armadillos. Paintings by Carl Buell (copyright John Gatesy).
Dasypus novemcinctus, C. hoffmanni and M. darwinii share one premature stop codon in exon 4 of PDE6C (TGA), and the former two share a stop codon in exon 5 (TGA; no BLAST results for M. darwinii). As inactivated PDE6C leads to rod monochromacy in vertebrates [41–43], these shared mutations suggest that rod monochromacy originated in an ancestor to Xenarthra. To test this hypothesis, we performed PCR and successfully sequenced exons 4 and 5 in 10 and 9 xenarthrans, respectively. PDE6C is inactivated in both Choloepus species, B. tridactylus, D. novemcinctus, E. sexcinctus, C. villosus, P. maximus and T. matacus (figure 1; electronic supplementary material, table S2), indicating rod monochromacy is present in all of these species. However, the putative shared mutations appear to be convergent as they are absent in all armadillos that were examined except D. novemcinctus (electronic supplementary material, table S2). Nonetheless, four sloth species share stop codons in PDE6C (figure 1; electronic supplementary material, table S3), and we estimate this gene was inactivated in the common ancestor of Pilosa (anteaters + sloths) shortly after this lineage diverged from cingulates near the Cretaceous–Palaeogene (K-Pg) boundary (figure 3; electronic supplementary material, table S4). This estimate predates the earliest unambiguous pilosan fossils (31.5 Ma, Pseudoglyptodon spp. [57]) and suggests that all known extinct and extant sloths and anteaters were/are rod monochromats (figure 3). However, exon 4 of PDE6C is intact in Cyclopes didactylus and M. tridactyla (electronic supplementary material, dataset S1), so complete sequences from anteaters will be required to test this hypothesis. No inactivating mutations in exons 4 and 5 of PDE6C were shared by all armadillos (electronic supplementary material, table S3), but our estimates for the inactivation of SWS1 (80.06 Ma), LWS (65.43 Ma), GNAT2 (59.55 Ma), PDE6C (45.7 Ma) and CNGB3 (43.65 Ma; electronic supplementary material, table S4) all predate crown armadillos (41.4 Ma [48]) and the earliest fossils of the two major extinct cingulate lineages: pampatheres (16 Ma, Scirrotherium; reviewed in [58]) and glyptodonts (48.6 Ma, Glyptatelus [10], except PDE6C and CNGB3). This suggests that rod monochromacy was/is present in all of these taxa (figure 3).
Rod monochromacy is characterized by the complete absence of cones and results in complete colourblindness with poor visual acuity in dim-light and total blindness in bright-light conditions. As a result, xenarthrans probably use vision only at night, twilight and in burrows, though species that dwell in the understory of South America's rainforests may experience low enough levels of light during the day to facilitate limited vision. Extinct glyptodonts might have compensated for their presumed inability to see approaching predators with their tough carapace and enormous size. Burrowing armadillos, ground sloths and pampatheres might have been pre-adapted to the low-light conditions underground. Additionally, as xenarthrans are frequently the victims of vehicular collisions [17], awareness of their degenerate vision should aid in their conservation.
4. The xenarthran subterranean bottleneck
Rod monochromacy represents an extreme retinal adaptation to dim-light conditions because rods, not cones, are activated when very few photons are available. Consistent with this hypothesis, it has only been discovered in deep-sea fishes [59], deep diving whales [6] and subterranean vertebrates [7,60]. Therefore a long history of extreme dim-light conditions is predicted to eliminate the function of cones via directional selection for a higher density of rods or relaxed selection on the maintenance of cones. We propose that the loss of SWS1 and GRK7 in stem xenarthrans, and the subseqent, independent loss of cones in pilosans and armadillos, respectively, is a consequence of early xenarthrans passing through a subterranean bottleneck.
