Abstract
• Premise of the study: Positional (or map-based) cloning is a common approach to identify the molecular lesions causing mutant phenotypes. Despite its large and complex genome, positional cloning has been recently shown to be feasible in maize, opening up a diverse collection of mutants to molecular characterization.
• Methods and Results: Here we outline a general protocol for positional cloning in maize. While the general strategy is similar to that used in other plant species, we focus on the unique resources and approaches that should be considered when applied to maize mutants.
• Conclusions: Positional cloning approaches are appropriate for maize mutants and quantitative traits, opening up to molecular characterization the large array of genetic diversity in this agronomically important species. The cloning approach described should be broadly applicable to other species as more plant genomes become available.
Keywords: bulked segregant analysis, maize, mapping, positional cloning, simple sequence repeat (SSR) marker
Forward genetics is a powerful tool to uncover the genetic mechanisms regulating biological processes. Moving from mutants to genes, or cloning, can be accomplished by a variety of methods. However, the ease and popularity of chemical mutagenesis, particularly ethyl methane sulfonate (EMS), has generated a large collection of mutants in diverse plant species for which a map-based or positional cloning approach is appropriate.
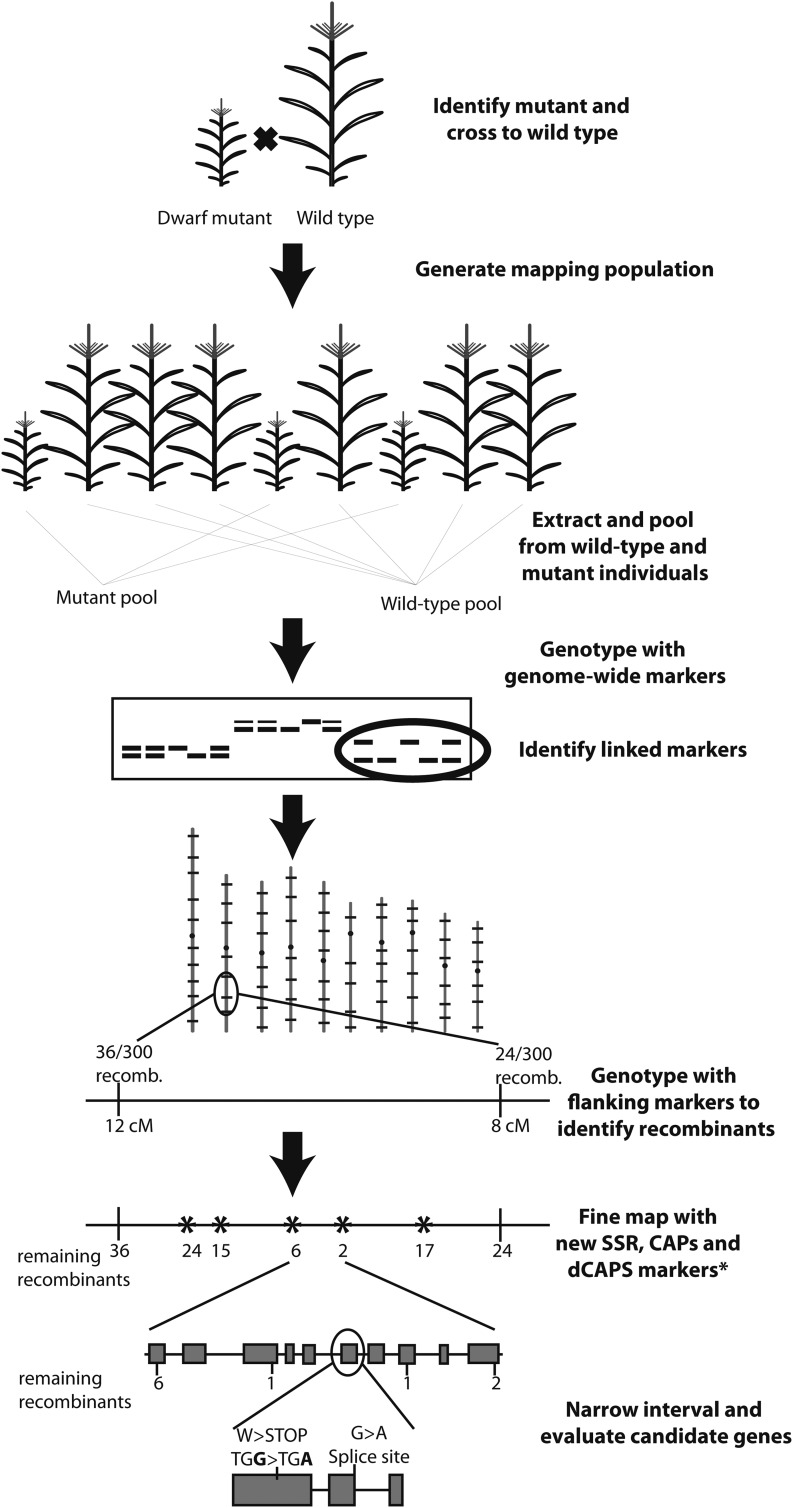
Map-based cloning (Fig. 1) uses the techniques of genetic mapping to define a progressively narrower chromosomal interval containing the mutant locus until the lesion causing the mutant phenotype is identified (Lukowitz et al., 2000). The strength of positional cloning is that it can be applied to mutations derived from almost any source (e.g., transposon, chemical, radiation, natural variation), as long as that mutation defines a locus that can be mapped.
Fig. 1.
Overview and summary of positional cloning approach.
Maize (Zea mays L.) has a long history of genetic research, and consequently a diverse collection of mutations affecting a broad range of biological processes has accumulated (Neuffer et al., 1997). Traditional cloning approaches in maize (e.g., candidate gene approaches, or transposon tagging) are labor intensive and risky. The large genome of maize and the lack of sequence information effectively discouraged any attempt at positional cloning, as it was assumed that extremely large mapping populations would be necessary to provide sufficient resolution. Undeterred, several laboratories have demonstrated that positional cloning in maize is generally no more difficult than in rice or even Arabidopsis (DC.) Heynh. (see, e.g., Wang et al., 2005; Bortiri et al., 2006; Gallavotti et al., 2008; Whipple et al., 2010). Although the maize genome is approximately 20 times larger than Arabidopsis, the majority of this size increase is the result of retrotransposon amplification, creating large nonrecombining heterochromatic seas interspersed with highly recombining, gene-rich euchromatic islands (SanMiguel et al., 1996; Fu et al., 2002). These poorly recombining tracts of retrotransposons effectively disappear during mapping, leaving a much smaller, gene-rich portion of the genome that is “seen” in a genetic map. With the recent completion of the maize genome sequence (Schnable et al., 2009), positional cloning is rapidly becoming routine, opening up the large and diverse collection of maize mutants to cloning. While positional cloning is appropriate for cloning any trait that can be genetically mapped, there are situations for which a positional cloning approach may not be the best option. The high resolution needed for fine mapping requires a reasonable recombination frequency, which may be suppressed for loci that map to centromeres or other poorly recombining regions. Similarly, mutations associated with chromosomal rearrangement can prove difficult to map.
A universal protocol for positional cloning in maize would be unwieldy because the steps taken will depend upon the particularities of the locus to be cloned. Consequently, we will focus here on the most likely case, a single locus recessive or dominant mutant, while at the same time describing some of the more common alternatives that may need to be considered. Recently, high-throughput sequencing technology has opened up the possibility of mutant mapping and cloning with a single sequencing run, dramatically reducing the time typically spent on map-based cloning (Williams-Carrier et al., 2010; Schneeberger and Weigel, 2011; Abe et al., 2012; Liu et al., 2012). These approaches and the rapidly decreasing cost of sequencing will likely open many maize mutants to cloning by sequencing in the near future. However, even with these advances some loci will be recalcitrant to a sequencing approach, and thus require the traditional approach described here.
