Abstract
Successful in vivo gene delivery mediated by nonviral vectors requires efficient extracellular and intracellular gene delivery, but few studies have given attention to the former. That is why numerous gene delivery systems have succeeded in vitro, while the expected clinical success has not come about. To realize efficient extracellular gene delivery, the stability of vectors and/or their complexes with genes in body fluids is first required, which prevents loaded genes from premature unloading and degradation. Furthermore, the storage stability of vectors under common conditions is important for their widespread applications. Lipopolysaccharide-amine nanopolymersomes (NPs), a gene vector developed by our group recently, have higher than 95% in vitro transfection efficiency in mesenchymal stem cells when delivering pEGFP, and induce significant angiogenesis in zebrafish when delivering plasmid encoding vascular endothelial growth factor deoxyribonucleic acid (pVEGF). To reveal their extracellular delivery ability and storage stability, in this study their stability in various simulant physiological environments and storage conditions was systematically studied by monitoring their changes in disassembly, size, zeta potential, and transfection efficiency. Additionally, damage to the mitochondria of mesenchymal stem cells was evaluated. Results show that NPs and plasmid deoxyribonucleic acid (pDNA)-loaded NPs (pNPs) have acceptable stability against dilution, anions, salts, pH, enzyme, and serum, presumably assuring their efficient extracellular delivery in vivo. Moreover, both the lyophilized NPs at room temperature and NP/pNP solution at 4°C have high storage stability, and pNPs show low damage to the mitochondria. The acceptable stability of NPs combined with compatibility and efficient gene transfection highlight their huge potential in the clinic as a gene delivery vector.
Keywords: nanopolymersomes, extracellular delivery stability, storage stability, toxicity, polyethyleneimine, alginate, cholesterol, mesenchymal stem cells
Introduction
As a potential panacea in treating various disorders such as cancer and chronic and genetic disorders, gene therapy has roused extensive attention and thus produced an explosion of related papers during past decades. So far, great progress has been made in this field, but the expected clinical success has not come about.1–5 One of the reasons is that most of the research has focused on in vitro gene delivery in cells, and numerous novel cationic polymer gene vectors have been designed to overcome intracellular barriers such as gene encapsulation, internalization, endo-/lysosome escape, gene dissociation, and nucleus entry, but the in vivo extracellular delivery barriers have often been neglected.4–6 In fact, it is a long, difficult journey for gene delivery systems before they reach target cells in vivo, where they often undergo danger of disassembly or aggregation, gene unloading, and degradation induced by dilution, enzymes, anions, and salts in body fluids, and only when they survive from this journey might a good efficacy be expected.2–6
To survive from this journey, the acceptable stability of gene vectors/polyplexes (complexes of cationic polymer and plasmid deoxyribonucleic acid [pDNA]) against the aforementioned inducing factors is first required. It is demonstrated that the stability of polyplexes could be improved by the functionality of cationic polymer vectors/polyplexes including PEGylation, alkylation, cholesterol (Cho) modification, and anionic polymer grafting.5,7 Among them, PEGylation is one of the most extensively used means that can offer shielding effects to minimize the undesired interactions with body fluids.4,5,8 However, it was reported that PEGylation caused a reduction in transfection efficiency, presumably because it decreased the surface charge, the cellular uptake, and subsequent endo-/lysosome escape of polyplexes, and notably the high stability of PEGylated polyplexes might compromise the release of loaded pDNA.5,9,10 Alternatively, cholesterol modification has been proved to be an effective strategy to increase both the stability and the gene transfection efficacy of polyplexes, which may be realized by alleviating serum inhibition, enhancing the cellular/nuclear uptake, and decreasing cytotoxicity.4,5,7,11–17 Additionally, introducing anionic polymer such as anionic polysaccharide is able to improve the polyplex stability and the transfection efficiency by conferring the charge shielding and biodegradability, aiding the rupture of endo-/lysosome and gene release and decreasing cytotoxicity.5,18–22
Based on these studies, we engineered a novel brush copolymer of lipopolysaccharide-amine (LPSA) as a cytosolic delivery vector by introducing anionic polysaccharide of oxidized alginate (OA) and Cho to polyethyleneimine (PEI) with molecular weight of 1.8k dalton (PEI 1.8k), where OA and Cho-graft-PEI act as the backbone and side chains, respectively. Encouragingly, LPSA can spontaneously and quickly self-assemble into nanopolymersomes (NPs) in water at a concentration higher than 1.38×10−3 mg/mL.23 Therefore, LPSA normally exists in an aggregate form, namely NPs. LPSA–NP vectors are expected to achieve acceptable colloid stability and gene transfection efficacy by the synergistic effects of PEI 1.8k, Cho, and anionic polysaccharide. We have demonstrated that, as a gene vector, NPs obtain higher than 95% transfection efficiency in mesenchymal stem cells (MSCs) in vitro23 and induce significant angiogenesis in zebrafish when delivering plasmid encoding vascular endothelial growth factor deoxyribonucleic acid (pVEGF).24 To the best of our knowledge, such transfection efficacy is outstanding among nonviral gene vectors,4,5,25 suggesting the huge potential of NPs in gene therapy. To explore whether their extracellular delivery stability and storage stability satisfy demands of future clinical applications, in this study we investigated the stability of empty or pDNA-loaded LPSA NPs (pNPs) under analogous physiological environments and different storage conditions by monitoring the changes in size, zeta potential, disassembly, and transfection efficiency. Additionally, considering that polycations can induce mitochondrially mediated apoptosis, the damage of pNPs to mitochondria was also evaluated.
