Abstract
In this report, we examine the hypothesis that the drivers of latitudinal selection observed in the eastern US Drosophila melanogaster populations are reiterated within seasons in a temperate orchard population in Pennsylvania, USA. Specifically, we ask whether alleles that are apparently favoured in northern populations are also favoured early in the spring, and decrease in frequency from the spring to autumn with the population expansion. We use SNP data collected for 46 metabolic genes and 128 SNPs representing the central metabolic pathway and examine for the aggregate SNP allele frequencies whether the association of allele change with latitude and that with increasing days of spring–autumn season are reversed. Testing by random permutation, we observe a highly significant negative correlation between these associations that is consistent with this expectation. This correlation is stronger when we confine our analysis to only those alleles that show significant latitudinal changes. This pattern is not caused by association with chromosomal inversions. When data are resampled using SNPs for amino acid change the relationship is not significant but is supported when SNPs associated with cis-expression are only considered. Our results suggest that climate factors driving latitudinal molecular variation in a metabolic pathway are related to those operating on a seasonal level within populations.
Keywords: natural selection, metabolic genes, clines, seasonal selection
1. Introduction
Patterns of geographical variation in molecular polymorphisms are commonly used to understand adaptive change. One of the most familiar patterns is the systematic spatial change of allele frequencies in concordance with climatic variables in the form of latitudinal clines. Over the past four decades, many geographical clines in molecular polymorphism have been reported in many species (e.g. [1–7]), and these observations are often taken prima facie as evidence of natural selection.
While geographical clines are an attractive argument for the action of natural selection, it is known that isolation-by-distance, colonization fronts and historical admixture are potential non-adaptive processes well capable of generating patterns of spatial change [8–10]. Rejecting these non-adaptive alternatives is nearly impossible, since we possess poor information on realistic selection coefficients, rates of migration, population sizes and changes, and the sequences of unique events involved in the historical founding of populations. What is needed is an independent parallel assessment of the action of selection, one that is free of these confounding issues. Our study of the polymorphism for diapause capability in Drosophila melanogaster [11] and the associated polymorphism in the couch potato gene [12] in the eastern US demonstrated a statistically significant seasonal change, where the northern diapause allele frequency was highest early in the season and dropped in frequency from the summer into the autumn [13]. The estimated selection intensity associated with this seasonal change in allele frequency was high.
One question is whether such seasonal change that is consistent with selection against northern favoured alleles is a general feature of other polymorphisms. To address this question, we examine the set of genes of central metabolism reported in a previous paper [14]. We hypothesize that latitudinal clines in SNPs result from overwintering selection favouring particular alleles in temperate northern populations. This overwintering selection increases in intensity northward along the latitudinal climatic gradient and is the causal force behind the many common clines seen in the USA. It is also believed that local temperate populations of D. melanogaster are reseeded from the remnants of local overwintering individuals because the local estimates from lethal allelism are in the several thousand [15,16]; and the surviving individuals should be northern genotypes. After the winter, these populations are then subject to selection for alleles that are favoured under summer conditions (i.e. the southern-associated alleles in the cline). Framed in this manner, a working hypothesis is that the signs of the latitudinal and seasonal (monthly) correlations with any arbitrary SNP allele frequency generally will be inverted.
In this report, we specifically address this hypothesis by studying these associations in enzymes representing the central metabolic pathway for which we possess detailed data on latitudinal variation [14], as well as seasonal collections from the same temperate population in Pennsylvania studied previously [13]. For 128 SNPs embedded in 46 genes, we pair for each SNP the correlation of latitudinal variation with the correlation of seasonal monthly change and test the overall aggregate rank correlation by random permutation. We find a highly significant negative association between those alleles increasing with latitude to the north and the change in frequency of those same alleles throughout the summer population expansion. Our results strongly suggest that natural selection across populations appears to be reiterated seasonally within this temperate population.