METHODS AND RESULTS
1. Genetic characterization of mutant
Poorly studied or recently isolated mutants will need to be genetically characterized, as the mapping strategy ultimately employed will depend on the nature of the mutation. The steps below outline a basic genetic characterization.
a. Determine the mode of inheritance. Cross the mutant to wild-type maize inbreds to create multiple F1s (Appendix 1, Note 1). If F1 progenies from these crosses display the mutant phenotype, the mutation is either dominant or semidominant. If all F1 progenies are phenotypically normal (hereafter, “wild type” will also be used to indicate phenotypically normal plants that can, however, carry polymorphisms compared to the B73 reference genome sequence), then the mutation is likely recessive. Self-pollinate the F1 plants from each distinct inbred cross to generate F2 populations. Recessive mutations should be seen segregating in these populations.
b. Determine the number of loci involved. Examine the frequency of the mutant phenotype in an F2 population. A population of approximately 100 F2 individuals from each F1 should be sufficient to determine the segregation ratio. A single recessive locus should conform to the expected 3:1 (normal:mutant) ratio. This can be confirmed by a χ2 goodness-of-fit test to the expected 3:1 ratio. Any significant deviation from this ratio could indicate that multiple loci contribute to the mutant phenotype (Appendix 1, Note 2).
c. Identify inbred lines that show consistent and strong expressivity (phenotype severity) and penetrance (i.e., plant phenotype consistently matches the genotype) of the mutant phenotype. Because the genetic background in maize can dramatically affect the severity, or even presence, of the mutant phenotype, F2 populations from crosses to inbreds where the mutant phenotype is consistent and easy to score will be very helpful. This will avoid problems when mapping, where it is imperative that the genotype at the mutant locus can be confidently determined from the plant phenotype (Appendix 1, Note 3).
2. Create a mapping population
After initial genetic characterization, an appropriate mapping strategy should be chosen. Because the creation of the mapping population is often the rate-limiting step in positional cloning, it is advisable to initiate multiple mapping populations as soon as possible. Ultimately, only polymorphic populations that are well-suited to your mutant of interest need to be used. The two most common and useful mapping strategies involve creation of F2 or backcross (BC) mapping populations. The steps in creating each mapping population are described below.
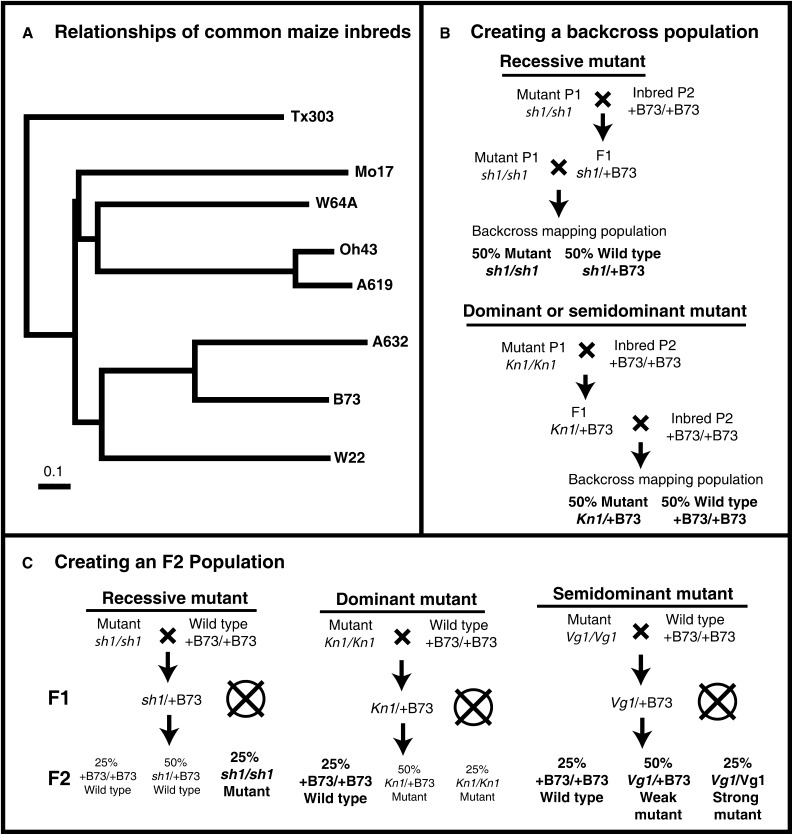
a. Cross the mutant to many diverse inbred lines (Appendix 1, Note 4). This is the first step for both BC and F2 populations. If possible, choose inbreds that are likely to maximize the number of polymorphic markers. Fig. 2A shows the relationships of common inbreds and can provide a rough guide for choosing lines that are more likely to be polymorphic. The crosses initiated during the genetic characterization can be used for this step.
b. Generate BC populations (Appendix 1, Note 5) by crossing the F1 back to one of the parental lines originally used to generate the F1. This backcross can be to either the mutant parent or to the wild-type inbred. The first case will generate progeny that are 50% homozygous mutant and 50% heterozygous, while a backcross to the inbred parent will create 50% heterozygous and 50% homozygous wild-type progeny. As a general rule, a backcross to the wild-type parent is appropriate for dominant/semidominant mutants, while recessive mutants must be backcrossed to the mutant parent to see the phenotype in the next generation (see Fig. 2B).
c. Generate F2 mapping populations (Appendix 1, Note 6) by selfing the F1 individuals (step 2a) to create segregating F2 populations. The phenotypic segregation of the mutant phenotype will depend on the nature of the mutation: recessive, dominant, or semidominant (Fig. 2C). In an F2 population for a recessive mutant, only the 25% mutant individuals will have a known genotype at the mutant locus, because the remaining wild-type plants can be either homozygous wild type or heterozygous. For a dominant mutant the situation is reversed, and only the 25% wild-type individuals will have a known genotype (homozygous wild type) because mutant individuals can be either homozygous or heterozygous for the dominant mutant allele. A semidominant mutant will segregate three phenotypic classes, and each class will have a known genotype.
Fig. 2.
(A) Relationships of common maize inbreds, as determined by shared alleles of 94 distinct SSR loci, redrawn and extracted from Liu et al. (2003). Inbreds from separate clades (e.g., Mo17 and A632) are much more likely to be polymorphic than closely related inbreds (e.g., Oh43 and A619). Scale bar represents 0.1 nucleotide substitutions per site. (B) Schematic representation of the steps for creation of a backcross (BC) mapping population for the recessive shrunken1 (sh1) mutant or the dominant Knotted1 (Kn1) mutant. (C) Schematic representation of the steps for creation of an F2 mapping population for the recessive sh1, dominant Kn1, or semidominant Vestigial glume1 (Vg1) mutants. Circles are placed around the phenotypic classes where the phenotype precisely predicts the genotype. These are the only useful individuals for fine mapping, unless the genotypes of unknown classes are determined by progeny tests (this requires an additional generation, but may be warranted if field space is not limiting). In the case of a semidominant mutant, all phenotypic classes are useful, so long as there is a clear phenotypic distinction between heterozygous and homozygous mutant individuals.
3. Bulked segregant analysis—rough mapping
In this section, we describe a general method for bulked segregant analysis (BSA), used to quickly identify molecular markers linked to any trait of interest (Michelmore et al., 1991). BSA involves creation of two pools by bulking individuals identical in phenotype (e.g., wild type and mutant) from a segregating population. The pooled DNA samples are then genotyped with molecular markers that span the entire genome. Markers that are not linked to the mutant will show no difference in the wild-type and mutant pools, while linked markers will show an overrepresentation of the mutant parent allele only in the mutant pool. Although a variety of markers have traditionally been used for BSA, new methodologies have been developed for the rapid detection of single nucleotide polymorphisms (SNPs) that provide a more efficient alternative for this preliminary mapping step. One example is the MassARRAY System (Sequenom, San Diego, California, USA), which has been optimized at Iowa State University for the simultaneous detection of approximately 1000 SNPs polymorphic between B73 and Mo17 inbreds (Liu et al., 2010). The same set of markers can also be used for mapping populations created with different genetic backgrounds, with the caveat that only a subset of the B73-Mo17 SNPs will be polymorphic. In our experience, MassARRAY has also succeeded with the following genetic backgrounds: A619 × B73, A632 × Mo17, and A632 × Oh43. The steps below are appropriate for BSA using either MassARRAY or more traditional simple sequence repeat (SSR) markers (Taramino and Tingey, 1996). Regardless of the marker type used in BSA, the preparation of the pooled samples is identical.
a. Score your mapping population for both mutant and wild-type individuals. Flag and number the mutant and wild-type plants for collecting tissue.
b. Collect tissue from each of the mutant and wild-type plants scored in step 3a for DNA extraction. Pool an equal amount of tissue (leaf disks or equivalent) from at least 10–20 individual plants for both wild-type and mutant pools (Appendix 1, Note 7). We generally use a standard single-hole paper punch modified so that a 15-mL Falcon tube can be attached for collection of the leaf disks (punches) of uniform size (Fig. 3; Appendix 1, Note 8). Punch the same leaf three times into the collection tube to ensure that enough tissue is obtained from each sample in the pool. Be consistent with all the samples collected, to minimize sample-to-sample variation in the amount of tissue pooled.
c. Independently, collect individual leaf samples from the same individuals used to prepare the pools, and store them at −80°C. These samples will be used to confirm linkage in step 3g.
d. Extract genomic DNA from each pool. General protocols for extracting genomic DNA are usually sufficient. If samples are to be prepared for the MassARRAY System, we use a protocol (Appendix 2) that would also be appropriate for a standard BSA using SSR markers.