Materials and methods
Materials
LPSA was prepared following our established method.23 Briefly, PEI-graft-Cho was first synthesized by the amidation reaction between chloroformates in cholesteryl chloroformate and primary amines in PEI 1.8k, and then PEI-Cho was grafted onto oxidized sodium alginate through the condensation of primary amine groups in PEI-Cho and aldehyde groups in OA. Thereafter, the formed graft copolymer was reduced by sodium borohydride to obtain the brush copolymer of LPSA. In LPSA, OA and PEI-Cho serve as backbone and side chains, respectively, and the nitrogen content and mass ratios of OA:PEI:Cho were controlled to be 11.29 mmol/g and 48.5:11.2:40.3, which were determined by elemental analysis and proton magnetic resonance.23 The plasmids expressing enhanced green fluorescence protein (EGFP) were purchased from FulenGene (Guangzhou, People’s Republic of China), and their amplification and purification followed our established method.23 Neonatal Sprague Dawley rats at the age of 1–3 days were obtained from the Laboratory Animal Center of Sun Yat-sen University (Guangzhou, People’s Republic of China). MSCs were obtained from these rats, and their isolation, purification, culture, and passage were performed according to our established method.23 All animal experiments were approved by the Animal Ethics Committee of Sun Yat-sen University and conducted according to Institutional Animal Care and Use Committee guidelines. The other reagents were purchased from local suppliers. All reagents were used as received without further purification.
Preparation of NPs and pNPs
NPs were prepared by directly dissolving LPSA in distilled water via mild mechanical stirring, and they were kept at 4°C as stock solutions for subsequent experiments. According to different N/P ratios (the molar ratio of nitrogen in LPSA to the phosphate in pEGFP), pEGFP solution (40 μg/mL) and an equal volume of NP stock solution with different concentrations were mixed and vortexed for 4 seconds, followed by standing at room temperature for 30 minutes. In this paper, all pNP solutions were freshly prepared in ultrapure water before use, and their N/P ratio was set at 60 unless otherwise noted. We chose this N/P ratio because it is optimal for serum-free gene transfection in MSCs.23,24
In vitro cell culture and transfection
In vitro cell culture and serum-free transfection were performed following our published method.23 Briefly, MSCs (passage 3–7) were seeded in 24-well plates at a density of 1×105 cells/well and cultured in 1 mL complete medium (HyClone Dulbecco’s Modified Eagle’s Medium/F12 1:1 supplemented with 10% fetal bovine serum [FBS], 100 μg/mL streptomycin, and 100 U/mL penicillin). When cell confluence reached ~70%, the medium was replaced with transfection medium that contained pNPs with predetermined N/P ratios and FBS with predetermined concentration. Then, cells were routinely cultured for 4 hours for serum-free transfection or for 24 hours for serum transfection, respectively. After that, cells were further incubated in normal fresh complete medium. At 48 hours posttransfection, all cells were washed thrice with PBS. Then, some cells were observed under a fluorescent microscope, and others were routinely treated for determination of transfection efficiency (percentage of GFP-positive cells) and the average fluorescence intensity per cell with a flow cytometer (BD Influx, Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Stability to dilution
Generally, for gene delivery systems, the first barrier of in vivo applications is dilution. Their stability to dilution is the prerequisite for gene protection and target delivery. The stability of pNP solutions to dilution was evaluated by critical micelle concentration (CMC) and their changes in size, size distribution, and structure integrity (the dissociation of pDNA from pNPs). CMC was measured via surface tension method.26,27 Briefly, aqueous pNP solutions with known LPSA concentrations ranging from 1×10−8 to 1 mg/mL in PBS (pH 7.4) were freshly prepared as mentioned previously, then placed in a thermostatic beaker at 25°C for 30 minutes to equilibrate the system, and their surface tension was determined using a platinum plate method on a surface tensiometer (K12, Krüss, Hamburg, Germany). The CMC was estimated by plotting the surface tension to the logarithm of LPSA concentration.
The size of diluted pNP solutions whose concentration ranged from 2 to 2000 μg/mL was measured by dynamic light scattering (DLS) technique on a Malvern Zetasizer Nano ZS90 at room temperature. PEI 25k/pDNA complex was used as a control. Furthermore, the possibility of dissociation of pNPs induced by dilution was estimated by agarose gel electrophoresis following our published method.22,23 The aliquots of diluted pNP solution were incubated with 20 mg/mL heparin solution at room temperature for 2 hours to completely dissociate pDNA from pNPs. Then, the diluted solution with or without heparin treatment was analyzed on gel electrophoresis.
