Abstract
• Premise of the study: Bacterial contamination is a major problem in plant tissue culture, resulting in loss of experimental strains or preventing use of field-collected isolates. Here we evaluated an agar embedding method for eliminating bacteria from experimental cultures of the mosses Ceratodon purpureus and Physcomitrella patens.
• Methods and Results: We blended moss protonema that had been inoculated with bacteria and embedded the cell fragments in antibiotic-containing, low-concentration agar. The plants were placed in a growth chamber and allowed to grow until the moss grew out of the media. The plants were then transferred to new plates and observed for contamination. The embedding method consistently outperformed standard procedures.
• Conclusions: The embedding method places moss in direct contact with antibiotics, arresting bacterial replication and allowing moss to outgrow contamination. We anticipate this method will prove valuable for other plants capable of clonal propagation by blending.
Keywords: antibiotics, Ceratodon, contamination, Physcomitrella, protonema, sterile tissue culture
The purpose of this paper is to describe a method for eliminating bacterial contamination from in vitro moss cultures. The mosses Physcomitrella patens (Hedw.) Bruch & Schimp. and Ceratodon purpureus (Hedw.) Brid. have recently emerged as model organisms for a variety of biological phenomena (Cove et al., 2009a). They are haploid, undergo gene targeting, and are easily cultured in vitro. However, microbial contamination is a major obstacle for researchers working with moss cultures. At worst, some microbial organisms can kill experimental cell lines, but even sublethal infections can alter growth patterns or introduce contaminant DNA or RNA in shotgun sequencing projects. Here we present a method that reliably eliminates bacterial contamination by regenerating blended moss tissue in an antibiotic-containing agar matrix.
Bacterial contamination can be introduced at many points during moss tissue culture. Field-collected samples often contain microorganisms from their local environment, and laboratory isolates may become contaminated through exposure to airborne microbes during sample handling or from inadequately sterilized equipment such as forceps or media (Leifert and Cassells, 2001). A commonly used practice for cleaning gametophyte tissue is plating on top of antibiotic-containing media. While this can be effective at controlling contamination, we have found it can have a limited effect once the moss is removed from the antibiotic. Obtaining axenic cultures is most easily achieved by the sterilization of mature, operculate sporophytes using sodium hypochlorite with subsequent culturing of the released spores (e.g., McDaniel et al., 2007; Vujičić et al., 2011; Sabovljević et al., 2012). However, for many species sporophytes are not readily available. Some authors have used sodium hypochlorite to sterilize gametophytic tissue, but fully sterile cultures are difficult to achieve with this method because the concentration necessary to kill all the microbial contaminants is generally lethal to the moss as well (Vujičić et al., 2011; Sabovljević et al., 2012). The juvenile moss gametophyte stage, protonema, is more challenging because it is more sensitive to antimicrobial agents like sodium hypochlorite (Duckett et al., 2004). The ability to generate clean protonemal tissue is particularly useful, because many laboratories maintain strains as protonema given the ease of manipulating plants at this stage. Additionally, all damaged haploid tissues regenerate as protonema.
Here we present an agar embedding method to generate bacteria-free cell lines of protonemal-stage moss tissue. This method takes advantage of the fact that many mosses regenerate from damaged haploid, gametophyte tissue and are capable of growth in antibiotic-containing agar suspension (e.g., see protoplast regeneration method described by Cove et al. [2009b]). To test this method, we first evaluated the minimum concentration needed for three antibiotics commonly used in plant tissue culture—cefotaxime (Gold Biotechnology, St. Louis, Missouri, USA), chloramphenicol (Gold Biotechnology), and vancomycin (Acros Organics, Geel, Belgium)—on eliminating the growth of four bacteria strains that we isolated from moss cultures in the laboratory. We also evaluated the effects of the antibiotics on moss growth. We next inoculated sterile moss tissue with our experimental bacteria and embedded blended, experimentally contaminated moss in low-concentration, antibiotic-containing agar. Lastly, we evaluated the efficacy of the embedding method and compared it to the currently existing protocol of sterilizing in sodium hypochlorite. Overall, we found the agar embedding method with vancomycin to significantly reduce the presence of bacterial contaminants from gametophyte tissue compared to currently existing protocols.