2. Material and methods
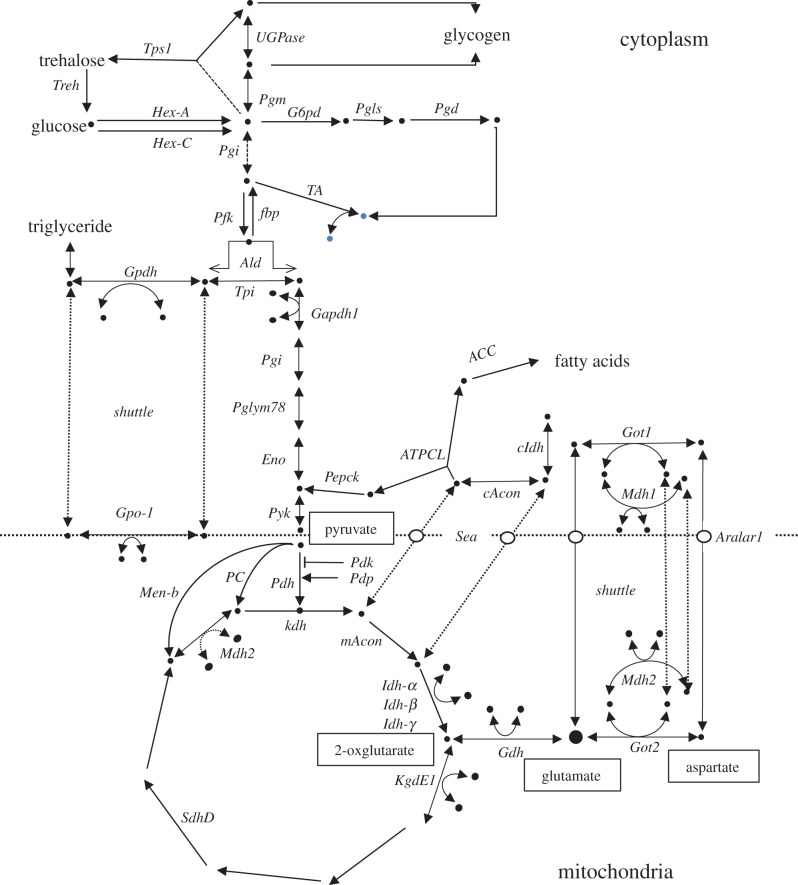
We already possess data on 128 SNPs in 46 central metabolic genes that have been screened via bulk pyrosequencing, and these are reported in the supplemental table in a previous paper [14]. The central metabolic pathway positions of the sampled genes are shown in figure 1. As a source for SNP identification, we used the 37 D. melanogaster genome sequences released in August 2010 by the D. melanogaster Genetic Reference Panel (DGRP) [18]. These sequences had already been assembled and annotated to the FlyBase reference sequence (v. 5.12). The identification of SNPs was carried out with gene-by-gene manual inspection of coding regions. In each case, the quality score for each base was assessed, and SNPs where the minority allele quality score was less than 30 were reset to the majority allele (most of these cases involve singletons). Finally, focal SNP polymorphisms for the cline were selected where the minority allele was more than 0.10 in the DGRP.
Figure 1.
The generalized central metabolic pathway adapted and modified from a previous study [14]. It shows the pathway context of all the genes reported in the study. The genes for Men, Men-b, Gpt and Adh have been omitted to reduce complexity. The positions of metabolites with high connectivity (pyruvate, glutamate, 2-oxglutare and aspartate) [17] are shown. (Online version in colour.)
Many SNPs could not be screened by pyrosequencing because flanking polymorphisms bias amplification in the bulk preparation, or the pyrosequencing step simply failed. Amino acid polymorphisms and SNPs that were diagnostic of important haplotypes seen in the DGRP collection were favoured. Attempts were made to minimize LD among sites by spacing SNPs at distances of more than 500 bp, when possible.
Bulk DNA purification was performed with Puregene Core Kit A (Qiagen) using 42–100 flies per population (see line sampling below). We used pyrosequencing in the bulk DNA preparations to estimate SNP frequency [19,20]. The precision of the method was evaluated by comparing the estimated frequency of each SNP after pyrosequencing to the expected frequency for the DGRP population based on the genome sequences (r = 0.99). Pyrosequencing was carried out using PyroMark MD and peak heights scored to estimate SNP frequency. Primers were designed using PyroMark Assay Design software.