Stability to pH
Since pH is different in different body fluids and different organelles, we evaluated the short-/long-term pH stability of pNPs. For the short-term case, pNP solutions containing 0.035 mg/mL LPSA and 2 μg/mL pDNA were prepared, and their pH levels were adjusted to the range of 1.75–12.0 by using 3 mol/L NaOH and 12 mol/L HCl. The application of concentrated acid/base was expected to minimize the change in solution concentration. After overnight equilibrium at 4°C, the solutions were taken for size and zeta potential determination by DLS and morphology observation on a transmission electron microscope (TEM) (JEM-1400, JEOL Ltd, Tokyo, Japan) using a phosphotungstic acid negative staining technique at room temperature. For the long-term pH stability, freshly prepared NP solutions with a concentration of 20 mg/mL at pH 4.5 and 7.4 were incubated in a water bath at 37°C. At 1 month postincubation, the solution was lyophilized and used for size measurement, pDNA-retarding capacity analysis by gel electrophoresis, and serum-free transfection.
Stability of pNPs to anionic heparin
The stability of pNPs to anionic macromolecules in body fluids was evaluated using a heparin competitive replacement assay. Briefly, pNP solution containing 2 μg/mL pDNA at N/P=60 was prepared as mentioned previously. Ten microliter aliquots of pNP solution were placed into Eppendorf tubes. Then, 10 μL of heparin solution of different concentrations was added into tubes, and the tubes were incubated at room temperature for a predetermined time. Thereafter, the samples were immediately analyzed by gel electrophoresis according to our previously reported method.22
Stability of pNPs to saline
Similar to the preparation of transfection solution, LPSA was first dissolved in water and then mixed with an equal volume of pDNA.23 Thirty minutes later, the complexes were diluted by 20-fold with 0.9% saline and then stored at room temperature. At determined time points, sample solutions were visually observed, and their size and zeta potential were analyzed by DLS. PEI 25k/pNDA (N/P=10) was used as a control.
Stability of pNPs to serum
In systemic delivery in vivo, pNPs were first exposed to a serum environment containing various anions and enzymes, where anions may dissociate pDNA from pNPs, and subsequently enzymes may degrade free pDNA. We performed two experiments to evaluate the stability of pNPs against serum. The first one was to analyze the stability of pNPs in 50% (v/v) serum using agarose gel electrophoresis. Briefly, a series of pNP solutions containing 50% serum with different N/P ratios were incubated at 37°C for different times, and then samples were incubated with heparin solutions to fully dissociate pDNA from pNPs. After that, samples were analyzed by gel electrophoresis. Naked pDNA treated by 50% serum was used as a control. The second experiment was to evaluate the in vitro transfection of pNPs in MSCs, which was performed according to the aforementioned method, where the transfection medium contained pNPs whose N/P ratios ranged from 60 to 130 and FBS with concentration ranging from 0% to 50%.
Storage stability
A successful gene vector needs not only transfection efficiency but also storage stability for at least 1–3 years. We evaluated the storage stability of NPs and pNPs (N/P=60) at −20°C, 5°C, and 25°C by monitoring their changes in size, zeta potential, and transfection efficiency with time. Briefly, freshly prepared NP or pNP solution containing 0.035 mg/mL LPSA or plus 2 μg/mL pDNA (N/P=60) was incubated at different temperatures for a predetermined time, and then samples were taken for determination of size, zeta potential, and transfection efficiency as mentioned previously. Specifically, samples stored at −20°C were ultrasonically treated for 10 minutes before assays.
Toxicity to MSCs
In our previous publications, the toxicity to MSCs and zebrafish was evaluated by cell viability via AlamarBlue method and zebrafish survival, and the results have shown that NPs and pNPs have low toxicity.23,24 Herein, we further studied the effects of pNP transfection on mitochondria according to the reported methods.28,29 Briefly, serum-free transfection was performed in MSCs as mentioned previously, then, at 48 hours posttransfection, rhodamine 123 solution was added to each well at a final concentration of 5 μg/mL, and cells were cultured for additional 0.5 hours. Subsequently, cells were washed thrice with PBS, then trypsinized and resuspended in PBS, and their average fluorescence intensity was determined by a flow cytometer.
Statistical analysis
Each experiment was performed at least three times. Data were expressed as mean ± standard deviation. A two-tailed Student’s t-test was used for statistical analysis. P<0.05 is considered statistically significant.