To our knowledge, Simpson [61] was the first to suggest that the last common ancestor of xenarthrans (‘edentates’) was fossorial. Molecular timetrees suggest that xenarthrans last shared a common ancestor near the K-Pg boundary [31,48]. Fossoriality in Mesozoic mammals is not without precedent and several lineages of Mesozoic synapsids are inferred to have exhibited burrowing behaviour [62–65]. Robertson et al. [66] suggested that mammals survived the mass extinction event at the K–Pg boundary, in part, by sheltering themselves from stressful conditions (e.g. infrared radiation resulting from the Chicxulub impact) in underground burrows. The earliest xenarthran fossils (Middle Palaeocene) show fossorial limb adaptations [67] and extant armadillos and many extinct xenarthrans display(ed) fossorial behaviours and/or adaptations [68–74]. Though extant anteaters and sloths are terrestrial to arboreal, xenarthran synapomorphies include features that reflect a fossorial ancestry: strongly curved claws; a secondary scapular spine, allowing for a stronger retraction of the humerus [75]; plus a synsacrum and lateral accessory articulations of the lumbar vertebrae, which help stabilize the body while digging [76,77]. Besides xenarthrans, the latter character is present only in the subterranean Mesozoic mammal Fruitafossor [64].
The morphological and palaeontological evidence of ancestral fossoriality, coupled with the loss of SWS1 and GRK7 in a stem xenarthran and rod monochromacy in early cingulates and pilosans, argues for a subterranean lifestyle in the earliest xenarthrans. We suggest that passage through this hypothesized subterranean bottleneck is a historical contingency that constrained xenarthran evolution. Specifically, rod monochromacy and modifications to the postcranium related to fossoriality probably prevented diversification into various locomotory types (e.g. gliding, flying, running) and feeding habits (e.g. active predation), and canalized tree sloths to convergently adopt a suspensory posture [77]. The presence of rod monochromacy in xenarthrans should be taken into account in future behavioural, ecological and conservation studies involving this enigmatic lineage of mammals.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank four anonymous reviewers for comments on an earlier stage of our manuscript. Choloepus hoffmanni, M. pentadactyla and P. macrocephalus genomes were generated by Richard K. Wilson, Wesley Warren and The Genome Institute, Washington University School of Medicine.
Data accessibility
New DNA sequences were deposited in GenBank (KP096697–KP096713). Accession numbers for all sequences can be found in the electronic supplementary material, table S6. DNA alignments uploaded as electronic supplementary material, dataset S1.
Funding statement
This research was funded by NSF Grant EF0629860 (M.S.S.) and an American Society of Mammalogists Grant-In-Aid of Research (C.A.E.).
References
- 1.Ahnelt PK, Kolb H. 2000. The mammalian photoreceptor mosaic-adaptive design. Prog. Retin. Eye Res. 19, 711–777. ( 10.1016/S1350-9462(00)00012-4) [DOI] [PubMed] [Google Scholar]
- 2.Davies WIL, Collin SP, Hunt DM. 2012. Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158. ( 10.1111/j.1365-294X.2012.05617.x) [DOI] [PubMed] [Google Scholar]
- 3.Jacobs GH. 2013. Losses of functional opsin genes, short-wavelength cone photopigments, and color vision: a significant trend in the evolution of mammalian vision. Vis. Neurosci. 30, 39–53. ( 10.1017/S0952523812000429) [DOI] [PubMed] [Google Scholar]
- 4.Hunt DM, Peichl L. 2013. S cones: evolution, retinal distribution, development, and spectral sensitivity. Vis. Neurosci. 31, 115–138. ( 10.1017/S0952523813000242) [DOI] [PubMed] [Google Scholar]