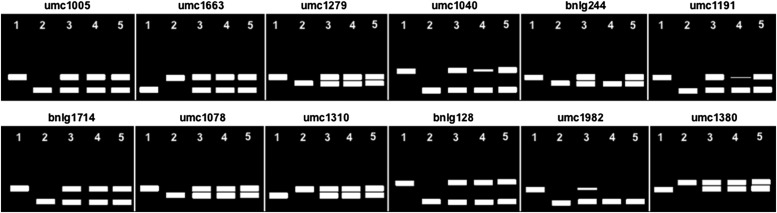
e. This and the following step (3f) can be skipped if BSA is performed using the MassARRAY System. If not using MassARRAY, analyze the mutant and wild-type pooled samples with SSR markers that are distributed across the whole genome and are polymorphic between the two parental lines used to generate the mapping population (Appendix 1, Note 9). It is also advisable to analyze, alongside the two pools, DNA from the parental lines and the corresponding F1 as controls. PCR-based SSR markers are highly polymorphic, inexpensive, and easy to use (Weber and May, 1989; Taramino and Tingey, 1996), and more than 1300 maize SSR loci have been mapped. To increase the likelihood of identifying a linked marker, select a polymorphic SSR every 1–2 bins (bins are arbitrary genetic units of maize chromosomes and correspond approximately to 20 cM). Mapped, polymorphic SSRs can be selected from those catalogued at the Maize Genetics and Genomics Database (MaizeGDB, http://www.maizegdb.org [Appendix 1, Note 9]). A list of polymorphic SSRs appropriate for BSA between A619 and B73 is provided in Table 1, and others have been identified for W22 × B73 (or Mo17) (Martin et al., 2010). Arrange the SSR set in a 96-well plate format (with forward and reverse primers for each SSR combined in a single well) to facilitate a high-throughput analysis of the DNA pools by PCR. These primers will then be used to set up five separate PCR reactions in a 96-well format using the following DNA templates: wild-type parent (1), mutant parent (2), wild-type pool from mapping population (3), mutant pool from mapping population (4), and F1 of mutant and wild-type parent (5). For PCR, use a touchdown protocol with the annealing temperature stepping down (−1°C/cycle) from 65°C to 55°C, followed by 30 cycles at 55°C, and a 45 s extension time for all cycles.
f. Analyze the PCR reactions on Agarose 3:1 HRB (AMRESCO, Solon, Ohio, USA) or MetaPhor Agarose (3–4%; Lonza, Basel, Switzerland) gels in 1× TBE, electrophoresed for 1–2 h at 100–120 V, or until all bands resolve. For each SSR marker, load each of the five PCR reactions set up in step 3e into adjacent lanes. Potentially linked SSRs will be different between the mutant and wild-type DNA pools (Fig. 4). For a recessive mutation, any linked SSR marker should show only the band corresponding to the mutant parent allele in the mutant pool sample, while the wild-type pool should carry both the wild-type and mutant parental alleles (Fig. 4), although there is often a faint wild-type band in the mutant pool (Appendix 1, Note 10).
g. Confirm the linkage of the polymorphic SSRs from step 3f by using the DNA extracted from each individual that formed the pools (step 3e). A linked marker should show an overrepresentation of the mutant parental allele in the mutant samples. Verify the significance of any potential linkage with a χ2 test. If using the MassARRAY System, select a few SSRs in the same chromosomal region for confirmation.
Fig. 3.
Modified single-hole punch to collect pooled leaf samples as described in Appendix 1, Note 8.
Table 1.
Simple sequence repeat (SSR) markers polymorphic between A619 and B73 for bulked segregant analysis (BSA). These SSR markers, ordered by chromosome (chr.) location, were selected uniquely based on gel patterns available at MaizeGDB. Note that for some bins no polymorphic SSR marker was available. By analyzing the bulk DNA samples with these markers, there is a good chance of obtaining a rough map location for the mutant of interest. We successfully mapped all five mutants that we attempted with this set. A similar list of polymorphic SSR markers useful for BSA in W22 × Mo17 and W22 × B73 mapping populations has been described (Martin et al., 2010).
| chr.1 | bin | chr.2 | bin | chr.3 | bin | chr.4 | bin | chr.5 | bin | chr.6 | bin | chr.7 | bin | chr.8 | bin | chr.9 | bin | chr.10 | bin |
| bnlg1014 | 1.01 | bnlg1338 | 2.01 | umc2071 | 3.01 | umc1232 | 4.00 | umc1097 | 5.00 | phi126 | 6.00 | bnlg1367 | 7.00 | umc1359 | 8.00 | umc1279 | 9.00 | umc1380 | 10.00 |
| umc1222 | 1.02 | bnlg1297 | 2.02 | bnlg1144 | 3.02 | umc2148 | 4.01 | umc2036 | 5.01 | bnlg1371 | 6.01 | umc1409 | 7.01 | umc1483 | 8.01 | umc1040 | 9.01 | umc1319 | 10.01 |
| umc1880 | 1.03 | bnlg2248 | 2.03 | umc2158 | 3.04 | umc1943 | 4.02 | umc1587 | 5.02 | umc1178 | 6.02 | bnlg1247 | 7.02 | umc1304 | 8.02 | bnlg244 | 9.02 | umc1337 | 10.02 |
| bnlg2295 | 1.04 | bnlg108 | 2.04 | bnlg1505 | 3.05 | umc2082 | 4.03 | umc1935 | 5.03 | umc1014 | 6.04 | bnlg434 | 7.03 | umc2146 | 8.03 | umc1191 | 9.03 | bnlg1037 | 10.03 |
| umc1689 | 1.05 | bnlg180 | 2.05 | umc1949 | 3.06 | bnlg490 | 4.04 | bnlg1208 | 5.04 | umc1805 | 6.05 | bnlg1666 | 7.04 | umc1172 | 8.04 | bnlg1714 | 9.04 | umc1053 | 10.04 |
| umc1590 | 1.06 | bnlg1396 | 2.06 | umc1404 | 3.07 | umc1303 | 4.05 | bnlg1237 | 5.05 | umc2170 | 6.06 | umc1154 | 7.05 | umc1562 | 8.05 | umc1078 | 9.05 | umc1506 | 10.05 |
| umc1356 | 1.07 | bnlg1045 | 2.07 | umc1521 | 3.08 | bnlg2291 | 4.06 | umc1524 | 5.06 | umc1248 | 6.07 | umc1724 | 8.06 | umc1310 | 9.06 | bnlg1028 | 10.06 | ||
| umc1991 | 1.08 | bnlg1316 | 2.08 | umc2152 | 3.09 | umc1847 | 4.07 | bnlg1346 | 5.07 | umc1521 | 6.08 | umc1268 | 8.07 | bnlg128 | 9.07 | bnlg1360 | 10.07 | ||
| umc1082 | 1.09 | bnlg1520 | 2.09 | umc2048 | 3.10 | umc1051 | 4.08 | umc1225 | 5.08 | umc1005 | 8.08 | umc1982 | 9.08 | ||||||
| bnlg1347 | 1.10 | umc1101 | 4.09 | umc1829 | 5.09 | umc1663 | |||||||||||||
| bnlg1055 | 1.11 | bnlg589 | 4.10 | 8.09 | |||||||||||||||
| umc1797 | 1.12 | bnlg2186 | 4.11 |
Fig. 4.
Hypothetical BSA of the sh1 mutant in A619. The bulked samples are derived from an F2 mapping population with the inbred line B73. The represented SSRs (see Table 1) span the genome from bin 8.08 to bin 10.00. (1) wild-type parent, (2) mutant parent, (3) wild-type pool, (4) mutant pool, (5) F1. Based on the SSR patterns, there are two potentially linked SSRs, bnlg244 and umc1982. It is important to note, however, that only bnlg244 is flanked by SSR markers on adjacent bins with an overrepresentation of one of the two alleles. This is usually additional evidence for potential linkage. In the case of umc1982, the F1 sample suggests preferential amplification of one of the two segregating alleles (see Appendix 1, Note 10).