Results and discussion
To develop a gene delivery system with high efficiency and safety, we have constructed a brush graft copolymer of LPSA, which combines three functional blocks of OA (a hydrophilic degradable natural polyanion), PEI with molecular weight of 1.8k dalton (PEI 1.8k, a hydrophilic polycation), and Cho (a hydrophobic natural membrane lipid), and thus possesses amphiphilicity and amphotericity.23,24 Due to the distinguished properties of three blocks, we have demonstrated that 1) through electrostatic, hydrophobic, and hydrogen bonding interactions, LPSA itself or with gene can spontaneously self-assemble into empty or gene-loaded NPs with hydrophobic Cho and neutralized PEI/OA or pDNA/PEI/OA interpolyelectrolyte complexes as the membrane layers, and with unneutralized PEI and/or OA as the inner and outer coronas; 2) NPs are degradable and have low toxicity to MSCs and zebrafish; and 3) NPs can overcome intracellular barriers such as endocytosis and endosome escape, and thus obtain high in vitro transfection efficiency in MSCs and trigger significant angiogenesis in zebrafish in vivo when delivering gene. All of these results suggest that NPs may have huge potential in gene therapy as a vector. But besides these, for successful gene delivery systems, their stability in extracellular delivery in body fluids is required, which prevents loaded gene from unloading and deactivation before they reach the expected sites. Additionally, long-term storage stability is critical for a successful gene vector. Therefore, in this study, the stability of NPs/pNPs to various analogous physiological environments such as dilution, polyanions, salts, enzyme, and serum and various storage conditions was investigated.
Stability to dilution
The key function of gene delivery systems is to deliver gene without losing bioactivity to the expected sites. When gene-loaded nanoparticles are used in vivo, they will be largely diluted by serum or other body fluids. Dilution often causes the disassembly or aggregation of gene-loaded nanoparticles, where the former results in the unloading of genes, and the latter leads to the precipitation of particles. Both hinder gene delivery to expected sites. Thus, a successful gene delivery system should at first overcome the barrier from dilution. That is, they should keep stable in size and structure integrity during in vivo delivery.
CMC was first determined because it is an index of thermostability of self-assembled nanoparticles against dilution. The plot of surface tension versus log of LPSA concentration shows three regions (Figure 1A): region A and region C, where the surface tension changes little with concentration, and region B, where the surface tension declines sharply with concentration. Normally, the turning point between region B and region C is taken as the CMC. After CMC, all added LPSA was removed from the solution to form NPs rather than to further lower the surface tension, and thus the plot keeps flat. In our case, the CMC of pNPs was estimated as 1.95×10−3 mg/mL, which is close to that of NP solution (CMC: 1.38×10−3 mg/mL) determined by pyrene probe method,23 suggesting that the two systems have similar aggregation behavior. It should be noted that pNPs have lower CMC than most reported vectors, such as PEGylated vectors, whose magnitude of CMC ~10−2 mg/mL,8,30,31 suggesting that pNPs are less susceptible to disassembly upon dilution. Thus, during in vivo applications, when subject to large dilution, pNPs can provide continuous protection to loaded genes, which ensures that genes can survive from enzyme digestion and successfully reach the expected sites with bioactivity.
Figure 1.
Stability of plasmid deoxyribonucleic acid (pDNA)-loaded lipopolysaccharide-amine (LPSA) nanopolymersomes (pNPs) against dilution.
Notes: (A) Plot of surface tension of pNP solution versus logarithm of LPSA concentration at 25°C for determination of critical micelle concentration (CMC), (B) changes in particle size of pNPs with LPSA concentration, (C) gel electrophoresis of diluted pNP (N/P=60) solution containing 15 μg/mL LPSA and 0.875 μg/mL pDNA without (lane 1) or with (lane 2) heparin dissociation treatment as well as 8.75 ng naked pDNA (lane 3).
Although CMC can give information on the critical concentration of disassembly of pNPs, it cannot reveal whether or not their size is changed, which is very important for circulation and cell uptake.6 Figure 1B shows that when pNP solution is diluted from 2,000 to 10 μg/mL, its size keeps constant at about 124 nm with a polydispersity index <0.5, whereas the size of pDNA/PEI 25k complexes diluted by 20-fold is about four-fold bigger than their initial size (data not shown), suggesting that dilution causes their aggregation. It should be noted that the size of pNPs at 2 μg/mL (close to CMC) is larger than that at 2,000 μg/mL with a polydispersity index of ~0.7, suggesting that data are not reliable, which may be caused by a too low concentration and limited particles in the solution.
In addition, since LPSA itself can self-assemble into NPs with similar size and CMC to pNPs, it is possible that, at a low concentration, pDNA may be dissociated from pNPs but cannot be detected using the aforementioned two methods. To clarify this, diluted solutions were analyzed on agarose gel electrophoresis. Figure 1C shows that no band of pDNA in diluted pNP solution containing 15.00 μg/mL of LPSA can be observed, but after dissociation by heparin, a pDNA band appears, suggesting that dilution does not dissociate pDNA from pNPs. We did not choose a lower pNP concentration close to its CMC, such as 2–3 μg/mL of LPSA, in this experiment. This is because the detection limit of agarose gel electrophoresis to pDNA is 8.75 ng according to our experiments (data not shown), whose corresponding concentration for pDNA and LPSA in pNP solution (N/P=60) is 0.875 and 15.00 μg/mL, respectively. All of these results indicate that pNPs have good stability in structure integrity and size against dilution, possibly meaning that pNPs can protect pDNA and transfect cells successfully even in large dilution in vivo.