- 5.Gerkema MP, Davies WIL, Foster RG, Menaker M, Hut RA. 2013. The nocturnal bottleneck and the evolution of activity patterns in mammals. Proc. R. Soc. B 280, 20130508 ( 10.1098/rspb.2013.0508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9, e1003432 ( 10.1371/journal.pgen.1003432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emerling CA, Springer MS. 2014. Eyes underground: regression of visual protein networks in subterranean mammals. Mol. Phylogenet. Evol. 78C, 260–270. ( 10.1016/j.ympev.2014.05.016) [DOI] [PubMed] [Google Scholar]
- 8.Murphy WJ, et al. 2001. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294, 2348–2351. ( 10.1126/science.1067179) [DOI] [PubMed] [Google Scholar]
- 9.Gaudin TJ, McDonald HG. 2008. Morphology-based investigations of the phylogenetic relationships among extant and fossil xenarthrans. In The biology of Xenarthra (eds Vizcaino SF, Loughry WJ.), pp. 24–36. Gainesville, FL: University Press of Florida. [Google Scholar]
- 10.McKenna MC, Bell SK. 1997. Classification of mammals above the species level. New York, NY: Columbia University Press. [Google Scholar]
- 11.Flynn JJ, Wyss AR. 1998. Recent advances in South American mammalian paleontology. Trends Ecol. Evol. 13, 449–454. ( 10.1016/S0169-5347(98)01457-8) [DOI] [PubMed] [Google Scholar]
- 12.Jones KE, et al. 2009. PanTHERIA: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 ( 10.1890/08-1494.1) [DOI] [Google Scholar]
- 13.Newman HH. 1913. The natural history of the nine-banded armadillo of Texas . Am. Nat. 47, 513–539. ( 10.1086/279370) [DOI] [Google Scholar]
- 14.Goffart M. 1971. Function and form in the sloth, vol. 34 Oxford, UK: Pergamon. [Google Scholar]
- 15.Mendel F, Piggins D, Fish D. 1985. Vision of two-toed sloths (Choloepus). J. Mammal. 66, 197–200. ( 10.2307/1380987) [DOI] [Google Scholar]
- 16.Eisenberg JF, Redford KH. 1999. Mammals of the Neotropics (volume 3): the Central Neotropics—Ecuador, Peru, Bolivia, Brazil. Chicago, IL: University of Chicago Press. [Google Scholar]
- 17.de Carvalho Oliveira L, Mendel S, Loretto D, de Sousa e Silva Junior J, Fernandes G. 2006. Edentates of the Saracá-Taquera National Forest, Pará, Brazil. Edentata 3–7.
- 18.de Sampaio C, Camilo-Alves P, de Miranda Mourão G. 2006. Responses of a specialized insectivorous mammal (Myrmecophaga tridactyla) to variation in ambient temperature. Biotropica 38, 52–56. [Google Scholar]
- 19.Wislocki GB. 1928. Observations on the gross and microscopic anatomy of the sloths (Bradypus griseus griseus Gray and Choloepus hoffmanni Peters). J. Morphol. 46, 317–397. ( 10.1002/jmor.1050460202) [DOI] [Google Scholar]
- 20.Walls GL. 1942. The vertebrate eye and its adaptive radiation. London, UK: Hafner Publishing Company. [Google Scholar]
- 21.Watillon M, Goffart M. 1969. The eye of the sloth (Choloepus hoffmanni Peters). Acta Zool. Pathol. Antverp. 49, 107–122. [PubMed] [Google Scholar]
- 22.Piggins D, Muntz WRA. 1985. The eye of the three-toed sloth. In The evolution and ecology of armadillos, sloths and vermilinguas, pp. 191–197. Washington, DC: Smithsonian Institution Press. [Google Scholar]
- 23.Tan Y, Li WH. 1999. Trichromatic vision in prosimians. Nature 402, 36 ( 10.1038/46947) [DOI] [PubMed] [Google Scholar]
- 24.Parry JWL, Bowmaker JK. 2002. Visual pigment coexpression in guinea pig cones: a microspectrophotometric study. Invest. Ophthalmol. Vis. Sci. 43, 1662–1665. [PubMed] [Google Scholar]
- 25.Zhao H, Rossiter SJ, Teeling EC, Li C, Cotton JA, Zhang S. 2009. The evolution of color vision in nocturnal mammals. Proc. Natl Acad. Sci. USA 106, 8980–8985. ( 10.1073/pnas.0813201106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Heled J. 2012. Geneious v. 5.6.5 See http://www.geneious.com.
- 27.Rambaut A. 1996. Se-Al: Sequence Alignment editor See http://tree.bio.ed.ac.uk/software/seal/.