METHODS AND RESULTS
Bacteria isolation and identification
Four phenotypically distinct bacterial specimens were isolated from existing contaminated moss cultures. The bacteria were streaked to isolate single colonies on solid Luria–Bertani (LB) medium and incubated at 37°C overnight. To identify the bacteria, we amplified the 16S ribosomal subunit using the forward primer 27F-16S (5′-AGAGTTTGATCMTGGCTCAG-3′) and the reverse primer 1525R-16S (5′-AAGGAGGTGWTCCARCC-3′) in 16-μL PCR reactions using GoTaq Green Master Mix 2× (Promega Corporation, Madison, Wisconsin, USA) with the following PCR conditions: 94°C for 2 min, 35 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 1.5 min, with a final extension of 72°C for 10 min. Sequencing was performed at the Interdisciplinary Center for Biotechnology Research at the University of Florida. Sequences were manually edited using UGENE (Okonechnikov et al., 2012). Comparisons to known sequences on GenBank (Benson et al., 2005) were made using BLAST (Altschul et al., 1990) and identification was based on maximum similarity. These included Paenibacillus taichungensis (PT), Paenibacillus humicus (PH), Bacillus aryabhattai or megaterium (BAM), and one unidentified bacterium (UNK) (Appendix 1). Based on the accession descriptions, we cannot definitively establish ecologically important attributes of any of these bacteria. However, given that the moss cultures from which these bacteria were isolated were at one point axenic, we do not believe that the collection site of the moss isolate and bacterial contaminant are the same.
Testing antibiotics on bacteria
We based the starting concentration for each antibiotic on the manufacturer’s recommended ranges for plant tissue culture and following Silva and Fukai (2001): vancomycin 1× = 50 μg/mL cefotaxime 1× = 100 μg/mL and chloramphenicol 1× = 15 μg/mL. To generate fresh bacterial growth prior to antibiotic exposure, we inoculated liquid LB with bacteria PT, PH, BAM, and UNK and incubated at 37°C, 225 rpm, for 16 h. One hundred microliters of bacteria culture was added to 3 mL of liquid LB and 0.5×, 1×, 2×, 4×, and 10× concentrations of antibiotic, replicated twice. To determine the effectiveness of these antibiotic concentrations at eliminating bacterial growth, every two days 5 μL from each antibiotic treatment was streaked onto solid LB plates and incubated. The resulting colonies were counted and compared over a 10-d period. We found the antibiotics had varying effects on suppressing the growth of bacteria (Table 1, Fig. 1). Vancomycin was the only antibiotic that suppressed the growth of all bacteria, and it remained effective at a concentration of 1× against the four bacteria that we isolated.
Table 1.
The minimum concentrations of antibiotics effective at killing the experimental bacteria.
| Bacteria | Chloramphenicol | Cefotaxime | Vancomycin |
| PT | 1× | — | 0.5× |
| PH | 4× | — | 1× |
| UNK | 2× | 10× | 1× |
| BAM | — | 1× | 0.5× |
Note: — indicates there was no effective dose tested.
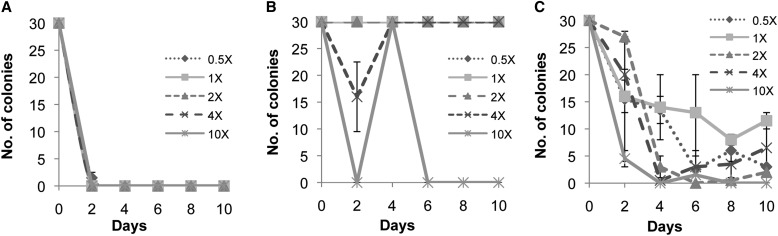
Fig. 1.
Representative graphs of the effects of antibiotics on experimental bacteria in log phase growth showing (A) the effects of vancomycin on PT, (B) cefotaxime on PH, and (C) chloramphenicol on BAM. Vancomycin was effective on all bacteria, and chloramphenicol and cefotaxime varied greatly among bacteria. The points at days 2–10 are colony counts after streaking 5 μL of bacteria and antibiotic solution on Luria–Bertani. At greater than 30, colonies were too numerous to count.