Stability against pH
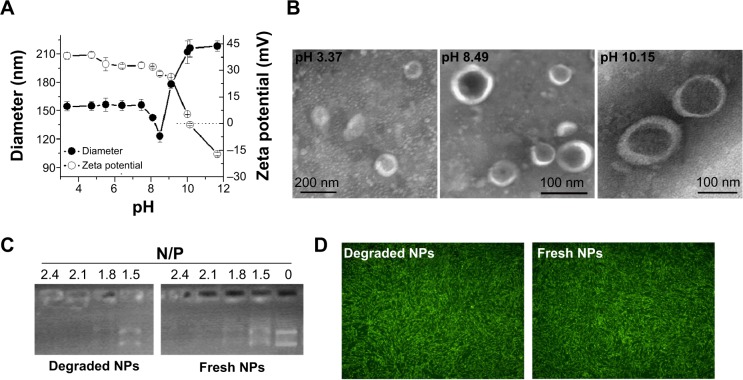
Since pH is different in different physiological environments (eg, it is ~pH 2 in the stomach, ~pH 4.5–5.0 in lysosome, ~pH 7.4 in blood and cytosol, ~pH 5.0–6.5 in endosome, and ~pH 8 in intestine), nanoparticles may be exposed to various pH conditions in biomedical applications, and their pH stability is certainly important for therapeutic efficacy. We first studied the short-term pH stability of LPSA NPs by monitoring their diameter, zeta potential, and morphology at different pH via DLS and TEM. The pH of NP solution was adjusted to 10.4–3.4 by adding NaOH or HCl, and the solutions were incubated overnight to equilibrate the system. The changes of NPs in size, zeta potential, and morphology with pH are presented in Figure 2A and B. Figure 2A shows that in the studied range, the change of size over pH can be divided into four phases: first at a plateau of ~55 nm at pH ≤7.45, then with a sudden drop to the minimum (123 nm) at pH 8.49 (this pH was obtained by directly dissolving LPSA in ultrapure water without any pH adjustment), after that with a sharp rise to the maximum (212 nm) at pH 10.01, and finally at a plateau again. Unlike the size, the zeta potential of NPs decreases with pH: at pH ≤9.01, the zeta potential drops off slowly from 38.3 mV to 26.3 mV, but at pH >9.01, the zeta potential decreases quickly from 26.3 mV to −17.1 mV. Noticeably, the zeta potential of zero is achieved at about pH 10.1, suggesting the isoelectric point of polyampholyte of LPSA. That is, at ~pH 10.1, in LPSA, all carboxyls from the OA block are deprotonated and possess negative charges, and some amine groups from the PEI block are protonated and possess positive charges, and the net charges are zero in the solution. Figure 2B visually displays that NPs exist in the form of vesicle in our studied pH range, proving that pH cannot disassemble them and they have pH stability in structure. Such stability ensures that loaded genes are protected by NPs no matter at which pH, which is very important for efficient gene transfection because the variation in particle structure, such as from NPs to micelles, or disassembly may lead to unexpected gene release and succedent enzyme digestion to genes.
Figure 2.
pH stability of lipopolysaccharide-amine nanopolymersomes (NPs).
Notes: (A) Changes of NPs in diameter and zeta potential with pH adjusted by concentrated HCl or NaOH, (B) transmission electron microscope images of dried NPs prepared from their aqueous solution with different pH (stained negatively with phosphotungstic acid), (C) retarding capacity of degraded NPs (prepared at pH 7.4) and fresh NPs, (D) serum-free transfection in mesenchymal stem cells mediated by degraded NPs (prepared at pH 7.4) and fresh NPs.
We believe that all of these changes arise from the variation in charges of LPSA, including cations from protonated amines in PEI and anions from deprotonated carboxyls in OA under different pH, which leads to the alteration in electrostatic interactions and hydrogen bonding, and these complicated interactions maintain their vesicle structure and make contrary contributions to the size. Theoretically, the electrostatic attractions from opposite charges may determine the compact degree of vesicle membrane and contribute to the size decrease, electrostatic repulsions from like charges may determine the stretched degree of the corona and contribute to the size increase, and hydrogen bonding from carboxyls and amines with themselves may contribute to the formation of vesicle membrane and thus decrease size, while hydrogen bonding of PEI and OA with solvent of water may increase the hydration of corona and thus make the size increase. Furthermore, it should be kept in mind that electrostatic interactions are normally stronger than hydrogen bonding, and no matter at which pH, all of these interactions coexist and accordingly cause the aforementioned variations in size and zeta potential. For example, at ~pH 8.49, according to their pKa (≤4.0 for carboxyl groups in OA, and ~9 for primary amines, ~8 for secondary amines, and ~6–7 for tertiary amines in PEI), and the mole ratio of carboxyl groups to primary amines, secondary amines, and tertiary amines in LPSA (about 7.55:10.5:21:10.5),23 almost all carboxyls are deprotonated and about 30% of amines are protonated; therefore 1) nearly all carboxyls are neutralized, generating the maximum electrostatic attractions and thus forming the most compact vesicle membrane combined with hydrophobic Cho and 2) the electrostatic repulsions from unneutralized amine cations and the hydrogen bonding from unprotonated amines oppositely contribute to the size, presumably forming a dense corona of vesicle, and the most compact membrane and corona bring about NPs with the minimum size. From this point of pH 8.49, no matter decreasing or increasing pH causes larger NPs because the aforementioned interactions increasing particle size play a leading role.