- 28.Edgar RC. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burset M, Seledtsov IA, Solovyev VV. 2000. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28, 4364–4375. ( 10.1093/nar/28.21.4364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, Springer MS. 2009. Molecular decay of the tooth gene Enamelin (ENAM) mirrors the loss of enamel in the fossil record of placental mammals. PLoS Genet. 5, e1000634 ( 10.1371/journal.pgen.1000634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meredith RW, et al. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science 334, 521–524. ( 10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 32.Invergo B, Montanucci L, Laayouni H, Bertranpetit J. 2013. A system-level, molecular evolutionary analysis of mammalian phototransduction. BMC Evol. Biol. 13, 52 ( 10.1186/1471-2148-13-52) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kohl S, Marx T, Giddings I, Jägle H, Jacobson SG, Apfelstedt-Sylla E, Zrenner E, Sharpe LT, Wissinger B. 1998. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nat. Genet. 19, 257–259. ( 10.1038/935) [DOI] [PubMed] [Google Scholar]
- 34.Reicher S, Seroussi E, Gootwine E. 2010. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics 95, 101–104. ( 10.1016/j.ygeno.2009.10.003) [DOI] [PubMed] [Google Scholar]
- 35.Johnson S, Michaelides M, Aligianis IA, Ainsworth JR, Mollon JD, Maher ER, Moore AT, Hunt DM. 2004. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J. Med. Genet. 41, 1–5. ( 10.1136/jmg.2003.011437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohl S, et al. 2000. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 9, 2107–2116. ( 10.1093/hmg/9.14.2107) [DOI] [PubMed] [Google Scholar]
- 37.Sidjanin DJ, Lowe JK, McElwee JL, Milne BS, Phippen TM, Sargan DR, Aguirre GD, Acland GM, Ostrander EA. 2002. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 11, 1823–1833. ( 10.1093/hmg/11.16.1823) [DOI] [PubMed] [Google Scholar]
- 38.Kohl S, Baumann B, Rosenberg T, Kellner U, Lorenz B, Vadalà M, Jacobson SG, Wissinger B. 2002. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am. J. Hum. Genet. 71, 422–425. ( 10.1086/341835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang B, Dacey MS, Hawes NL, Hitchcock PF, Milam AH, Atmaca-Sonmez P, Nusinowitz S, Heckenlively JR. 2006. Cone photoreceptor function loss-3, a novel mouse model of achromatopsia due to a mutation in Gnat2. Invest. Ophthalmol. Vis. Sci. 47, 5017–5021. ( 10.1167/iovs.05-1468) [DOI] [PubMed] [Google Scholar]
- 40.Akhmedov NB, Piriev NI, Pearce-Kelling S, Acland GM, Aguirre GD, Farber DB. 1998. Canine cone transducin-gamma gene and cone degeneration in the cd dog. Invest. Ophthalmol. Vis. Sci. 39, 1775–1781. [PubMed] [Google Scholar]
- 41.Stearns G, Evangelista M, Fadool JM, Brockerhoff SE. 2007. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J. Neurosci. 27, 13 866–13 874. ( 10.1523/JNEUROSCI.3136-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thiadens AAHJ, et al. 2009. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am. J. Hum. Genet. 85, 240–247. ( 10.1016/j.ajhg.2009.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang B, et al. 2009. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc. Natl Acad. Sci. USA 106, 19 581–19 586. ( 10.1073/pnas.0907720106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohl S, et al. 2012. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am. J. Hum. Genet. 91, 527–532. ( 10.1016/j.ajhg.2012.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikonov SS, et al. 2013. Cones respond to light in the absence of transducin β subunit. J. Neurosci. 33, 5182–5194. ( 10.1523/JNEUROSCI.5204-12.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keers R, et al. 2011. Variation in GNB3 predicts response and adverse reactions to antidepressants. J. Psychopharmacol. 25, 867–874. ( 10.1177/0269881110376683) [DOI] [PubMed] [Google Scholar]