4. Fine mapping
While BSA (step 3) provides a rough chromosomal location, it is still necessary to substantially refine the map position of the mutant of interest for the eventual cloning of the gene. This will require developing new molecular markers and transitioning from a genetic map to a physical map. Keep in mind that the resources available for fine mapping are quickly evolving and constantly updated (a list of useful databases is provided in Table 2). Furthermore, with the release of the first assembled genomic sequence for maize (Schnable et al., 2009), a wealth of additional information became available. For these reasons, we recognize that there are multiple, yet equally valid, ways to proceed. Here we describe a procedure that has worked successfully for us in the past.
Table 2.
Available online resources.
| Database | Website URL | Function |
| Maize Genetics and Genomics Database (Lawrence et al., 2004) | http://www.maizegdb.org | SSRs; SNPs; genetic maps; gene predictions; genome browser; physical map |
| National Center for Biotechnology Information | http://www.ncbi.nlm.nih.gov | Alignment software; BLAST; BAC sequences |
| Phytozome | http://www.phytozome.org | Sorghum (Paterson et al., 2009), maize, rice, and Brachypodium genome browsers |
| Gramene (Ware et al., 2002) | http://www.gramene.org | Rice genomic sequence browser |
| Panzea (Zhao et al., 2006) | http://www.panzea.org | SNPs |
| PlantGDB (Dong et al., 2004) | http://www.plantgdb.org | Gene predictions; genome browser |
| Primer3 (Rozen and Skaletsky, 2000) | http://bioinfo.ut.ee/primer3-0.4.0/primer3/ | Primer design for PCR amplification of genic regions |
| dCAPS markers (Neff et al., 2002) | http://helix.wustl.edu/dcaps/dcaps.html | Identify CAPS markers; design dCAPS primers |
Before proceeding, it is important to understand that the maize physical map is constructed of contigs and bacterial artificial chromosomes (BACs). A contig is a set of overlapping (contiguous) DNA fragments. BACs are bacterial plasmids carrying large fragments (∼100–150 Kb) of genomic DNA that have been used to assemble the sequence of the maize genome. Large contigs are usually assembled by overlapping sequences of BAC clones. Genetic markers often define a unique sequence that has been localized to a specific BAC, and thus to a specific contig. These markers are said to be “anchored” and make it possible to transition from the genetic map to a physical map location. Anchoring information for individual markers is usually available at MaizeGDB (http://www.maizegdb.org; Table 2).
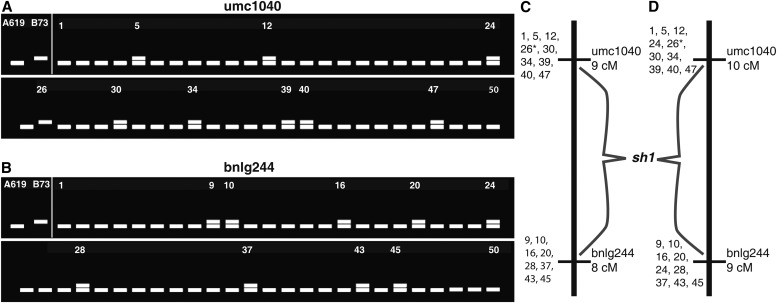
a. Identify polymorphic markers flanking the mutation (i.e., markers located proximal and distal to the mutation). These markers will define the interval on a genetic map containing the mutant locus. Using the map location determined in step 3g, select additional polymorphic SSRs in flanking bins. Genotype each individual from the mutant pool with these new markers (if dealing with a BC population, both mutant and wild-type individuals should be genotyped). Identify individuals that are recombinant between these SSRs and the mutant locus. Recombinants are individuals whose SSR genotype does not match the genotype at the mutant locus. These recombinant individuals make it possible to order the SSR loci relative to the mutant. Any markers that are on the same side of the mutation should have recombination in the same F2 or BC individuals, with closer markers showing recombination in a progressively smaller subset of the F2 or BC individuals. Markers that are on the opposite sides of the mutation should show recombination in different F2 or BC individuals (Appendix 1, Note 11). Identify two SSR markers located on the opposite sides of the mutant locus (Fig. 5) and calculate the recombination frequency between the mutant locus and the SSR markers to establish an ordered genetic map. If possible, find flanking markers that are each ∼5 cM or less from the mutant to reduce double recombination events that will complicate later mapping.
b. Extract genomic DNA from all the mutant samples of a large (approximately 500 chromosomes, see Appendix 1, Note 12) F2 mapping population. If using a BC population with either a recessive or a dominant mutant, extract also all wild-type samples (see Fig. 2B). A “quick-and-dirty” protocol (Appendix 2) for DNA extraction of the entire mapping population is suggested to reduce time. Most of these samples will not be used as only the recombinants are informative for fine mapping. Store numbered leaf samples from each individual at −80°C as these will provide a backup source of DNA and allow for a more “clean” DNA extraction from just the recombinant individuals.
c. Genotype each sample from step 4b with the flanking SSR markers determined in step 4a. Identify recombinant individuals with each flanking marker; these should comprise a largely nonoverlapping set (Fig. 5). If there are more available polymorphic SSRs within the region defined by the flanking markers, use these to genotype only the recombinants and further narrow the interval. Ultimately, a set of recombinant individuals for each of your closest flanking markers will be identified (Appendix 1, Note 13). Because these recombinants will be genotyped with several more markers in subsequent steps, we suggest extracting DNA from the leaf samples set aside earlier at −80°C (step 4b).
d. Analyze the corresponding physical map of the genetic interval defined by the flanking SSR markers. Anchor these markers by identifying BACs, or sequence contigs containing them, at MaizeGDB (http://www.maizegdb.org) or Gramene (http://www.gramene.org), taking care to search the most recent genome assembly; these will define the physical interval containing your gene. Determine the ratio of the genetic:physical distances for your interval, by dividing the genetic distance (in cM) by the physical distance (in mega base pairs [Mb]). This analysis (cM/Mb) provides an estimate of the recombination rate in your interval, and can help determine if the number of recombinants remaining will narrow the region to a reasonable number of genes to evaluate in step 5 (Appendix 1, Note 14).
e. After exhausting all of the polymorphic SSR markers in the region, if the physical region is still too large (see step 4f below) it will be necessary to develop new markers to further restrict the interval. There are multiple ways to proceed as there are different types of molecular markers. This step requires the use of information from different databases (Table 2). Depending on the background of the mapping population, there may already be information available on polymorphic SNPs, SSRs, and insertion-deletions (indels) (Qu and Liu, 2013; Xu et al., 2013; Settles et al., 2014) (Appendix 1, Note 15). If using backgrounds for which SNP information is not available, new markers must then be developed using sequence polymorphisms found in genes within the interval. Identify several predicted unique genes in the mapping interval. To avoid retrotransposon or other potentially repetitive sequence, we generally select genes that have been annotated with a predicted function, excluding all repetitive elements and genes of unknown function. Design primers to amplify 1–2-kb portions of these predicted genes, and preferably design primers that bind to exons and amplify across introns to maximize the likelihood of finding polymorphisms. Amplify this region from both parents of the mapping population, isolate the bands, and send these samples to be sequenced. Align the parental sequences with blast2seq (at the National Center for Biotechnology Information [NCBI]) to identify useful polymorphisms, including indels or SNPs. If indels are present (of at least 8 bp or more), these can be resolved on 3–4% Agarose 3:1 HRB (or MetaPhor) gels after designing a new set of primers to amplify a smaller region (∼100–300 bp) across the indel (Fig. 6). If only SNPs are identified, these are often easy to develop as cleaved amplified polymorphic sequence (CAPS) markers or as derived cleaved amplified polymorphic sequence (dCAPS) markers (Konieczny and Ausubel, 1993; Neff et al., 1998). If the SNP creates a polymorphic restriction site, a CAPS marker can be created by PCR amplification of a short fragment containing the polymorphic site, followed by restriction digest of the PCR product (Fig. 6). If SNPs do not generate a restriction site polymorphism, it is often still possible to generate a dCAPS marker. One of the dCAPS primers artificially introduces one or more point mutations to create a restriction enzyme site that distinguishes the mutant from the wild-type allele (Fig. 6; Appendix 1, Note 16). The dCAPS Finder website (http://helix.wustl.edu/dcaps/dcaps.html) can be used to design primers. Following a standard PCR reaction, the samples are digested with the restriction enzyme. We use 5–8 μL of the PCR reaction in a 20-μL restriction digest with 10–20 units of enzyme for 1–2 h (buffer and incubation temperatures vary by enzyme). It is necessary to amplify a small fragment (150–250 bp) because the restriction polymorphism will often result in only small length differences following digest that must then be resolved on 3–4% agarose.
f. If the estimated distance between the SSR markers and the mutant locus is sufficiently small (<10 cM), proceed to analyze only the recombinant samples from step 4c with the newly developed indel, CAPS, or dCAPS markers. If the interval is >10 cM, it is advisable to genotype the whole population to avoid mis-scoring double recombinants as nonrecombinants, and thus losing informative recombinant samples.
g. Reiterate steps 4e–f to narrow the interval until all recombinants are exhausted or the interval is small enough to proceed to step 5 (see criteria below). These iterations are often laborious, with progress limited by the number of recombinants available (the more, the better), and by the frequency of polymorphisms among the parental lines (the more diverse the parents, the better; see step 2a).