Based on this analysis, NPs should be more stable at pH 8.49 than at other pH because the compact structure slows the entrance of water and subsequent degradation. Our experiment substantiated this. At 4°C, 1 month’s storage at pH 8.49 slightly affects the size, zeta potential, and transfection efficiency of NPs, while 1 month’s storage at pH 4.5 makes the size of NPs decrease by 35.8% and the zeta potential unchangeable (data not shown), which is beneficial for circulation and transfection.
For gene vectors, their long-term stability in vivo is not expected. After they finish their task, they should be degraded and metabolized to avoid accumulation in the body, which often results in long-term toxicity. We previously demonstrated that for NPs in water (pH 8.49) degraded at 37°C, the degradation was realized by the hydrolysis of glucosidic bonds and ester bonds in LPSA, and their final products can be metabolized.23 Here, by simulating physiological environments in endo-/lysosome, cytosol, and blood, respectively, the stability of NPs at pH 4.5 and 7.4 was evaluated by determining the changes in size. The diameter for NPs incubated at pH 4.5 or 7.4 for 1 month is about 42.7 nm, which is about 55% of the initial (~120 nm), suggesting their degradation in all pH environments and that the degradation is pH dependent.
Besides pH-dependent degradation, for in vivo application, it is ideal for gene vectors to keep the capacity in retarding pDNA and delivering them to transfect cells even during degradation. In view of this, we performed pDNA-retarding and transfection experiments using fresh NPs and degraded NPs, which were prepared under pH 7.4 and 4.5. Excitingly, degradation does not affect NPs to retard and deliver pDNA to transfect cells. The retarding capacity to pDNA of degraded NPs is the same as fresh NPs, and they both completely retard pDNA at N/P of 2.1 (Figure 2C); the transfection efficiency (percentage of GFP-positive cells) is about 95% no matter whether degraded NPs or fresh NPs are used as a vector, and their fluorescence images further prove the successful transfection in MSCs (Figure 2D). It should be pointed out that only those data from NPs degraded under pH 7.4 were presented in Figure 2C and D, because results under pH 7.4 and 4.5 are almost the same. Additionally, we performed in vitro transfection by using the mixture of reduced OA and Cho-graft-PEI 2k (the raw materials for LPSA preparation) as a vector, and similar transfection efficiency was obtained. These data will be reported after a further investigation. Based on these results, we conclude that the stability of NPs is pH dependent, their in vivo delivery is accompanied by degradation, and the degradation causes the decrease in size of NPs but does not disassemble them and impair their retarding and delivery capacity to pDNA. We believe that this is especially helpful in decreasing the long toxicity of gene vectors and simultaneously preserving the good therapeutic effects.
Stability of pNPs to anionic heparin
For vector/pDNA complexes, because they will be exposed to a polyanionic environment during in vivo application, their relative stability to polyanions is very important. That is, in view of pDNA protection, it should not be displaced from pNPs by polyanions before reaching cytosols or cell nucleus, which requires relative stability, but, after that, it should be released completely to exert its biofunctions, which requires instability (reversibility). We used heparin as a polyanion model to evaluate the stability of pNPs to polyanions. Figure 3A shows that when the incubation time is 15 minutes, with increasing heparin concentration, the amount of displaced pDNA increases (the brightness and longitudinal width of pDNA bands increase gradually), and the complete displacement occurs at heparin concentration ≥20 mg/mL, equivalently the complete displacement concentration of ~10 mg heparin/μg pDNA (~1,000 IU heparin/μg pDNA), which is much higher than the value of PEGylated PEI derivatives (0.4 IU heparin/μg pDNA).32 At the same time, we explored the effect of incubation time on displacement of pDNA at a heparin concentration of 10 mg/mL, and Figure 3B shows that complete displacement occurs at 120 minutes in our studied range. These results indicate that resistance of pNPs to competitive displacement of polyanions is concentration and time dependent, which may exactly meet transfection demands for stability to polyanions because pNPs obtain successful transfection in MSCs and zebrafish.23
Figure 3.
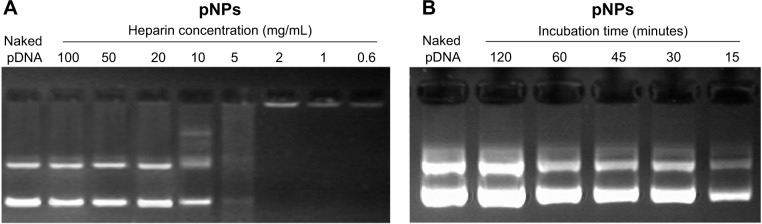
Stability of plasmid deoxyribonucleic acid (pDNA)-loaded lipopolysaccharide-amine nanopolymersomes (pNPs) to heparin replacement.