- 47.Kumar R, Kohli S, Alam P, Barkotoky R, Gupta M, Tyagi S, Jain SK, Pasha MAQ. 2013. Interactions between the FTO and GNB3 genes contribute to varied clinical phenotypes in hypertension. PLoS ONE 8, e63934 ( 10.1371/journal.pone.0063934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delsuc F, Superina M, Tilak M-K, Douzery EJP, Hassanin A. 2012. Molecular phylogenetics unveils the ancient evolutionary origins of the enigmatic fairy armadillos. Mol. Phylogenet. Evol. 62, 673–680. ( 10.1016/j.ympev.2011.11.008) [DOI] [PubMed] [Google Scholar]
- 49.Carleton KL, Spady TC, Cote RH. 2005. Rod and cone opsin families differ in spectral tuning domains but not signal transducing domains as judged by saturated evolutionary trace analysis. J. Mol. Evol. 61, 75–89. ( 10.1007/s00239-004-0289-z) [DOI] [PubMed] [Google Scholar]
- 50.Davies WIL, et al. 2012. Next-generation sequencing in health-care delivery: lessons from the functional analysis of rhodopsin. Genet. Med. 14, 891–899. ( 10.1038/gim.2012.73) [DOI] [PubMed] [Google Scholar]
- 51.Naash MI, Hollyfield JG, Al-Ubaidi MR, Baehr W. 1993. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc. Natl Acad. Sci. USA 90, 5499–5503. ( 10.1073/pnas.90.12.5499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dryja T, McGee T, Reichel E, Hahn L, Cowley G, Yandell D, Sandberg M, Berson E. 1990. A point mutation in the rhodopsin gene in one form of retinitis pigmentosa. Nature 343, 364–366. ( 10.1038/343364a0) [DOI] [PubMed] [Google Scholar]
- 53.Kaushal S, Khorana HG. 1994. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 33, 6121–6128. ( 10.1021/bi00186a011) [DOI] [PubMed] [Google Scholar]
- 54.Krebs MP, Holden DC, Joshi P, Clark CL, Lee AH, Kaushal S. 2010. Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. J. Mol. Biol. 395, 1063–1078. ( 10.1016/j.jmb.2009.11.015) [DOI] [PubMed] [Google Scholar]
- 55.Heesy CP, Hall MI. 2010. The nocturnal bottleneck and the evolution of mammalian vision. Brain. Behav. Evol. 75, 195–203. ( 10.1159/000314278) [DOI] [PubMed] [Google Scholar]
- 56.Hall MI, Kamilar JM, Kirk EC. 2012. Eye shape and the nocturnal bottleneck of mammals. Proc. R. Soc. B 279, 4962–4968. ( 10.1098/rspb.2012.2258) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenna M, Wyss A, Flynn J. 2006. Paleogene pseudoglyptodont xenarthrans from central Chile and Argentine Patagonia. Am. Museum Novit. 3536, 1–18. ( 10.1206/0003-0082(2006)3536[1:PPXFCC]2.0.CO;2) [DOI] [Google Scholar]
- 58.Scillato-Yané GJ, Carlini AA, Tonni EP, Noriega JI. 2005. Paleobiogeography of the late Pleistocene pampatheres of South America. J. South Am. Earth Sci. 20, 131–138. ( 10.1016/j.jsames.2005.06.012) [DOI] [Google Scholar]
- 59.Douglas RJ, Partridge JH, Hope AC. 1995. Visual and lenticular pigments in the eyes of demersal deep-sea fishes. J. Comp. Physiol. A 177, 111–122. ( 10.1007/BF00243403) [DOI] [Google Scholar]
- 60.Mohun SM, Davies WL, Bowmaker JK, Pisani D, Himstedt W, Gower DJ, Hunt DM, Wilkinson M. 2010. Identification and characterization of visual pigments in caecilians (Amphibia: Gymnophiona), an order of limbless vertebrates with rudimentary eyes. J. Exp. Biol. 213, 3586–3592. ( 10.1242/jeb.045914) [DOI] [PubMed] [Google Scholar]
- 61.Simpson G. 1931. Metacheiromys and the Edentata. Bull. Am. Museum Nat. Hist. 59, 295–381. [Google Scholar]
- 62.Groenewald G. 1991. Burrow casts from the Lystrosaurus–Procolophon Assemblage-zone, Karoo Sequence, South Africa. Koedoe-African Prot. Area Conserv. Sci. 34 13–22. [Google Scholar]