Testing antibiotics on moss tissue
To investigate whether the antibiotics had a negative effect on protonemal growth, we tested one C. purpureus isolate (12-2-4) in 1×, 2×, and 4× vancomycin, 1× and 10× cefotaxime, and 4× and 8× chloramphenicol. Protonemal tissue was submerged in 6 mL of sterile water and antibiotics and mixed on an aliquot mixer. After 1, 3, and 6 d in the antibiotics, two replicates of protonema were clonally propagated onto a solid medium containing the necessary nutrients for moss growth (BCDA; following Cove et al. [2009c]) without antibiotics. These were grown in a growth chamber with 16-h lighting at 25°C. We found no qualitative effect of antibiotics on moss tissue regrowth. Both replicates of moss, once removed from the antibiotic-containing solution, grew similarly to controls regardless of the concentration of antibiotic used or the duration of exposure. Thus, while low concentrations of antibiotics may be sufficient to inhibit bacterial growth in the embedding method, higher concentrations are unlikely to have a deleterious effect on moss growth (i.e., green, healthy tissue as opposed to dead, white tissue that failed to regenerate after several days) should they be required.
Agar embedding method
The logic of the agar embedding method is to place all bacterially contaminated moss tissue in contact with an antibiotic that inhibits the growth of the bacteria, thereby allowing the moss to outgrow the contamination. To test the embedding method, we grew seven strains of C. purpureus and one strain of P. patens (Appendix 2), clonally replicated three times, for 14 d after which each replicate was inoculated with a single bacterial culture PT, UNK, or BAM. Bacteria pH was not used due to loss of the pure culture. After 20 d of growth, contaminated moss tissue was ground at 1700 rpm in a Geno/Grinder (SPEX SamplePrep, Metuchen, New Jersey, USA) in 2-mL tubes with 0.7 mL of sterile water, 1 μL/mL CaCl2, 1× vancomycin, and a steel grinder ball. The ground tissue was mixed with 0.5833 mL of 0.4% protoplast regeneration medium top layer (PRMT) without D-mannitol (Cove et al., 2009b) and plated on top of two wells of BCDA media supplemented with 50 μg/mL vancomycin. The plates were placed in a growth chamber with 16-h lighting at 25°C for 12 d, allowing the moss to grow out of the top layer of agar. For each isolate, protonemal filaments, which had grown out of the PRMT, were isolated and moved onto regular BCDA media (without antibiotics). This was replicated three times per isolate. Cultures were observed to assess the growth of the tissue and the presence or absence of bacteria.
To test the results of the embedding treatment, a full factor ANOVA analysis comparing vancomycin treatment, moss strain, and bacteria was run using JMP 9.0.2 (SAS Institute, Cary, North Carolina, USA). The controls consisting of tissue embedded in agar without vancomycin generated tissue visibly contaminant free in only 12.5% of the replicates (18/144). The agar embedding method using 1× vancomycin generated samples free of visible bacterial contamination in 90% of the clonal replicates (129/144). All clonal replicates survived in both the antibiotic and antibiotic-free embedding treatments. We found a strong effect of the antibiotic treatment (P ≤ 0.0001), indicating the agar embedding method with 1× vancomycin is more effective at generating moss tissue free of bacterial contamination than controls (Fig. 2; Table 2). We did find that some strains were slightly more likely to remain contaminated after treatments, which is most likely a result of strain-specific growth features (e.g., linear growth causing matting) that caused us to transfer plants with small amounts of agar that contained viable bacteria (Appendix S1 (265.6KB, pdf) ). The protocol for the agar embedding method is provided in Appendix 3.
Fig. 2.
The effect of vancomycin vs. control in the agar embedding method on the number of contaminated clonal isolates of tissue contaminated by bacteria PT, UNK, and BAM. Using vancomycin significantly increases the number of clean isolates (P ≤ 0.001). BAM = Bacillus aryabhattai or megaterium; LSM = least square mean; PT = Paenibacillus taichungensis; UNK = unidentified bacterium.
Table 2.