The geographical collections are 20 local populations sampled along a latitudinal gradient in the eastern USA from Florida to Maine [14] during the period of 2005–2012 (but largely in 2009 and 2010). The seasonal data are from 11 collections sampled in the Linvilla orchard population in Media, Pennsylvania, USA in 2009–2011 in the months of June to November. Sample sizes and collection dates for the seasonal collections are given by Cogni et al. [13]. For each collection, males were preserved in 95% ethanol and stored at −70°C. Females were allowed to oviposit, preserved and stored. The progeny from the F1 generation were preserved in EtOH, and two female progeny were sampled from each preserved line and pooled with the earlier collected males for bulk pyrosequencing. By sampling two progeny per line in the F1 generation, we are sampling 2–4 independent chromosomes from the population per line, with an average of 3. The expected number of autosomal chromosomes per bulked sample is therefore three times the number of female lines plus twice the number of wild-collected males. The average number of independent genomes pooled per population sample was n = 114.7. The entire database thus consisted of 2524 genomes.
To assess latitudinal clines, allele SNP frequencies were used in a linear regression against degrees latitude and the Pearson product-moment correlation (r) was computed. To assess importance, given multiple testing, the entire set was tested for set-wide significance using q-values [21], and 4% was assigned as a cutoff as support of a cline in each SNP. The q-value cutoffs were used to reduce the dataset to the SNPs most likely to represent latitudinal clines.
For seasonal data, we computed the Pearson correlation of the same SNP allele against days after 31 May. Finally, recognizing the non-normality of the distribution of the Pearson correlation coefficients, especially in the reduced subsets, we computed the Spearman rank correlation of the gene-wise paired latitude and seasonal correlations using the ‘corr’ program in R. The significance of each Spearman rank correlation is evaluated in all cases by full random permutation of the elements of the seasonal vector, and computation of this null correlation (against latitude) 10 000 times.
Previously [14], we explored the latitudinal–seasonal correlation for SNPs that were associated with cis-expression effects. This used the whole-adult Affymetrix Drosophila 2.0 array expression data (accession number E-MEXP-1594) reported by Ayroles et al. [22] for the 37 sequences in the Genetic Reference Panel [18]. Probes with underlying SNPs were removed or masked [23,24], the sex-effect for each gene was removed, and the residuals were rescaled to standardized deviates. A nested ANOVA is carried out on the standardized residuals to estimate SNP allele effects using the JMP program (JMP-SAS).
3. Results
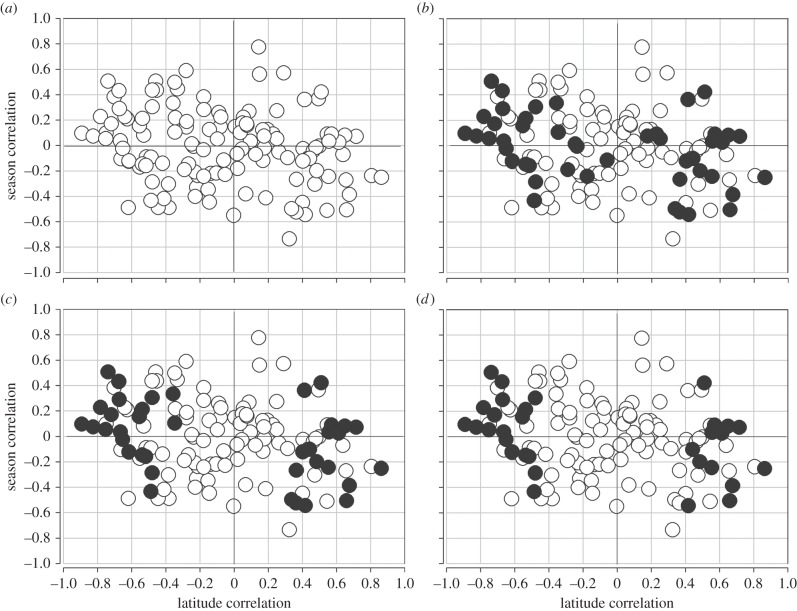
There are statistically significant latitudinal clines with p-values of less than 0.05 in 53 SNPs in 31 of 46 genes [14]. Electronic supplementary material, table S1, provides the observed seasonal SNP frequencies, and electronic supplementary material, table S2, provides the full dataset with chromosome localization, SNP functional state and inversion association. We find only two SNPs that show significant Pearson product–moment correlations with day of collection after 31 May (June to November) in the Pennsylvania orchard, the level we might expect under simple type 1 error of α < 0.05. The seasonal correlations are based on fewer collections (n = 11) than the cline correlations (n = 20), and thus in comparison the seasonal results have a reduced power to detect allele frequency changes. Moreover, in each case we expected the seasonal allele change over about six generations to be smaller than the overall change with latitude. Seasonal changes in allele frequencies average about half the overall clinal changes. Nevertheless, in aggregate we see a pattern where SNP alleles that increase in frequency with increasing latitude also decrease in frequency during the summer period. Figure 2a shows the correlation for all 128 SNPs surveyed in the study. The overall Spearman rank correlation of seasonal- and latitudinal-frequency correlations is r = −0.222 with p < 0.007.