- 63.Damiani R, Modesto S, Yates A, Neveling J. 2003. Earliest evidence of cynodont burrowing. Proc. R. Soc. Lond. B 270, 1747–1751. ( 10.1098/rspb.2003.2427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo Z-X, Wible JR. 2005. A Late Jurassic digging mammal and early mammalian diversification. Science 308, 103–107. ( 10.1126/science.1108875) [DOI] [PubMed] [Google Scholar]
- 65.Fernandez V, Abdala F, Carlson KJ, Cook DC, Rubidge BS, Yates A, Tafforeau P. 2013. Synchrotron reveals early Triassic odd couple: injured amphibian and aestivating therapsid share burrow. PLoS ONE 8, e64978 ( 10.1371/journal.pone.0064978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robertson DS, McKenna MC, Toon OB, Hope S, Lillegraven JA. 2004. Survival in the first hours of the Cenozoic. Geol. Soc. Am. Bull. 116, 760–768. ( 10.1130/B25402.1) [DOI] [Google Scholar]
- 67.Bergqvist L, Abrantes E, Avilla L. 2004. The Xenarthra (Mammalia) of São José de Itaboraí Basin (Upper Paleocene, Itaboraian), Rio de Janeiro, Brazil. Geodiversitas 26 323–337. [Google Scholar]
- 68.Bargo M, Vizcaíno S, Archuby FM, Blano RE. 2000. Limb bone proportions, strength and digging in some Lujanian (Late Pleistocene–Early Holocene) mylodontid ground sloths (Mammalia, Xenarthra). J. Vertebr. Paleontol. 20, 601–610. ( 10.1671/0272-4634(2000)020) [DOI] [Google Scholar]
- 69.Vizcaíno S, Zárate M. 2001. Pleistocene burrows in the Mar del Plata area (Argentina) and their probable builders. Acta Palaeontol. Pol. 46 289–301. [Google Scholar]
- 70.Vizcaíno SF, Bargo MS, Kay RF, Milne N. 2006. The armadillos (Mammalia, Xenarthra, Dasypodidae) of the Santa Cruz Formation (early–middle Miocene): an approach to their paleobiology. Palaeogeogr. Palaeoclimatol. Palaeoecol. 237, 255–269. ( 10.1016/j.palaeo.2005.12.006) [DOI] [Google Scholar]
- 71.Dondas A, Isla FI, Carballido JL. 2009. Paleocaves exhumed from the Miramar Formation (Ensenadan Stage-age, Pleistocene), Mar del Plata, Argentina. Quat. Int. 210, 44–50. ( 10.1016/j.quaint.2009.07.001) [DOI] [Google Scholar]
- 72.Blanco RE, Rinderknecht A. 2012. Fossil evidence of frequency range of hearing independent of body size in South American Pleistocene ground sloths (Mammalia, Xenarthra). Comptes Rendus Palevol 11, 549–554. ( 10.1016/j.crpv.2012.07.003) [DOI] [Google Scholar]
- 73.Genise JF, Farina JL. 2012. Ants and xenarthrans involved in a Quaternary food web from Argentina as reflected by their fossil nests and palaeocaves. Lethaia 45, 411–422. ( 10.1111/j.1502-3931.2011.00301.x) [DOI] [Google Scholar]
- 74.Toledo N, Bargo MS, Cassini GH, Vizcaíno SF. 2012. The forelimb of early Miocene sloths (Mammalia, Xenarthra, Folivora): morphometrics and functional implications for substrate preferences. J. Mamm. Evol. 19, 185–198. ( 10.1007/s10914-012-9185-2) [DOI] [Google Scholar]
- 75.McDonald HG. 2003. Xenarthran skeletal anatomy: primitive or derived? Senckenb. Biol. 83 5–17. [Google Scholar]
- 76.Gaudin TJ, Biewener AA. 1992. The functional morphology of xenarthrous vertebrae in the armadillo Dasypus novemcinctus (Mammalia, Xenarthra). J. Morphol. 214, 63–81. ( 10.1002/jmor.1052140105) [DOI] [PubMed] [Google Scholar]
- 77.Nyakatura JA. 2012. The convergent evolution of suspensory posture and locomotion in tree sloths. J. Mamm. Evol. 19, 225–234. ( 10.1007/s10914-011-9174-x) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New DNA sequences were deposited in GenBank (KP096697–KP096713). Accession numbers for all sequences can be found in the electronic supplementary material, table S6. DNA alignments uploaded as electronic supplementary material, dataset S1.