Abstract
In January, 2014, increased mortality was reported in piglets with acute diarrhea on an Ontario farm. Villus atrophy in affected piglets was confined to the small intestine. Samples of colon content were PCR-positive for porcine epidemic diarrhea virus (PEDV). Other laboratory tests did not detect significant pathogens, confirming this was the first case of PED in Canada.
Résumé
Premier cas de diarrhée épidémique porcine au Canada. En janvier 2014, une mortalité accrue a été signalée chez des porcelets atteints de diarrhée aiguë dans une ferme de l’Ontario. L’atrophie des villosités chez les porcelets touchés a été confinée au petit intestin. Des échantillons du contenu du côlon étaient positifs par RCP pour le virus de la diarrhée épidémique porcine (VDEP). D’autres tests de laboratoire n’ont pas détecté d’agents pathogènes importants, ce qui confirme qu’il s’agit du premier cas de DEP au Canada.
(Traduit par Isabelle Vallières)
Porcine epidemic diarrhea virus (PEDV) is an Alphacoronavirus that was first associated with clinical disease of pigs in the United Kingdom in 1971 (1). The disease subsequently spread to many European countries, and also from Europe to Asia where it has become endemic in China, Japan, Korea, and Thailand (2). Since PED was first recognized in the United States in April 2013 (3), the disease has spread rapidly across the continental US and caused over 7111 confirmed cases in 30 states by May 31, 2014 (4). Early reports regarding genetic composition of the virus found that the US PEDV was very similar to a predominant PEDV strain/lineage circulating in China (5). However, a “variant” PEDV with < 90% nucleotide identity in the spike (S1) protein gene was also detected in Ohio (6). The Ohio “variant” PEDV was over 99% identical to a “variant” PEDV strain/lineage that emerged in China in 2010 (7).
Because of the explosive spread of PED in the US and the Canadian industry’s connectedness to the US markets, in July 2013 the University of Guelph Animal Health Laboratory (AHL) developed a PCR test to rapidly detect PEDV in cases of acute enteritis in swine. The purpose of the work described herein is to document the first Canadian identification of PEDV as a cause of acute diarrhea affecting a swine herd in Ontario.
Case description
History
In January of 2014, piglets from a 500 sow farrow-to-finish herd in southwestern Ontario experienced diarrhea, vomiting, and mortality over a 24-hour period. In the first 24 h, the clinical signs began in 1 farrowing room and spread to include almost all of the litters in this room. Affected piglets were < 1 wk of age. The other farrowing rooms were not yet affected. The herd had a past history of sporadic Escherichia coli diarrhea in piglets < 5 d of age. Injectable tylosin (200 mg/mL, Elanco, Greenfield, Indiana, USA) had been successfully used at 0.25 mL/piglet, once daily for 1 to 2 d. The diagnostic rule-outs by the referring veterinarian also included transmissible gastroenteritis (TGE) and PED. The producer was instructed to submit live, acutely affected piglets to the AHL.
The diarrhea did not respond to antibiotic treatment, and had escalated in 24 h to include most of the piglets in the affected farrowing room. The producer had noted vomiting in some litters. Piglet morbidity and mortality were 100%.
Autopsy, histopathology, and immunohistochemistry (IHC)
Four live piglets with acute diarrhea were submitted to the AHL and euthanized for autopsy. They ranged in age from 2 to 5 d, and in weight from 0.7 to 0.84 kg. Samples of stomach, small intestine, and colon were rapidly fixed in 10% buffered formalin for histologic examination. These were routinely processed, sectioned at 5 μm, stained with hematoxylin and eosin, and examined by light microscopy. For TGEV IHC, deparaffinized sections were stained using monoclonal antibody FIPV3-70 (Custom Monoclonals International, Sacramento, California, USA).