Fig. 5.
Hypothetical scoring of the two SSRs identified in the BSA of Fig. 4 in an F2 mapping population generated by crossing the sh1 mutant in A619 background with B73. All samples analyzed are homozygous mutants. In (A) the total number of recombination events is 10 (samples no. 5-12-24-26-30-34-39-40-47). Note that in sample 26 both chromosomes are recombinant for umc1040. In (B) the total number of recombination events is 9 (samples no. 9-10-16-20-24-28-37-43-45). The estimate of the genetic distance between the mutant locus and umc1040 (A) is 10 cM (10 recombination events/100 chromosomes, because each individual has two potentially recombinant chromosomes), while bnlg244 (B) is 9 cM. Only one sample is recombinant for both SSR markers (sample 24); therefore, we can confidently assume that the two SSRs are on the opposite side of the mutant locus (flanking markers). Individuals similar to sample 24 that appear to be recombinant with both flanking markers are not uncommon, and this could be due to either double recombination or a mis-scored phenotype (see Appendix 1, Note 11). The position of both of these markers relative to the sh1 mutant is illustrated in (C) and (D), with individual recombinants indicated to the left of each marker, which will be genotyped with subsequent markers to fine map. Part C shows the map under the assumption that individual 24 is mis-scored, while part D assumes it is truly recombinant with both markers. Individual 26 is marked (*) because it is recombinant with umc1040 on both chromosomes, and thus contributes two recombination events.
Fig. 6.
Developing molecular markers for fine mapping. Examples of indel (A), CAPS (B), and dCAPS (C) markers. Three hypothetical scenarios after sequencing the same genomic regions in A619 and B73. In (A) a deletion is present in B73 but not in A619. Designing primers flanking the deletion allows the detection of a size difference in PCR products. (B) In this scenario, an SNP (in bold, underlined) is present between the two inbred lines. The SNP creates a restriction enzyme site (boxed) in A619 (TCGA, TaqI). A size difference will be detectable upon digestion of the PCR product with TaqI. In (C) the SNP (in bold, underlined) does not create a restriction site, but can be used to develop a dCAPS marker. By using the italicized oligonucleotide carrying an artificially introduced mutation (in bold) as a forward primer for PCR, the PCR product will contain a restriction enzyme site (boxed) in B73 (GAATTC, EcoRI), but not in A619. A size difference will be detectable following a restriction enzyme digest of the PCR product.
5. Evaluating candidate genes in the interval and confirmation that the correct gene has been identified
At this point, the mutation should be localized to a minimal interval flanked by markers, each showing recombination with at least one individual from the mapping population. When the physical distance delineated by your closest flanking markers is localized to a single contig and narrow enough to contain a reasonable number of genes, it is possible to begin evaluating genes within this interval.
a. Anchor your closest flanking markers to the genome and determine the number of predicted genes within the minimal interval. This is simply a matter of examining the genes (as annotated by MaizeGDB [http://www.maizegdb.org] or Phytozome [http://www.phytozome.org]) that lie within this interval. If the number of genes is sufficiently small (Appendix 1, Note 17), evaluate each gene as a possible candidate (step 5b). If the region proves too large, identify new recombinants to narrow it by increasing the mapping population (Appendix 1, Note 18).
b. Evaluate each of the genes in the interval, and make an ordered list of good candidates (Appendix 1, Note 19).
c. Analyze candidate gene sequences for lesions. Design primers and PCR amplify candidate genes from both mutant and wild type with high-fidelity Taq (Appendix 1, Note 20). Sequence these PCR products and compare wild-type and mutant sequences, looking for sequence differences that are consistent with the nature of the mutation. EMS tends to produce point mutations (more than 99% are G/C to A/T transitions) (Greene et al., 2003), although deletions are also possible, and recessive mutants should result from deleterious mutations such as premature stop codons, splice-site mutations, or changes in conserved amino acid positions. Mutants from other sources (e.g., transposon active populations or naturally occurring mutations) could be caused by a variety of possible lesions including point mutations, insertions, deletions, and chromosomal rearrangements (Appendix 1, Note 21).
d. Confirm the candidate gene. If sequencing identifies a change relative to the reference B73 sequence in a candidate gene, this does not guarantee that this change causes the mutant phenotype. It could be a naturally occurring polymorphism or a linked mutation. Naturally occurring polymorphisms can be ruled out by sequencing the known progenitor of the mutant, if available (Appendix 1, Note 22). Ruling out a linked mutation is more difficult, and it is common practice in maize to sequence multiple independent alleles, which should each have distinct lesions in the same gene. Complementation of the mutant phenotype by transgenic insertion of the wild-type copy is an alternative approach, although less common given the time and expense of generating transgenic plants and performing the complementation crosses (Appendix 1, Note 23).
CONCLUSIONS
The preceding protocol has proven successful for positional cloning a number of maize mutants, and should be effective for a large number of the maize mutants that currently exist. As the genomes of more plant species become available, we anticipate that this protocol can be broadly applied with appropriate modifications, opening up a large number of novel species to positional cloning approaches.
Appendix 1.
Notes to the text.
1. It is advisable to cross the mutant of interest to multiple different inbreds (see Fig. 2). These will be useful when creating mapping populations for positional cloning. We generally cross the mutant to at least Mo17 and B73, as these are the most well-characterized in terms of genetic markers.
2. A mutant phenotype controlled by two recessive loci would have a 15:1 (wild type:mutant) segregation ratio. Other, more complex inheritance patterns are also possible. Even if the segregation ratio does not indicate single-gene inheritance, mapping is still possible, often using the mapping strategies described here. However, it may also be necessary to map each locus separately using F3 populations. Mapping a double mutant with F3 populations involves selfing multiple normal F2 individuals and examining their F3 progeny. It is expected that 4/15 of the normal F2 individuals will be homozygous for one of the mutations, but heterozygous at the other mutant locus; of these, half (2/15) will be homozygous for one mutation or the other. Each selfed F2 will generate a distinct F3 family which can then be scored. After screening ∼30 F3 families, you would expect to identify eight families segregating the double mutant phenotype in a 3:1 ratio. Each of these F3 families is fixed (homozygous) for one of the mutant loci, but segregating the other. Although you cannot know beforehand which mutant locus is fixed in any given family, if all eight are used for rough mapping, there is a good likelihood that each mutant locus will be segregating on its own in at least one of the eight families. An F3 family segregating each locus individually can then be used for fine mapping.
3. If the mutation does not show full expression or penetrance in any inbred background, it is still possible to proceed with mapping as long as the analysis is limited to plants that can be confidently genotyped at the mutant locus based on their phenotype.
4. The ideal situation is to start with a mutation in a uniform inbred background. If the background is not inbred or is unknown, it is best to introgress the mutation into a standard inbred background such as B73 or Mo17 by crossing to that inbred several times. This introgression will provide a more uniform genetic background at most loci that are not linked to the mutation, which will help during initial mapping because marker polymorphisms will be known for the inbred. As mapping progresses, however, even a well introgressed mutant will still retain a significant portion (likely ≥5 cM) of the progenitor genome at the mutant locus. Thus it is imperative to use the introgressed mutant rather than the inbred when developing new markers for fine mapping. This can be done by simply isolating DNA from a fully introgressed homozygous mutant, and using this for all marker development. Although a known and uniform genetic background is preferred, introgressions take a long time, and in practice this step can often be skipped. If introgression is not performed, it is necessary to keep in mind an important caveat. When the mutant is from a non-inbred (e.g., hybrid) background, the mapping populations generated from each individual F1 (steps 2a–c) should be analyzed separately, as they will be segregating differently for any markers that were heterozygous in the mutant parent. In addition, DNA should be extracted from each F1 individual for use as a control when genotyping its mapping progeny. In this way, it will be clear which of the alleles was inherited from heterozygous loci of the mutant parent. This is particularly important when doing the initial bulked segregant mapping (step 3). If the mutant parent line is not inbred, the individuals used for BSA should be the progeny of a single F1 individual. Furthermore, when the progenitor background is unknown it is even more important to create mapping populations with multiple inbreds and, as the mapping progresses, to focus on the mapping population that shows the highest degree of polymorphism.