Notes: pNP solution incubated with (A) heparin at a different concentration for 15 minutes and (B) 10 mg/mL heparin for a different time.
Stability of pNPs to saline
After 1 hour’s incubation at room temperature, for PEI/pDNA in saline, floc appeared in the solution, suggesting that saline causes their aggregation. Correspondingly, their diameter reached 2,678±969 nm, which is much bigger than the size of PEI/pDNA in water (160.2±10.7 nm), and their zeta potential was 8.08±2.06 mV, which is much lower than the value of PEI/pDNA in water (31.67±2.89 mV), indicating that PEI/pDNA complexes are unstable to saline, which is one of the barriers for its clinical application. But for pNPs in saline, after 1 hour, the solution was clear and transparent, their diameter was 121.3±2.15, which is the same as their initial value and the size of pNPs in water, and their zeta potential was 28.33±0.76 mV. After 1 week, pNP solution was still clear, its diameter slightly decreased to 110.23±8.58 nm, and its zeta potential slightly decreased to 25.64±4.29 mV, suggesting its degradation and stability to saline.
Stability of pNPs against serum
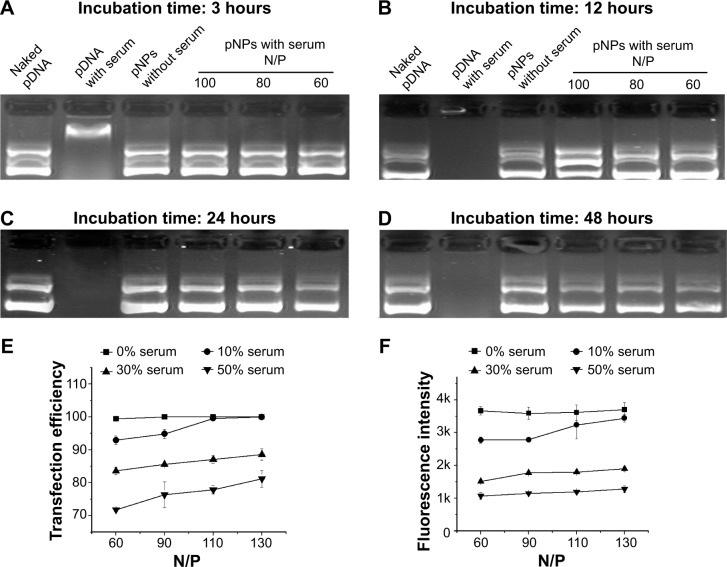
Although we evaluated the stability of pNPs against dilution, polyanions, DNase (published data: NPs can provide complete protection to pDNA for at least 6 hours when incubated with DNase I24), and saline, respectively, conditions in serum may be totally different because of the combined effects of all these factors. In view of the enzyme digestion and polyanion replacement in blood during in vivo application, we first studied the protection of NPs to pDNA from serum by incubating pNPs with 50% serum for different times. Figure 4A–D shows that naked pDNA is degraded into fragments with low molecular weight in 3 hours, and its band completely disappears in 12 hours. For pNPs, they can resist serum enzyme digestion in at least 48 hours. Such results suggest that pNPs have stability to serum and can provide protection to pDNA in vivo, and that the protection span may meet the needs of extracellular delivery, which will ensure efficient transfection in vivo.
Figure 4.
Stability of plasmid deoxyribonucleic acid (pDNA)-loaded lipopolysaccharide-amine nanopolymersomes (pNPs) to serum.
Notes: Gel electrophoresis of pNPs after incubation with 50% serum for a different time followed by complete dissociation by heparin (A–D), the transfection efficiency (E) and average fluorescence intensity (F) of mesenchymal stem cells transfected by pNPs in serum with different concentration.