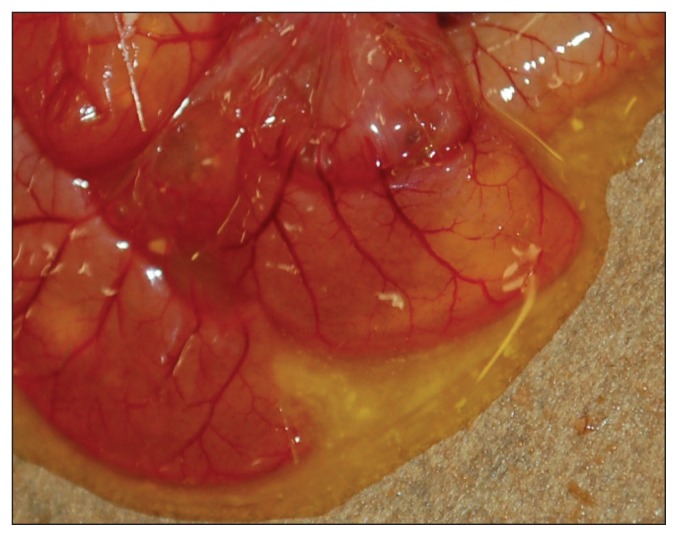
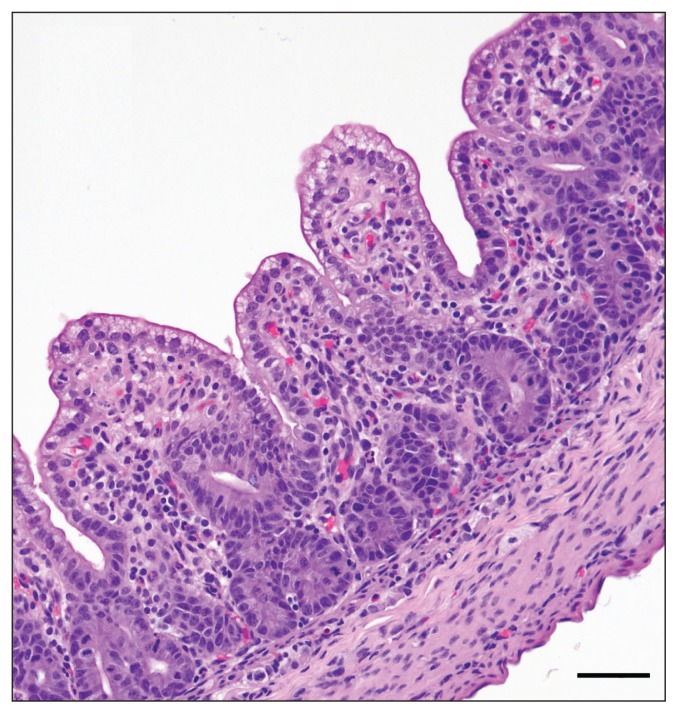
Gross pathological findings were similar in all 4 piglets. The piglets were transported together in a ventilated box and were mostly (80% of body area) covered in watery feces when they were received. Body condition appeared adequate for their age with signs of minimal dehydration. Stomachs contained milk, and the small and large intestine were distended with watery yellow translucent content (Figure 1). Intestinal serosae were congested. Significant lesions were confined to the small intestine, wherein all 4 pigs had moderate to marked atrophy and fusion of intestinal villi (Figure 2). Enterocytes on villi were sometimes attenuated and occasional neutrophils or pyknotic nuclear debris could be seen in the superficial lamina propria. In some piglets, villus tip epithelium was columnar but vacuolated. Occasionally at villus tips, clumps of 2 or 3 rounded epithelial cells could be seen exfoliating. No syncytial cells were seen. TGEV IHC was negative.
Figure 1.
Small intestine of a PED infected pig, showing typical watery yellow intestinal content and congestion of serosal blood vessels.
Figure 2.
Histology of small intestine in a PED infected pig, showing marked villus atrophy and villus fusion. Bar = 50 μm
Bacterial culture and serotyping
Small intestinal samples from all piglets were submitted for E. coli F4, Salmonella spp., and Clostridium perfringens cultures. For E. coli and Salmonella spp. isolations, samples were plated onto MacConkey and Hektoen agars and incubated overnight at 35°C in atmospheric conditions. In addition, tetrathionate broth, incubated overnight at 42°C in atmospheric conditions, was used as a Salmonella enrichment medium followed by plating onto brilliant green sulfa agar plates. For isolation of C. perfringens, phenylethylalcohol plates were incubated at 37°C for 48 h in a Concept 400 anaerobic workstation (Ruskinn Technologies, Sanford, Maine, USA). Bacterial identifications were performed using matrix assisted laser desorption ionization time-of-flight mass spectroscopy. All morphologically different E. coli isolates were tested for the presence of F4 by slide agglutination using F4 specific antiserum purchased from the OIE reference laboratory for E. coli at the University of Montreal.