5. BC populations are particularly useful for dominant mutants that are either male or female sterile, or cannot be selfed for some other reason. BC populations also tend to be more uniform than F2s, which may help the scoring of some mutant phenotypes. Furthermore, when there is good penetrance and expressivity of the mutant phenotype, the genotype for the mutant locus can be determined for all of the progeny from a BC population, which means that all the progeny can be used for fine mapping (step 4). However, because there is no informative recombination in the parental BC line, the mapping resolution is reduced by 50% relative to an F2 population, and thus the number of individuals analyzed from a BC mapping population must be twice that of an F2 population to have the same resolution.
6. Both chromosomes of individuals from an F2 mapping population are potentially recombinant (as opposed to just one in a BC population), giving more mapping power per individual. However, for dominant or recessive mutants only 25% of the individuals in the mapping population can be used for fine mapping (step 4) because the other individuals have an ambiguous genotype at the mutant locus (see Fig. 2C). Consequently, more field space is necessary to grow large F2 mapping populations. If space is not an issue, however, only the informative phenotypes need to be analyzed, and these provide twice the mapping power compared to a BC population.
7. When using the MassARRAY System for BSA, pools of at least 20 or more individuals are required to provide enough power for detection of linkage (30 or more are suggested). Although it requires more work, it is possible to pool extracted DNA samples rather than tissue. Pooling tissue reduces the number of DNA extractions required and avoids the inevitable errors associated with pooling individual DNA samples.
8. To modify the single-hole paper punch to accommodate a 15-mL Falcon tube, we usually cut a hole in the cap of the tube, large enough to fit the punch when pressed. Then a notch is cut on the side of the cap so that the bottom part of the punch slips into the cap. We secure the cap to the punch using glue. The bottom of the cap is left intact and is ready to hold the Falcon tubes for harvesting the leaf punches (Fig. 3).
9. To select polymorphic SSRs to use for BSA, a valuable resource is MaizeGDB (http://www.maizegdb.org). It is possible to browse through gel pictures of SSRs in several different inbred lines (A619, A632, B73, Mo17 are usually included, among others). Select the SSRs polymorphic between the inbred genetic backgrounds used for creating your mapping population. If the genetic background is unknown, select the SSRs that are highly polymorphic across the inbred panel to increase the chances of identifying a polymorphic marker. It is also advisable to arrange and test the selected SSRs in order, from the top of chromosome 1 to the bottom of chromosome 10. This allows rapid confirmation of SSR linkage with the mutant, because markers in nearby bins should also show linkage.
10. Occasionally, SSRs show preferential amplification of alleles from certain genetic backgrounds. Consequently, it is often helpful to analyze a DNA sample from the F1 used for creating the mapping population. This will help to identify false positives in the linkage analysis. It is also important to remember that quantitative differences in band intensity can also indicate linkage. In particular if this occurs with different SSRs in the same chromosomal region, it is a possible indication of linkage (Fig. 4).
11. When an individual appears to be recombinant with both flanking markers (e.g., #24 in Fig. 5A), it should send up a red flag as a potential mis-scored phenotype. Depending on the mutant phenotype, scoring is not always completely clear, and even when the phenotype is unambiguous, labeling errors can happen. However, the individual may also be legitimately recombinant on both sides, and thus informative. Thus, it is important to distinguish a mis-scored or mis-labeled individual, which should be ignored, from an informative double recombinant individual. Markers that are on the opposite sides of the mutation will show recombination in the same individuals if recombination events happened between both flanking markers and the mutation. Double recombination is unlikely to occur between markers <10 cM apart on a single chromosome in a mapping population of a few hundred chromosomes, due to cross-over interference during meiosis. Thus BC populations where only one parental chromosome can be recombinant are not likely to generate individuals that are legitimately recombinant with close flanking markers. However, cross-overs can occur independently without interference on the maternal and paternal chromosomes in an F2 population. The chance of a double recombination event (i.e., both parental chromosomes independently recombinant) can be estimated in an F2 population by calculating the product of probabilities for each cross-over. For example, if the genetic distance of each marker is 10 cM from the mutation, the chance of a double recombination event is 1% (0.1 × 0.1 × 100). Under this scenario, if you have an F2 population of 50 individuals (100 chromosomes) then you would expect at least one individual in that population to be recombinant with both flanking markers. The only way to distinguish a mis-scored individual from a double recombinant is to keep that individual in the set of recombinants used for fine mapping. If it is indeed a double recombinant, then as you test more closely linked markers these will eventually show linkage. However, a mis-scored individual should never show linkage even when genotyped at the causative locus.
12. The size of your mapping population will determine how effective the fine mapping and the positional cloning effort will be. It is not possible to know a priori how many samples will be needed, but try to minimize the number of samples to analyze. The size of the mapping population is mainly a function of the chromosomal region where the mutation is located, because the recombination frequency is not uniform in the genome. Recombination rates have been calculated across the maize genome and should be consulted (see Gore et al., 2009: Figures 2 and S4) to see if the mutant locus is in a region of low recombination, in which case increasing the mapping population could be necessary). Dividing the recombination frequency between flanking markers calculated in step 4a by the physical distance between those markers in the maize genome will provide a rough estimate of recombination rate around your mutation. For example, flanking markers that are 20 cM apart and span a physical distance of 28 Mb would indicate that recombination in this region is 0.71 cM/Mb, which is slightly higher than the average in maize of 0.67 cM/Mb (Farkhari et al., 2011). In this situation, a mapping population of 500 chromosomes would generate ∼100 total recombinants, which in turn corresponds to a recombinant every 280 kb, about the size of an average BAC. Thus, you could expect to localize the mutation to a single BAC with a recombination rate of 0.71 cM/Mb and a population of 500 chromosomes. Keep in mind that these calculations are only a rough guide (also see Note 14). Other considerations, such as the availability of field space to grow large mapping populations, should also be taken into account. In our experience, a good starting point is a mapping population of 500 chromosomes (either from 250 homozygous mutant F2 plants or 500 BC1 plants). More samples may ultimately be needed (and it is a good idea to generate several thousand seed that could be used if needed), but in several cases smaller populations were successfully used for cloning maize genes.
13. Be careful in organizing the data from the scoring of your mapping populations, and the map position of each marker. This is crucial to recognize potential mistakes in the population, such as contaminant samples, and prevents problems in the following map-based cloning step. We generally use an Excel sheet with a row of markers in order of genetic position across the top, and a column of each recombinant along the side. The genotype for each of these recombinants can then be scored for each of the markers.
14. A high cM/Mb ratio (i.e., more than the maize average of 0.67 cM/Mb) indicates high recombination, and is good news for positional cloning. You can also count the number of genes in this interval and divide by the number of remaining recombinants to get a sense of the number of genes to be expected inside the interval defined by your closest recombinants (although this is a very imprecise prediction because recombination is not expected to be evenly distributed across the interval). It is also important to note that often the genomic region flanked by the SSR markers is made of multiple nonoverlapping contigs. In this case, the cM/Mb ratio is likely to be overestimated, given that the gap between two contigs is of unknown size. An accurate estimate of the cM/Mb ratio is only possible if the flanking markers are on a single contig. Therefore, it is recommended to identify new molecular markers that restrict the region of interest onto a single physical contig before making this calculation.