Figure 2.
Correlations between latitudinal change and seasonal change in the same allele. (a) All 128 SNPs across 46 genes (r = −0.222, p < 0.007). (b) Correlation using just the highest latitudinal correlation SNP in each of n = 46 genes (r = −0.398, p < 0.0042). (c) Correlation using clines considered significant at q < 0.10, n = 38 (r = −0.402, p < 0.0076). (d) Correlation using the clines considered significant at q < 0.05, one per gene, n = 31 (r = −0.479, p < 0.0032).
Most genes possess multiple SNPs in the screen, and because of linkage disequilibrium some SNPs within genes are not independent. To establish the average correlation for datasets of only a single SNP sampled at random per gene, we reduced the data to sets of 46 SNPs and assembled 10 000 such datasets, recognizing that these sets are not independent. The average of these 10 000 correlation sets was r = −0.283. About 50% of these sets would be significant at α < 0.05.
To emphasize those genes that represent the strongest supported cases of latitudinal selection, we sampled the SNP at each gene with the highest correlation with latitude, irrespective of statistical significance. There is no reason to expect that selecting the SNP with the highest latitudinal correlation in each gene (n = 46) should favour those SNPs with the strongest seasonal change. We see a highly significant Spearman rank correlation with this reduced set, r = −0.398 (p < 0.0042; figure 2b). We also considered the case where we selected the SNP from each gene with the highest seasonal Pearson correlation. The Spearman correlation for this set (n = 46) was r = −0.311 (p < 0.0159). Finally, to reduce the set to those genes that provide yet the best support for true latitudinal clines, we selected only those SNPs (one per gene) that were statistically significant for latitudinal clines given q-value thresholds [13,21], where q < 0.10 (n = 38) and q < 0.05 (n = 31) (figure 2c,d). For the former, the Spearman rank correlation is now r = −0.402 (p < 0.0076) and for the latter r = −0.479 (p < 0.0032). The pattern remains as we increase the evidence of latitudinal clines (yet reduce the degrees-of-freedom and power). It appears that alleles that are significantly favoured in northern populations are favoured early after the winter in the PA orchard and decline with the summer into the autumn season.