Full factorial ANOVA evaluating the effect of vancomycin treatment, bacteria, and moss strain on the success of agar embedding method with 1× vancomycin.
| Source | Degrees of freedom | Sum of squares | F ratio | P value |
| Control or treatment | 1 | 128.34 | 821.4 | <0.0001 |
| Bacteria | 2 | 2.25 | 7.2 | 0.0018 |
| Control or treatment*bacteria | 2 | 3.25 | 10.4 | 0.0002 |
| Moss strain | 7 | 6.66 | 6.09 | <0.0001 |
| Control or treatment*moss strain | 7 | 2.74 | 2.5 | 0.0281 |
| Bacteria*moss strain | 14 | 18.25 | 8.34 | <0.0001 |
| Control or treatment*bacteria*moss strain | 14 | 10.92 | 4.99 | <0.0001 |
Testing the embedding method against sodium hypochlorite
To compare the agar embedding method to the widely used sodium hypochlorite method, we submerged bacterially contaminated protonema and field-collected gametophores in 0.2475% and 0.825% sodium hypochlorite (3% and 10% commercial bleach, respectively) for 1 min and 3 min. The plants were agitated, and then rinsed three times in autoclaved water. In all sodium hypochlorite treatments, the protonema died (turned white within a minute in the sodium hypochlorite), although we saw less contamination following the use of higher sodium hypochlorite concentration and longer durations of submersion. However, we could not find a combination of sodium hypochlorite concentration and duration that did not kill the protonema but did kill the microbial contaminants. We and others (Vujičič et al., 2011; Sabovljević et al., 2012) have tried using the sodium hypochlorite method to remove contaminants from field-collected gametophores, and in some cases we were able to remove algae and fungi but not bacterial contaminates. However, we have not found a sodium hypochlorite protocol that reliably kills the contaminants without also killing the moss (with the exception of spores inside an operculate sporophyte).
CONCLUSIONS
Bacterial contamination is a recurrent problem in moss tissue culture. Here we describe an agar-embedding method that is reliable, rapid, and relatively inexpensive, using materials that are readily available in most laboratories that conduct sterile tissue culture. The key advance of the embedding method is that the contaminated moss tissue remains in direct contact with the antibiotic, which suppresses the growth of the bacteria but not the growth of the moss. Provided the moss is given sufficient time to grow, new, noncontaminated protonemal filaments emerge from the agar. We did find that particular moss strains were slightly more likely to remain contaminated with particular bacteria (i.e., a significant moss strain × bacteria × vancomycin treatment effect; Appendix S1 (265.6KB, pdf) ). We have noted that even within a species our strains contain variation in protonemal growth rate and general growth habit, including filament strength (i.e., how likely the filament was to break above vs. below the agar). The two strains that showed a small amount of residual contamination had the greatest tendency to retain small quantities of agar when the tissue was transferred; although vancomycin is bactericidal, the agar used to embed contaminated moss may in some cases still contain viable bacteria. One way to avoid transferring any of the bacteria-containing agar is to take subsamples of tissue only after the moss has grown well away from the agar, which may take longer than for the strains tested here (Appendix 3). Of course, because one generally needs to isolate only a single clean isolate from each contaminated strain, a success rate considerably lower than the 90% that we report for the toughest strain would still be effective.
Mosses are well suited for experimental biology because many species can regenerate from blended gametophyte tissue (Shaw, 1986). For applications where axenic tissue is essential, however, additional sterilization procedures are necessary. We (and others) have used a sodium hypochlorite treatment on operculate sporophytes with success. Nevertheless, sporophytes are not always available, and while sodium hypochlorite is occasionally effective at removing or killing fungi and algae from gametophores, it is not an effective treatment of prokaryotic contamination. The embedding method adds a new technique that can be used in conjunction with sodium hypochlorite and vigilant sterile technique to initiate or maintain axenic laboratory cultures of mosses. We should point out that we discourage the prophylactic use of antibiotics in routine culture, as this can lead to the evolution of resistant bacterial strains that are more difficult to eradicate. We also encourage the proper disposal of bacteria that survive contact with antibiotics. Additionally, the eradication of contaminants other than those we studied here may require the use of different antibiotics. Despite these caveats, we anticipate that the embedding method will be applicable to more species than the two studied here, as well as for other plants that can be grown in tissue culture.
Supplementary Material
Appendix 1.
Putative identification of experimental bacteria and GenBank accession numbers of 16S sequences.
| Bacteria NCBI result | Result GenBank accession no. | Point mutations | Indels | Sequence GenBank accession no. |
| Paenibacillus taichungensis strain A80 | JX010963.1 | 2 | 1 single nucleotide | KJ725141 |
| Paenibacillus humicus strain 5002BRRJ | JF309272.1 | — | 3 single nucleotides | KJ725139 |
| *Unidentified bacterium | — | — | — | — |
| Bacillus megaterium strain BAB-2927; Bacillus aryabhattai strain ZZ12 | KF853118.1; KF436619.1 | 1 | — | KJ725140 |
Note: NCBI = National Center for Biotechnology Information.