15. Databases and marker resources are always changing. In particular, with the release of the draft of the B73 genome sequence (Schnable et al., 2009), and subsequent resequencing of diverse maize lines, a number of resources have been developed. If using a B73 × Mo17 mapping population, a valuable resource to develop indel or dCAPS markers is the SNP data set based on Mo17 resequencing at MaizeGDB (http://www.maizegdb.org; Table 2). This Mo17 sequence also includes insertion-deletion polymorphisms (indels) that can be rapidly developed into a polymorphic PCR product if the indel is 8 bp or greater. Settles et al. (2014) have conveniently catalogued these indels (see their Table S1) and shown that 22–31% of these indels are also polymorphic with B73 × W22 and Mo17 × W22 populations, respectively. Resequencing of diverse maize inbred lines has produced more than 100,000 potentially polymorphic SSR loci that have been catalogued (Qu and Liu, 2013; Xu et al., 2013). These markers were designed to be polymorphic among a broad diversity of maize lines (including tropical inbreds), and thus may not be polymorphic among the much more closely related inbreds commonly used for forward genetics. However, given the ease of genotyping with SSR markers, this is a resource that should be consulted before developing labor-intensive CAPS and dCAPS markers.
16. When developing a new marker from a predicted gene at MaizeGDB (http://www.maizegdb.org) or Phytozome (http://www.phytozome.org), confirm the physical location of the gene using more than just one database. Validate the physical location by searching for the genomic region of the predicted gene in other databases, such as the NCBI High-Throughput Genomic Sequences (HTGS) database (http://www.ncbi.nlm.nih.gov/genbank/htgs). This search usually identifies the BAC clones where the locus is mapped. Occasionally mistakes occur in databases. It is also helpful to compare syntenic regions in maize and sorghum. For dCAPS markers, preference should be given to primers with only one mismatch, not located at the 3′ end of the primer. When possible, choose a restriction enzyme already available in your laboratory.
17. The number of genes to be considered “sufficiently small” is largely up to the individual and his or her willingness to go through the laborious work of sequencing each candidate gene in the region to identify causative lesions. With some luck, there will be an obvious candidate that can be sequenced first, such as an ortholog of a mutant that has a similar phenotype in another species. If polymorphic, any interesting candidate that is sequenced can also be used as a new marker to narrow the interval in the mapping population. It is advisable to evaluate the genes in the interval once it spans 50 or fewer annotated genes. In highly recombinant and polymorphic regions of the genome, it is often possible to define a region of 10 or fewer genes using a mapping population of ∼300 chromosomes. However, in other regions it may not be possible to define such a narrow window even with a much larger population. In these cases, it is necessary to consider the time it would take to increase the mapping population and identify new recombinants compared to the time it would take to systematically sequence each candidate gene in the interval.
18. It is possible that the closest flanking markers will be located on separate contigs and thus span a gap of unknown size. Even if this is the case, it is possible to have a general idea if there are any genes present in the gap by comparing syntenous regions of other grass genomes. A careful comparison of the Sorghum, rice, and Brachypodium genomes will reveal if they have other genes that may have been missed in the maize assembly. Often the maize sequence for these “missing” genes can be obtained by a BLAST search of the NCBI HTGS databases (Table 2). Alternatively, several contigs have not yet been placed within the assembled maize chromosomes. These can also be BLASTed with orthologs from the syntenous region of other grasses. If a sequence is identified in this way, markers can be developed for these genes as described in step 4e. However, because these markers are not anchored with the genetic map, they must be tested for linkage on several mutants before proceeding to test just the informative recombinants.
19. What constitutes a good candidate gene depends on the nature of the mutant. Developmental mutants, for example, are frequently transcription factors or signaling molecules. If similar mutants have been cloned, consider homologous genes present in the interval. However, do not place too much confidence in the automated gene prediction programs that have produced these gene annotations. Frequently genes are wrongly annotated, or missed entirely. Using the genome browser at Phytozome (http://www.phytozome.org) or MaizeGDB (http://www.maizegdb.org), it is possible to check for expressed sequence tag (EST) sequences that align to each annotated gene. Check these for genes expressed in tissues that make sense for the mutant phenotype. Also look for ESTs aligning to regions of the interval without predicted gene models as this may indicate the presence of a gene that is not annotated. Another method to confirm proper annotation of maize genes is to examine the syntenous region of the Sorghum genome for an orthologous gene. However, lack of a Sorghum ortholog in this region is not sufficient indication that the gene is improperly annotated in maize.
20. If the candidate gene is large, it is often helpful to design primers that amplify smaller overlapping regions, which will speed the sequencing process. Furthermore, primers can be designed to exclude large introns, which generally do not need to be sequenced, so long as exon-intron junctions are included.
21. Chromosomal inversions will result in a suppression of recombination for markers internal to the inversion. If there is an unusual decrease in the cM/Mb ratio as the locus is narrowed, an inversion becomes likely. If a deletion is causing the mutation, it should not be possible to amplify the gene from the mutant, but it should be possible to do so from the wild-type progenitor.
22. If the wild-type progenitor of the mutant is available, this sample should be used to obtain the wild-type sequence. This will help avoid confusing naturally occurring sequence polymorphisms with mutations that may be causative. If the progenitor is not available, it may help to sequence multiple diverse inbreds to see if the same change is present as a naturally occurring polymorphism.
23. If there is only one allele of a recessive mutant, new alleles can generally be created by crossing wild-type pollen treated with EMS (Neuffer, 1994) onto mutant ears, and screening for progeny that show the mutant phenotype. These individuals should carry the original mutant allele from the maternal parent and a new EMS-induced allele from the paternal parent.
Appendix 2.
DNA isolation protocols and materials list.
A. Genomic DNA isolation to generate bulked segregant pools (see Methods and Results section: “Bulked segregant analysis—rough mapping,” step 4).
1. Grind pooled tissue in liquid nitrogen to a fine powder using a mortar and pestle.
2. Transfer the frozen powder to 15-mL Falcon tubes and add 2 mL of extraction buffer, pre-warmed to 65°C. Do not allow the tissue to thaw until the extraction buffer has been added.
3. Quickly vortex the samples and place them in a water bath at 65°C for 10 min.
4. Add 800 μL of 3 M sodium acetate, mix and place in ice for 10 min.
5. Add an equal volume of phenol:chloroform:isoamylalcohol and vortex. Spin in a centrifuge at 3000 × g at room temperature for 15 min.
6. Transfer the supernatant (aqueous phase) to a new tube. Add an equal volume of chloroform:isoamylalcohol, vortex, and incubate the samples at room temperature for 3 min.
7. Centrifuge at 3000 × g for 10 min. Transfer the supernatant (aqueous phase) to a new tube.
8. Add an equal volume of isopropanol, gently mix, and incubate for 10 min at room temperature. At this point, a cloudy DNA precipitate should be visible.
9. Centrifuge at 3000 × g for 10 min. Remove supernatant, leaving the DNA pellet undisturbed.
10. Wash the pellet with 1 mL of 70% ethanol. Centrifuge at 3000 × g for 5 min.
11. Remove the supernatant and air dry the DNA pellets. Take care not to over-dry the DNA pellets at this point (or at the final precipitation, step 18) or they will become very difficult to resuspend.
12. Carefully resuspend the DNA in 400 μL of 0.1× TE and transfer to a small 1.5-mL tube.
13. Add 4 μL of RNase A to the samples and incubate for 30 min at 37°C.
14. Add 0.4 volumes of 3 M sodium acetate and place in ice for 10 min.
15. Add an equal volume of phenol:chloroform:isoamylalcohol, briefly vortex, and incubate at room temperature for 3 min.
16. Centrifuge at maximum speed for 15 min.
17. Transfer supernatant (aqueous phase) to a new tube, add 2 volumes of 100% ethanol (ice cold), and gently mix. Incubate at −20°C for 2 h or overnight.
18. Spin at 14,000 rpm for 15 min. Remove supernatant, leaving the DNA pellet undisturbed.
19. Wash pellet with 500 μL of 70% ethanol. Spin at 14,000 rpm for 5 min.
20. Remove the supernatant and air dry the DNA pellet, then resuspend in 50–100 μL of deionized sterile water. For the MassARRAY System at Iowa State University, the required DNA concentration is 25 ng/μL and the total amount of DNA is 2.5 μg per pool.
B. Genomic DNA isolation for individual leaves for fine mapping (see Methods and Results section: “Fine mapping,” step 2).
1. Collect a small piece of leaf in a 1.5-mL tube.
2. Add 400 μL of extraction buffer (pre-warmed at 65°C), and grind in buffer at room temperature with a small pestle.
3. Put in a water bath at 65°C for 10 min.
4. Add 150 μL of 3 M sodium acetate and put on ice for 10 min.
5. Add 500 μL of chloroform:isoamylalcohol, vortex for 15 s, then let the sample rest for 3 min at room temperature.
6. Spin at 14,000 rpm for 10 min.
7. Transfer 350 μL of the aqueous (top) phase to a new 1.5-mL tube. Add 300 μL of isopropanol, gently mix, and let sit at room temperature for 10 min.