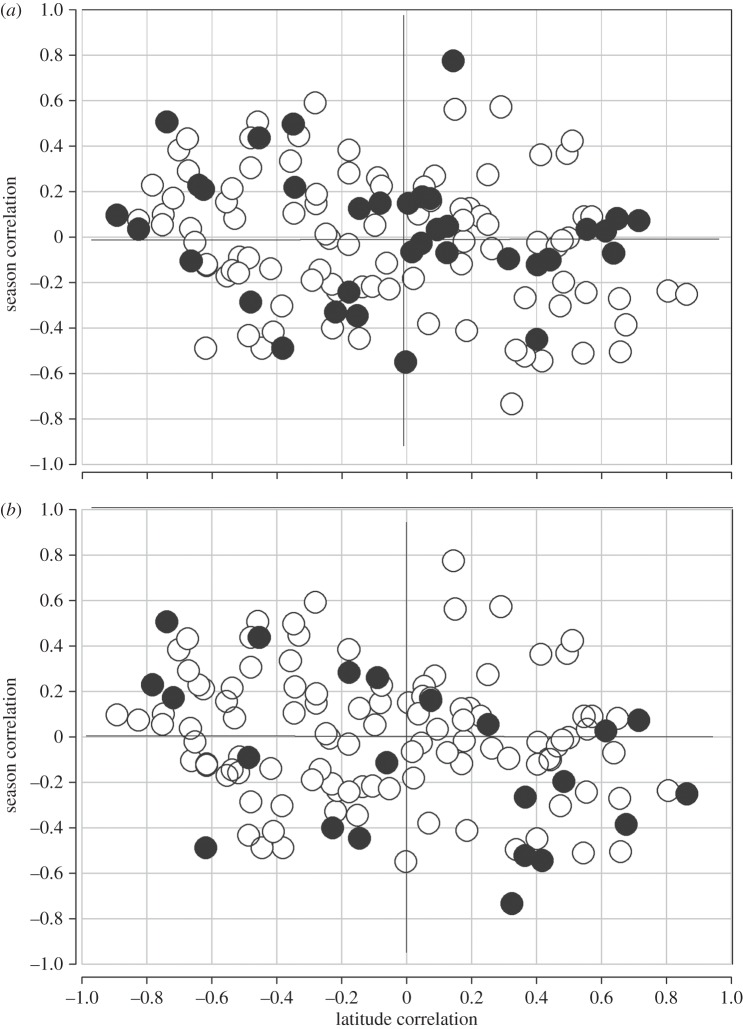
Finally, to functionally explore this feature, we partitioned the data into subsets based on two properties. The first set included those clines associated with amino acid polymorphism, and the second included those SNPs shown to have significant associations with transcript expression [14,22,25]. The average correlation using all 35 amino acid polymorphisms was r = −0.196 and is non-significant (n = 35, p < 0.22, figure 3a). The correlation among the 22 significant cis-expression-associated SNPs (figure 3b; from [14]) is r = −0.408 (p < 0.0336), but this is not significantly different from a sampling of 22 genes at random from the overall set (p < 0.0918).
Figure 3.
Correlation between latitudinal change and seasonal change in the same allele. (a) All amino acid polymorphisms (n = 35, r = −0.196, p < 0.220). (b) SNPs with significant cis-expression effects, one per gene (n = 22, r = −0.408, p < 0.0336).
Some of the genes lie within cosmopolitan inversions and seasonal cycling of inversions, if present, could drive the correlation. Inversions are uncommon at this latitude [1], but we have examined the Spearman correlations for genes inside and outside inversions. This further reduces the dataset; however, we see that we have 30 genes outside and 16 within inversion spans. The Spearman correlation is r = −0.325, (p < 0.0366) when the 16 inversion-localized genes are excluded. The correlation for genes inside inversions is r = −0.6147 (p < 0.0043).
We wished to determine what genes appear to have the greatest contribution to the observed Spearman correlation in the most stringent set, those 31 genes possessing significant clines at q < 0.05. To evaluate this, we removed each SNP one by one and recomputed the Spearman correlation. The top six with the largest weakening in the correlation were taken as those most strongly associated with the relationship. These genes were Pyk, Tpi, UGPase, Idh-β, Gpdh and Pdk. The direction of change in expression in these genes, with the exception of Gpdh, suggests increases in gene expression in the glycolytic pathway and TCA pathways through the season.