UNK was unable to be identified with any degree of certainty. The closest BLAST hit was a Physcomitrella patens subsp. patens predicted protein (accession no. XM_001764528.1) with eight point mutations in the alignment. UNK was likely not able to be identified due to poor results of Sanger sequencing.
Appendix 2.
Locations where moss strains were originally collected and the strain’s abbreviation.
| Moss straina | Provenance of moss strain | Abbreviation |
| Physcomitrella patens | ||
| PPX progeny | Progeny of cross between the Villersexel and Gransden lab strains | PPX |
| Ceratodon purpureus | ||
| DUR2013 | Durham County, North Carolina, USA | DUR |
| E13_E3 | Otavalo, Ecuador | ECU |
| ANT1 | Casey Base, Antarctica | ANT |
| ALK | Fairbanks Co., Alaska, USA | ALK |
| 12_2_4 | Tolland Co., Connecticut, USA | CCT |
| R40 | Rensselaer County, New York, USA | NYS |
| CHL1 | Chile | CHL |
All moss strains listed are maintained as live cultures in the McDaniel laboratory, Department of Biology, University of Florida, Gainesville, Florida, USA.
Appendix 3.
Protocol for Petri dish–sized embedding method.
This procedure is designed to generate bacteria-free clonal propagations of Ceratodon purpureus or Physcomitrella patens. This protocol makes two 100-mm standard Petri dishes of antibiotic-embedded, bacterially contaminated moss tissue. The moss tissue is then propagated to non-antibiotic-containing BCDA medium without contamination. All steps must be done in a laminar hood and utilizing sterile techniques.
Recipes:
Recipe 1:
BCDA (adapted from BCD; Cove et al., 2009c)
| Reagent | Quantity (for 1 L) | Final concentration |
| Agar (Sigma-Aldrich A9799) | 7 g | 0.7% (w/v) |
| *CaCl2 | 111 mg | 1 mM |
| FeSO4·7H2O | 12.5 mg | 45 μM |
| Solution B for moss media | 10 mL | 1 mM MgSO4 |
| Solution C for moss media | 10 mL | 1.84 mM KH2PO4 |
| Solution D for moss media | 10 mL | 10 mM KNO3 |
| Hoagland’s A-Z trace element solution | 1 mL | Trace |
| Diammonium tartrate | 92 g | 5 mM |
| *50 mg/mL vancomycin | 1 mL | 50 μg/mL |
Do not add until after autoclaving.
Recipe 2:
Top agar (adapted from PRMT; Cove et al., 2009b)
| Reagent | Quantity (for 1 L) | Final concentration |
| Agar (Sigma-Aldrich A9799) | 4 g | 0.4% (w/v) |
| *CaCl2 | 1.1 g | 10 mM |
| Diammonium tartrate | 920 mg | 5 mM |
| BCD medium, liquid | to 1 L |
Do not add until after autoclaving.
Materials:
• PowerGen 125 (Fisher Scientific, Hampton, New Hampshire, USA)
• Tissue homogenizers (OMNI-INC 3_750; Omni International, Kennesaw, Georgia, USA)
• Forceps
• Bunsen burner
• Sterile water
• Two sterile Petri dishes
• Pipetter and micropipetter
• Bacterially contaminated protonemal-stage moss
• Sterile cellulose (not required)
• 50 mg/mL vancomycin
• 200 proof ethanol
• Micropore surgical tape (3M, St. Paul, Minnesota, USA)
Additional points prior to beginning protocol:
• Distilled water, BCDA, top agar, cellulose, pipette tips, and tissue homogenizers must be sterilized using an autoclave prior to use.
• If vancomycin-containing BCDA medium is kept in cool, dark location, allow the medium to come to room temperature. This allows the molten top agar to spread evenly and not immediately cool upon application.
• Sterile cellulose overlays may be placed on the vancomycin BCDA medium to facilitate transferring the blended moss tissue to another plate if desired.
• Forceps should be ethanol dipped and flame sterilized prior to use.