No E. coli F4 positive or Salmonella spp. were isolated from any of the samples tested. Moderate numbers of C. perfringens were isolated from 1 piglet and 1 C. perfringens colony was isolated from another piglet. Clostridium perfringens was not isolated from the 2 remaining piglets.
Nucleic acid extraction and polymerase chain reaction (PCR)
Colon samples were stab-swabbed and agitated in virus transport medium (VTM). Total nucleic acids were extracted from 50-μL aliquots of VTM using the MagMAX-96 Viral RNA Isolation Kit in a MagMAX Express-96 Magnetic Particle Processor (Applied Biosystems, Foster City, California, USA). Samples were tested by a triplex real-time PCR assay designed to detect PEDV, TGEV, and generic porcine coronavirus (PCoV). Primers and probes (Table 1) for PEDV nucleocapsid protein gene and conserved 3′ end of PCoV were designed for this study, while primers and the probe targeting the TGEV spike protein gene were previously described (8). A porcine deltacoronavirus (PDCoV) real-time PCR assay targeting the conserved 3′ end of the Deltacoronavirus genus was also developed for this study (Table 2), and samples were also tested for PDCoV at a later date. Real-time PCR assays were carried out in 25-μL reactions in a Light Cycler 480 (Roche, Laval, Quebec) using Ag Path-ID One-Step RT-PCR Kit (Applied Biosystems) under standard conditions (Table 3). Samples with crossing points lower than 37 were considered positive.
Table 1.
Primer sequences for PEDV, TGEV, and PCoV
| Target | Primer/probe name | Reporter | Primer/probe sequence |
|---|---|---|---|
| PEDV-NC | PEDV_1_NC_130524_Pr | FAM | AGCAATGCTGCAACATTTGGTGC |
| PEDV-NC | PEDV_1_NC_130524_F | n/a | TATGCTCAGATCGCCAGT |
| PEDV-NC | PEDV_1_NC_130524_R | n/a | AGCCACATTACCACCAAAG |
| TGEV-S | TGEV_Pr | HEX | YAAGGGCTCACCACCTACTAC |
| TGEV-S | TGEV_F | n/a | TCTGCTGAAGGTGCTATTATATGC |
| TGEV-S | TGEV_R | n/a | CCACAATTTGCCTCTGAATTAGAAG |
| PCoV 3′ | PCoV_1_130524_Pr | CY5 | AAAGATCCGCTACGACGAGCCAAC |
| PCoV 3′ | PCoV_1_130524_F | n/a | GGAGGTACAAGCAACCCTATT |
| PCoV 3′ | PCoV_1_130524_R | n/a | CCAGACGTTAGCTCTTCCATT |
PEDV-NC — porcine epidemic virus nucleocapsid protein gene; TGEV-S — transmissible gastroenteritis spike protein gene; PCoV — porcine coronavirus; n/a — not available.
Table 2.
Primer sequences for porcine deltacoronavirus (PDCoV)
| Target | Primer/probe name | Reporter | Primer/probe sequence |
|---|---|---|---|
| PDCoV 3′ | PDCoV _140214-3 _Pr | FAM | ATGCAAACTAGGGCTGGCTACTCT |
| PDCoV 3′ | PDCoV _140214-3 _F | n/a | AGCCAGAGAGCCAGTCA |
| PDCoV 3′ | PDCoV _140214-3 _R | n/a | TGTACCCTCGATCGTACTCC |
n/a — not available.
Table 3.
Cycling parameters for PCR assays
| Step | Temperature | Hold (h:mm:ss) | Ramp | Number of cycles |
|---|---|---|---|---|
| RT | 45°C | 0:10:00 | 4.4°C/s | 1 |
| Activation | 95°C | 0:10:00 | 4.4°C/s | 1 |
| Denaturation | 94°C | 0:00:05 | 2.2°C/s | 45 |
| Annealing/Extension | 60°C | 0:01:00 | 2.2°C/s | |
| Cool | 40°C | 0:10:00 | 2.2°C/s | 1 |
Four colon samples were positive for PEDV with crossing points of 20.47, 25.96, 22.92, and 21.85. Samples were negative for TGEV, PCoV, and porcine rotavirus (PoRV) A, B, and C. The tests for PDCoV were negative.