4. Discussion
Local northern temperate populations of D. melanogaster maintain some genetic continuity between years [15,26]; they are not simply recolonized from southern source populations every year. As such, they face regular seasonal challenges associated with winter survival, and these challenges, such as winter duration, increase continuously along a north–south axis. We might expect the same environments that vary latitudinally to also seasonally change within populations. This leads to the expectation that early in the season populations will also bear genetic features that are adaptive to northern populations, and these features should shift to those of southern populations as the season progresses through summer and the populations grow over about six generations. We first observed such a shift in our study of the couch potato gene, as the northern-favoured diapause-associated allele dropped in frequency in accordance with the drop in the seasonal incidence in the diapause trait [13]. We now observe this pattern in our data for the central metabolic genes. Our results for the entire 128 SNPs in a set of 46 metabolic genes in aggregate significantly support this argument because alleles showing positive correlations with latitude generally possess negative associations with collection date in the same Pennsylvania population and vice versa. This observation advances the premise that local seasonally based selection, varying with latitude, may maintain variation in many metabolic genes. The reduction of this set into only those genes with significant latitudinal correlations shows a stronger negative correlation (albeit we are selecting from the extremes of a significant correlation), and emphasizes that it is the genes with significant latitudinal variation that most strongly show the seasonal effect (figure 2c,d).
We should emphasize that this is a pattern that only emerges as the full data are considered in aggregate. While there are many significant latitudinal clines, there are only two cases where seasonal change with month of collection is statistically significant. We feel this reflects the general difference in statistical power associated with the two measures, and difficulty of detecting an allelic change that requires very strong selection to produce a statistically significant seasonal change [13].
It is unlikely that many of the SNPs themselves are the direct targets of natural selection. In some cases, we are looking at non-conservative amino acid polymorphisms of unknown catalytic potential, but most SNPs are synonymous polymorphisms. Nevertheless, in general a large percentage of the silent polymorphisms have strong statistical associations with cis-acting expression variation [14,25] and many are linked to SNPs that have yet stronger expression effects. In only a few cases have we screened SNPs outside coding regions (usually because the coding region possessed little polymorphism), and so we have probably missed important cis-acting polymorphism.
Bergland et al. [27] recently addressed a similar question using next-generation sequencing technology (NGS) and a very different structural database. They screened 500 000 SNPs for seasonal variation across three successive seasons in the same Linvilla orchard, and time period, as our study and from five collection localities from Florida to Maine. They used a conservative statistical filter to identify 1750 candidate seasonally oscillating SNPs and reported a number of features of these genes that were enriched in this filtered group. It is important to emphasize that they found the large majority of latitudinally clinal SNPS are not seasonally cycling; however, with respect to the set of cycling SNPs they found a significant tendency for the winter-favoured allele to be the more common in Maine than Florida and vice versa, which is consistent with our observations. They also reported the enrichments of old balanced polymorphism and proposed a role for seasonally based marginal overdominance in the maintenance of oscillating polymorphisms. Our paper is fundamentally different because we focus on a small but specific set of genes selected for the metabolic pathway, and draw on the entire dataset of 46 genes and 128 SNPs to test our proposition. Our sample possesses more seasonal samples and latitudinal collections. The results of the two analyses are more similar than dissimilar. Where we find limited evidence for seasonal selection per se, consistent with low statistical power for each gene, this is nonetheless sufficient to create the association where alleles that increase in frequency with latitude decrease in frequency through the season. It is possible that the central metabolic pathway genes, with their specific role in metabolism and energy homeostasis and their very high expression levels, may be prone to clinal variation and seasonal cycling.
Drosophila melanogaster populations also harbour chromosome inversion polymorphisms [1,28], and Dobzhansky [29] observed seasonal cycling for the common inversions in Drosophila pseudoobscura. Our inversions are clinal in frequency, and if SNPs are in linkage disequilibrium with these inversions, and the inversions cycle seasonally, this could drive this observation. Both sets incorporating genes either within or outside inversions show significant Spearman correlations, but these are not different from each other. Furthermore, at the latitude of the Pennsylvania population most cosmopolitan inversions are found at very low frequencies of only a few per cent [1], so it is unlikely that inversions drive this observation. Bergland et al. [27] also examined this possibility in this same population and found no notable association of seasonal genes with inversions, and no evidence that inversions even cycle appreciably in this population.