• If top agar was made prior to the protocol and is solidified, it should be melted in a microwave before use and stored in a 55°C hot water bath to remain molten.
Procedure:
1. In a sterile tissue culture tube, add 1.5 mL of sterile water.
2. Add 4 μL of 50 mg/mL vancomycin (final concentration will be 1 μL/mL after addition of top agar).
3. Add 4 μL of 1 M CaCl2 to blended moss tissue (final concentration will be 1 μL/mL after addition of top agar).
4. Using sterilized forceps, transfer contaminated moss tissue to culture tube containing antibiotic water.
5. Blend moss tissue using a tissue homogenizer attached to the PowerGen 125 (Fisher Scientific).
6. Add 2.5 mL of melted 0.4% top agar.
7. Pipette mix solution.
8. Pipette 1.5–2 mL of solution to BCDA plate containing 50 μg/mL vancomycin and optional cellulose overlay.
9. Carefully tilt the plate until top agar is spread on plate and allow to solidify.
10. Once top agar is solidified, tape plate using micropore surgical tape.
11. Grow at light and temperatures normally used for optimal growth of the strain/species used.
12. Allow protonema to grow out of the top agar. This is time dependent on the moss strain/species used (10–14 d for C. purpureus and P. patens).
13. Remove protonema that is not in contact with agar to a new BCDA plate.
14. Autoclave embedded plate to reduce the risk of harboring antibiotic-resistant bacteria.
LITERATURE CITED
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L. 2005. GenBank. Nucleic Acids Research 33: D34–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. J., Perroud P. F., Charron A. J., McDaniel S. F., Khandelwal A., Quatrano R. S. 2009a. The moss Physcomitrella patens: A novel model system for plant development and genomic studies. Cold Spring Harbor Protocols 2009: 10.1101/pdb.emo115. [DOI] [PubMed] [Google Scholar]
- Cove D. J., Perroud P. F., Charron A. J., McDaniel S. F., Khandelwal A., Quatrano R. S. 2009b. Isolation and regeneration of protoplasts of the moss Physcomitrella patens. Cold Spring Harbor Protocols 2009: 10.1101/pdb.prot5140. [DOI] [PubMed] [Google Scholar]
- Cove D. J., Perroud P. F., Charron A. J., McDaniel S. F., Khandelwal A., Quatrano R. S. 2009c. Culturing the moss Physcomitrella patens. Cold Spring Harbor Protocols 2009: 10.1101/pdb.prot5136. [DOI] [PubMed] [Google Scholar]
- Duckett J. G., Burch J., Fletcher P. W., Matcham H. W., Read D. J., Russell A. J., Pressel S. 2004. In vitro cultivation of bryophytes: A review of practicalities, problems, progress and promise. Journal of Bryology 26: 3–20. [Google Scholar]
- Leifert C., Cassells A. 2001. Microbial hazards in plant tissue and cell cultures. In Vitro Cellular & Developmental Biology. Plant 37: 133–138. [Google Scholar]
- McDaniel S. F., Willis J. H., Shaw A. J. 2007. A linkage map reveals a complex basis for segregation distortion in an interpopulation cross in the moss Ceratodon purpureus. Genetics 176: 2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M., the UGENE team 2012. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics (Oxford, England) 28: 1166–1167. [DOI] [PubMed] [Google Scholar]
- Sabovljević A., Vujičić M., Skorić M., Sabovljević M. 2012. Axenically culturing the bryophytes: Establishment and propagation of the pleurocarpous moss Thamnobryum alopecurum Nieuwland ex Gangulee (Bryophyta, Neckeraceae) in in vitro conditions. Pakistan Journal of Botany 44: 339–344. [Google Scholar]
- Shaw J. 1986. A new approach to the experimental propagation of bryophytes. Taxon 35: 671–675. [Google Scholar]
- Silva J. D., Fukai S. 2001. The impact of carbenicillin, cefotaxime and vancomycin on chrysanthemum and tobacco TCL morphogenesis and Agrobacterium growth. Journal of Applied Horticulture 3: 3–12. [Google Scholar]
- Vujičić M., Sabovljević A., Sabovljević M. 2011. Axenically culturing the bryophytes: Establishment and propagation of the moss Hypnum cupressiforme Hedw. (Bryophyta, Hypnaceae) in in vitro conditions. Botanica Serbica 35: 71–77. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.