8. Spin at 14,000 rpm for 10 min to precipitate DNA.
9. Remove supernatant. Wash with 70% ethanol.
10. Spin at 14,000 rpm for 6 min.
11. Remove ethanol and air dry the pellet for approximately 15 min.
12. Resuspend in 200 μL of deionized sterile water. The sample is then ready for PCR amplification.
C. Materials list (corresponds to the Methods and Results sections: “Genetic characterization of mutant” and “Create a mapping population”).
1. Maize mutant to be cloned.
2. Maize inbred lines (see Fig. 2A), at a minimum B73 and Mo17.
Bulked segregant analysis—rough mapping
1. Single-hole paper punch (Fig. 3).
2. DNA extraction buffer: 100 mM Tris-HCl, pH 8.0, 50 mM EDTA, 500 mM NaCl, 1% (w/v) SDS, 1% (v/v) β-mercaptoethanol.
3. Liquid nitrogen.
4. 15-mL Falcon tubes.
5. 65°C water bath.
6. 3 M sodium acetate.
7. Phenol:chloroform:isoamyl alcohol 25:24:1 (v:v:v), pH 8.2.
8. Centrifuge with rotor (or adaptor) for 15-mL Falcon tubes.
9. Chloroform:isoamylalcohol 24:1 (v:v).
10. RNase A 10 mg/mL. Store at −20°C.
11. Tris-borate EDTA buffer (TBE). Make a 10× stock solution: 0.89 M Tris base, 0.89 M boric acid, 20 mM EDTA pH 8.0.
12. Agarose 3:1 HRB (AMRESCO, Solon, Ohio, USA) or MetaPhor (Lonza, Basel, Switzerland) agarose.
13. PCR buffer, 5× stock: 0.25 M Tris-HCl, pH 8.5, 0.1 M KCl, 12.5% (w/v) Ficoll 400, 0.25% (w/v) bovine serum albumin, 0.1% (w/v) xylene cyanol. Store at −20°C.
14. PCR reaction: 1× PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 0.4 μM oligonucleotides, 1 unit Taq polymerase, 20–100 ng of genomic DNA. Loading dye is not necessary as it is included in 5× PCR buffer.
15. 0.1× Tris EDTA (TE) buffer: 1 mM Tris-HCl, 0.1 mM EDTA, pH 8.0.
Fine mapping
1. DNA extraction buffer: 100 mM Tris-HCl, pH 8.0, 50 mM EDTA, 500 mM NaCl, 1% (w/v) SDS.
2. Primers for CAPS and dCAPS markers. Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/; Rozen and Skaletsky, 2000) is a useful website for primer design.
3. High-fidelity Taq polymerase, such as Phusion Taq (New England Biolabs, Ipswich, Massachusetts, USA).
4. Tris-acetate EDTA buffer (TAE). Make a 20× stock solution: 0.4 M Tris base, 0.2 M acetic acid, 10 mM EDTA pH 8.0.
5. QIAGEN gel extraction kit (QIAGEN, Valencia, California, USA), or equivalent, for gel purification of PCR products. Follow manufacturer’s instructions.
6. For online sequence alignment, use the “align 2 sequences” BLAST tool at NCBI (www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi).
7. Restriction enzymes for CAPS/dCAPS.
LITERATURE CITED
- Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., et al. 2012. Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology 30: 174–178. [DOI] [PubMed] [Google Scholar]
- Bortiri E., Chuck G., Vollbrecht E., Rocheford T., Martienssen R., Hake S. 2006. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Schlueter S. D., Brendel V. 2004. PlantGDB, plant genome database and analysis tools. Nucleic Acids Research 32: D354–D359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkhari M., Lu Y. L., Shah T., Zhang S. H., Naghavi M. R., Rong T. Z., Xu Y. B. 2011. Recombination frequency variation in maize as revealed by genomewide single-nucleotide polymorphisms. Plant Breeding 130: 533–539. [Google Scholar]
- Fu H., Zheng Z., Dooner H. K. 2002. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proceedings of the National Academy of Sciences, USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Barazesh S., Malcomber S., Hall D., Jackson D., Schmidt R. J., McSteen P. 2008. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proceedings of the National Academy of Sciences, USA 105: 15196–15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M. A., Chia J. M., Elshire R. J., Sun Q., Ersoz E. S., Hurwitz B. L., Peiffer J. A., et al. 2009. A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Greene E. A., Codomo C. A., Taylor N. E., Henikoff J. G., Till B. J., Reynolds S. H., Enns L. C., et al. 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F. M. 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant Journal 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Lawrence C. J., Dong Q., Polacco M. L., Seigfried T. E., Brendel V. 2004. MaizeGDB, the community database for maize genetics and genomics. Nucleic Acids Research 32: D393–D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Goodman M., Muse S., Smith J. S., Buckler E., Doebley J. 2003. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics 165: 2117–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen H. D., Makarevitch I., Shirmer R., Emrich S. J., Dietrich C. R., Barbazuk W. B., et al. 2010. High-throughput genetic mapping of mutants via quantitative single nucleotide polymorphism typing. Genetics 184: 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Yeh C. T., Tang H. M., Nettleton D., Schnable P. S. 2012. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C. S., Scheible W. R. 2000. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiology 123: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Dailey S., Settles A. M. 2010. Distributed simple sequence repeat markers for efficient mapping from maize public mutagenesis populations. Theoretical and Applied Genetics 121: 697–704. [DOI] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R. V. 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences, USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M. M., Neff J. D., Chory J., Pepper A. E. 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant Journal 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Neff M. M., Turk E., Kalishman M. 2002. Web-based primer design for single nucleotide polymorphism analysis. Trends in Genetics 18: 613–615. [DOI] [PubMed] [Google Scholar]
- Neuffer M. G. 1994. Mutagenesis. In M. Freeling and V. Walbot [eds.], The maize handbook, 212–218. Springer-Verlag, New York, New York, USA. [Google Scholar]
- Neuffer G. M., Coe E. H., Wessler S. R. 1997. Mutants of maize. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., Gundlach H., Haberer G., et al. 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Qu J., Liu J. 2013. A genome-wide analysis of simple sequence repeats in maize and the development of polymorphism markers from next-generation sequence data. BMC Research Notes 6: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. In S. Misener and S. A. Krawetz [eds.], Methods in molecular biology, vol. 132: Bioinformatics methods and protocols, 365–386. Humana Press, Totowa, New Jersey, USA. [DOI] [PubMed] [Google Scholar]
- SanMiguel P., Tikhonov A., Jin Y. K., Motchoulskaia N., Zakharov D., Melake-Berhan A., Springer P. S., et al. 1996. Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768. [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S., Liang C., et al. 2009. The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Weigel D. 2011. Fast-forward genetics enabled by new sequencing technologies. Trends in Plant Science 16: 282–288. [DOI] [PubMed] [Google Scholar]
- Settles A. M., Bagadion A. M., Bai F., Zhang J., Barron B., Leach K., Mudunkothge J. S., et al. 2014. Efficient molecular marker design using the MaizeGDB Mo17 SNPs and Indels track. G3 (Bethesda) 4: 1143–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G., Tingey S. 1996. Simple sequence repeats for germplasm analysis and mapping in maize. Genome 39: 277–287. [DOI] [PubMed] [Google Scholar]
- Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y., Faller M., Bomblies K., et al. 2005. The origin of the naked grains of maize. Nature 436: 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware D. H., Jaiswal P., Ni J., Yap I. V., Pan X., Clark K. Y., Teytelman L., et al. 2002. Gramene, a tool for grass genomics. Plant Physiology 130: 1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L., May P. E. 1989. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain-reaction. American Journal of Human Genetics 44: 388–396. [PMC free article] [PubMed] [Google Scholar]
- Whipple C. J., Hall D. H., DeBlasio S., Taguchi-Shiobara F., Schmidt R. J., Jackson D. P. 2010. A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D. B., Monde R. A., Coalter R., Barkan A. 2010. Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant Journal 63: 167–177. [DOI] [PubMed] [Google Scholar]
- Xu J., Liu L., Xu Y., Chen C., Rong T., Ali F., Zhou S., et al. 2013. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Research 20: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Canaran P., Jurkuta R., Fulton T., Glaubitz J., Buckler E., Doebley J., et al. 2006. Panzea: A database and resource for molecular and functional diversity in the maize genome. Nucleic Acids Research 34: D752–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]