The population structure of D. melanogaster consists of local metapopulations where numbers in orchards reach high abundances during the summer and autumn, only to collapse during the winter. It is generally understood that individuals, mostly females, survive in association with human habitation. These remnants reseed local populations in the spring and the populations expand followed by local migration. In contrast to the hypothesis of long-distance dispersal, one possibility is that early spring populations in orchards are recolonized from local urban refugia. For example, the diapause trait, which also cycles in orchards, is found at much lower levels in urban fruit markets in Philadelphia [30]. Thus, the larger regional population structure might be described as a mosaic, with pockets of urban and rural genotypes. However, Cogni et al. [13] in discussing the seasonal pattern in the couch potato gene polymorphism rejected the idea that large-scale southern migration plays a role in the seasonal change, and Bergland et al. [27] used a series of model simulations to discount the role of local refugia recolonization in creating their consistently oscillating SNPs. Irrespective of how much selection or local migration play in the recovery of southern allele frequencies during the season, the seasonal trend within the PA population must always involve natural selection operating over the overwintering period to reset the allele frequencies with a bias towards northern frequencies. Drift as a source of seasonal change is unrealistic and requires founder sizes much smaller than the estimates based on model simulations [27] and estimates from lethal allelism [15,31]. Finally, long-range migration is insufficient as a general cause of summer rebound in frequencies because, as Bergland et al. [27] point out, most latitudinal SNPs do not cycle and would be expected to do so if such migration were a universal alternative mechanism.
Finally, the study by Lavington et al. [14] placed the study of latitudinal gene expression variation in the context of the pathway and proposed several emergent features. They used a principal component analysis of the latitudinal changes in SNP-associated standardized gene expression to identify the genes in the pathway and its associated branches that are the most probably targets of natural selection. They showed that the targets are concentrated in the genes of the upper glycolytic pathway and pentose shunt, those controlling glycerol shuttle activities, and finally those enzymes associated with the utilization of glutamate and pyruvate. Both metabolites possess high connectivity [17], and thus may be the points where flux balance can be best shifted. They proposed that these points are conserved points associated with coupling energy homeostasis and energy-sensing in mammals. Here, we identified the top six genes that contribute to the negative correlation of latitudinal and seasonal selection to be Pyk, Tpi, UGPase, Idh-β, Gpdh and Pdk. These genes possess both strong clines as well as notable, albeit non-significant, changes within season. With the exception of Pyk, these are among the top genes showing expression variation across the pathway and are proposed as targets of latitudinal selection.
Supplementary Material
Acknowledgements
We would like to thank Thomas Merritt for supplying the lines from Sudbury, ON; and John True and Joe Lachance for three additional collections from New York. We would like to thank Eugene Brud and two anonymous reviewers who made constructive comments on the earlier version.
Funding statement
This study was supported by a National Institutes of Health grant (GM090094) to W.F.E. and John True; and Collaborative National Foundation Science grants (DEB0921372) to W.F.E. and (DEB0542859 and DEB0921307) to P.S.S.
References
- 1.Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu CT, Verrelli BC, Eanes WF. 2004. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics 168, 923–931. ( 10.1534/genetics.104.027649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry A, Kreitman M. 1993. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics 134, 869–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt PS, et al. 2008. Ecological genetics in the North Atlantic: environmental gradients and adaptation at specific loci. Ecology 89, S91–S107. ( 10.1890/07-1162.1) [DOI] [PubMed] [Google Scholar]
- 4.Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlotterer C, Flatt T. 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21, 4748–4769. ( 10.1111/j.1365-294X.2012.05731.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187, 245–260. ( 10.1534/genetics.110.123059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD. 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197, 361–373. ( 10.1534/genetics.114.161463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umina PA, Weeks AR, Kearney MR, Mckechnie SW, Hoffmann AA. 2005. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science 308, 691–693. ( 10.1126/science.1109523) [DOI] [PubMed] [Google Scholar]