Sequencing
Conventional N and S gene PCR followed by sequencing was used to confirm the real-time PCR results. The primers used to amplify the partial N and S genes of PEDV were based on those obtained from National Veterinary Services Laboratory (NVSL), Ames, Iowa, with minor modifications. Forward primer PEDN253 — 5′-GGCATTTCTACTACCTCGGA-3′ and reverse primer PEDN992 — 5′-ATAGCCTGACGCAT CAACAC-3′ and forward primer PEDS218 — 5′-GCTAGTGG CGTTCATGGTAT-3′ and reverse primer PEDS442 — 5′-TAGGCAATTACGACCTGTTG were used to amplify 739 bp and 224 bp fragments of the N and S1 genes, respectively. These were then sequenced directly, and gel-purified or cloned prior to sequencing. Sanger sequencing reactions were performed using Big Dye Terminator chemistry version 3.1 and the products were resolved on an Applied Biosystems 3130×l genetic analyzer.
N and S gene conventional PCR assays produced amplicons of the predicted size for all 4 colon samples. Sequencing of these PCR products showed that this Ontario PEDV was 99% identical to recent PEDV isolates from the USA and China.
Discussion
The etiology of neonatal diarrhea in pigs involves various pathogens. An agent-specific diagnosis requires laboratory testing because clinical signs and autopsy are most often non-specific, and multiple pathogens may be involved (9). A sudden incursion of PEDV and extensive economic damage related to PED in the US has raised awareness and sensitized farmers, practitioners, and diagnostic laboratories in Canada.
There is extensive truck traffic that transports early-weaned piglets, market hogs, and cull sows from Ontario to the USA. Empty trucks then return to Ontario, with or without being washed, disinfected, and dried. The transport process has been shown to be a source of transmission of PEDV in the USA (10). Porcine epidemic diarrhea virus is transmitted by the fecal-oral route, and the original fear was that contaminated trucks would transfer the virus into Canada. However, the index case and the next 10 Ontario cases, as well as an isolated case in Prince Edward Island, were suspected to have a common link via contaminated feed, not transport. Bioassay testing of feed and spray-dried porcine plasma (SDPP) determined that piglets inoculated with feed did not shed PEDV, but piglets inoculated with SDPP excreted relatively high levels of PEDV. Therefore feed still could not be ruled out as a potential source of infection (11).
In the case presented in this report, clinical signs, autopsy, and histopathology were compatible with viral infection. The clinical signs, in conjunction with the lesions seen and supporting laboratory test results allowed us to conclude this was the first documented and laboratory-confirmed case of PED in Ontario and in Canada.
Acknowledgments
The authors acknowledge financial support from the Ontario Ministry of Agriculture, Food and Rural Affairs, and the many clerical and technical staff at the AHL on whom we relied during this busy period of increased surveillance and infection control.
Footnotes
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office (hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.Chasey D, Cartwright SF. Virus-like particles associated with porcine epidemic diarrhoea. Res Vet Sci. 1978;25:255–256. doi: 10.1016/S0034-5288(18)32994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song D, Park B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes. 2012;44:167–175. doi: 10.1007/s11262-012-0713-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevenson GW, Hoang H, Schwartz KJ, et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 4.USDA-APHIS. Swine enteric coronavirus disease testing summary report. Jun 19, 2014. [Last accessed December 9, 2014]. pp. 1–29. Available from: http://www.aphis.usda.gov/animal_health/animal_dis_spec/swine/downloads/swine_report_6_19_14.pdf.
- 5.Huang YW, Dickerman AW, Pineyro P, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. 2013;4:e00737–13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Byrum B, Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg Infect Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Huo JY, Chen L, et al. Genetic variation analysis of reemerging porcine epidemic diarrhea virus prevailing in central china from 2010 to 2011. Virus Genes. 2013;46:337–344. doi: 10.1007/s11262-012-0867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vemulapalli R, Gulani J, Santrich C. A real-time TaqMan RT-PCR assay with an internal amplification control for rapid detection of transmissible gastroenteritis virus in swine fecal samples. J Virol Methods. 2009;162:231–235. doi: 10.1016/j.jviromet.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moeser AJ, Blikslager AT. Mechanisms of porcine diarrheal disease. J Am Vet Med Assoc. 2007;231:56–67. doi: 10.2460/javma.231.1.56. [DOI] [PubMed] [Google Scholar]
- 10.Lowe J, Gauger P, Harmon K, et al. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg Infect Dis. 2014;20:872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasick J, Berhane Y, Ojkic D, et al. Investigation into the role of contaminated feed as a source of the porcine epidemic diarrhea outbreak in Ontario, Canada. Transbound Emerg Dis. 2014;61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]