- 8.Vasemagi A. 2006. The adaptive hypothesis of clinal variation revisited: single-locus clines as a result of spatially restricted gene flow. Genetics 173, 2411–2414. ( 10.1534/genetics.106.059881) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchen P, Zivkoviç D, Hutter S, Stephan W, Laurent S. 2013. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193, 291–301. ( 10.1534/genetics.112.145912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright S. 1943. Isolation by distance. Genetics 28, 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt PS, Matzkin L, Ippolito M, Eanes WF. 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59, 1721–1732. ( 10.1111/j.0014-3820.2005.tb01821.x) [DOI] [PubMed] [Google Scholar]
- 12.Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF. 2008. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 105, 16 207–16 211. ( 10.1073/pnas.0805485105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cogni R, Kuczynski K, Koury S, Lavington E, Behrman EL, O'Brien KR, Schmidt PS, Eanes WF. 2013. The intensity of selection acting on the Couch Potato gene—spatial–temporal variation in a diapause cline in Drosophila melanogaster. Evolution 68, 538–548. ( 10.1111/evo.12291) [DOI] [PubMed] [Google Scholar]
- 14.Lavington E, Cogni R, Kuczynski K, Koury S, Behrman EL, O'Brien KR, Schmidt PS, Eanes WF. 2014. A small system—high resolution study of metabolic adaptation in the central metabolic pathway to temperate climates in Drosophila melanogaster. Mol. Biol. Evol. 31, 2032–2041. ( 10.1093/molbev/msu146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Band HT, Ives PT. 1963. Genetic structure of populations. I. On the nature of the genetic load in the South Amherst population of Drosophila melanogaster. Evolution 17, 198–215. ( 10.2307/2406466) [DOI] [Google Scholar]
- 16.Ives PT. 1970. Further genetic studies of the South Amherst population of Drosophila melanogaster. Evolution 24, 507–518. ( 10.2307/2406830) [DOI] [PubMed] [Google Scholar]
- 17.Wagner A, Fell DA. 2001. The small world inside large metabolic networks. Proc. R. Soc. Lond. B 268, 1803–1810. ( 10.1098/rspb.2001.1711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackay TFC, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482, 173–178. ( 10.1038/nature10811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavebratt C, Sengul S. 2006. Single nucleotide polymorphism (SNP) allele frequency estimation in DNA pools using pyrosequencing. Nat. Protoc. 1, 2573–2582. ( 10.1038/nprot.2006.442) [DOI] [PubMed] [Google Scholar]
- 20.Doostzadeh J, Shokralla S, Absalan F, Jalili R, Mohandessi S, Langston JW, Davis RW, Ronaghi M, Gharizadeh B. 2008. High throughput automated allele frequency estimation by pyrosequencing. PLoS ONE 3, e2693 ( 10.1371/journal.pone.0002693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl Aad. Si. USA 100, 9440–9445. ( 10.1073/pnas.1530509100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayroles JF, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41, 299–307. ( 10.1038/ng.332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benovoy D, Kwan T, Majewski J. 2008. Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic Acids Res. 36, 4417–4423. ( 10.1093/nar/gkn409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Page GP, Mehta T, Feng R, Cui XQ. 2009. Single nucleotide polymorphisms affect both cis- and trans-eQTLs. Genomics 93, 501–508. ( 10.1016/j.ygeno.2009.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massouras A, et al. 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8, e1003055 ( 10.1371/journal.pgen.1003055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ives PT. 1954. Genetic changes in American populations of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 40, 87–92. ( 10.1073/pnas.40.2.87) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergland AO, Behrman EL, O'Brien KRO, Schmidt PS, Petrov DA. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10, e1004775 ( 10.1371/journal.pgen.1004775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knibb WR, Oakeshott JG, Gibson JB. 1981. Chromosome inversion polymorphisms in Drosophila melanogaster. 1. Latitudinal clines and associations between inversions in Australasian populations. Genetics 98, 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobzhansky T. 1943. Genetics of natural populations IX. Temporal changes in the composition of populations of Drosophila pseudoobscura. Genetics 28, 162–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt PS, Conde DR. 2006. Environmental heterogeneity and the maintenance of genetic variation for reproductive diapause in Drosophila melanogaster. Evolution 60, 1602–1611. ( 10.1111/j.0014-3820.2006.tb00505.x) [DOI] [PubMed] [Google Scholar]
- 31.Ives PT, Band HT. 1986. Continuing studies on the South Amherst Drosophila melanogaster natural population during the 1970's and 1980's. Evolution 40, 1289–1302. ( 10.2307/2408954) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.