Although pDNA can be protected by NPs in serum, pNPs in serum may have poor transfection ability. To explore this, we further performed in vitro serum transfection experiments. Figure 4E shows that, in our studied range, the transfection efficiency increases with the increase of N/P ratio and, at the same N/P ratio, the transfection efficiency generally decreases with the increase of serum concentration, but the maximum transfection efficiency for each serum concentration is higher than 80%. The changes of fluorescence intensity with serum concentration further confirmed data of transfection efficiency (Figure 4F). These results suggest that serum concentration doses affect the transfection, but a similar transfection may still be achieved by using a higher N/P ratio. Our previous transfection experiments have demonstrated this. In vitro, optimal transfection has been achieved in MSCs at N/P of 60,23 while, in vivo, maximum angiogenesis has been induced in zebrafish at N/P ratio of 90.24
Storage stability
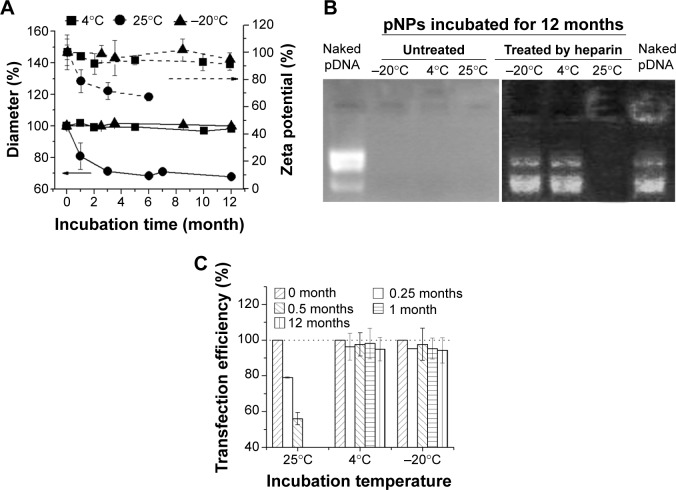
Storage stability under common conditions means a relative low cost, long shelf life, and wide application. Physicochemical properties of lyophilized NPs will not be impaired for at least 3 years if they are stored away from moisture at room temperature. The NP solution at 4°C is also stable for at least 2 years (a test for longer times has not been tried), which means their particle size, zeta potential, and gene delivery ability remain unchanged or slightly changed during storage (data not shown). Although lyophilized NPs are easily homogeneously resuspended in water through stirring and complexed with pDNA through simple mixing, it is more convenient to directly use prepared gene-loaded NPs (pNPs) in the clinic. Therefore, storage stability of pNPs at −20°C, 4°C, and 25°C was investigated according to International Conference on Harmonization of Technical Requirements for Registration of Pharmaceuticals for Human Use guidelines.33,34 Figure 5A shows that, at 4°C and −20°C, the changes of pNPs in size and zeta potential are very slight, suggesting their storage stability at these temperatures. But, at 25°C, they decrease significantly, which may be caused by the gradual degradation of NPs and pDNA. To verify this, we used agarose gel electrophoresis to detect the pDNA in pNPs with or without heparin treatment. The gel electrophoresis images (Figure 5B) show that, at 12 months poststorage, no bands are observed for all pNP solutions, but, after heparin treatment, pDNA bands similar to those of naked pDNA appear in agarose gel for solutions stored at 4°C and −20°C, while nothing emerges for solution stored at 25°C, suggesting the existence of intact pDNA compacted by NPs for the former but completely degraded pDNA for the latter. We further used transfection experiments to confirm these results. Consistently, the transfection efficiency for pNPs stored at 4°C and −20°C is slightly affected during 12 months’ storage, but it gradually decreases to zero for pNPs stored at 25°C. Based on these results, considering energy consumption and convenience, we believe that all are good choices: pNP solution stored at 4°C for 1 year’s use, NP solution stored at 4°C for 2 years’ use, and lyophilized NPs stored away from moisture at room temperature for long-term use.
Figure 5.
Storage stability of plasmid deoxyribonucleic acid (pDNA)-loaded lipopolysaccharide-amine nanopolymersomes (pNPs).
Notes: Changes of pNPs in (A) diameter and zeta potential, (B) protection to pDNA, and (C) transfection efficiency with time at a different temperature.
Taken together, all of these results show that NPs and pNPs have excellent stability against varied conditions, including physiological and storage environments. Furthermore, to the best of our knowledge, the stability of pNPs may be superior to that of other gene delivery vectors like PEGylated vectors,8,10,19,30,32,34–36 which is manifested through their stable size, zeta potential, pDNA-retarding capacity, structure, and the high transfection efficacy after the aforementioned treatments. But because related data were obtained under different experimental conditions and characterization methods, such comparison may be unscientific. We believe that their excellent stability is ascribed to the distinct synergy of three blocks in LPSA to overcome the encountered barriers. Firstly, the three blocks of OA, PEI 1.8k, and Cho confer LPSA amphiphilicity and amphotericity, which provide three coexisting but environment-dependent driving forces, including hydrophobic, electrostatic, and hydrogen bonding interactions for LPSA itself or with pDNA to spontaneously self-assemble to robust NPs and pNPs with suitable size. These multiple driving forces make them always maintain the intact structure in any stringent conditions but with pH-dependent compactness degree, which allows them to remain stable during extracellular delivery. Secondly, Cho imparts stability to nanoparticles during extracellular delivery by improving membrane stability and inhibits bindings of proteins and opsonins from serum. Thirdly, anionic polysaccharide of OA offers steric shielding effects to minimize undesired nonspecific interactions, and, at the same time, it imparts the temperature/pH-dependent degradability to NPs, and thus they can be degraded and metabolized at 37°C to avoid the toxicity induced by their accumulation.
Damage of pNPs to mitochondria of MSCs after transfection
Acceptable stability along with efficacy and safety are key factors for gene therapy. Cell viability is commonly used to evaluate the toxicity of gene delivery systems, but their effects on cellular organelles have been little studied. It has been reported that polycations such as poly(ethylenimine) and poly(l-lysine) can induce mitochondrially mediated apoptosis.37 PEI-mediated phase I cytotoxicity is induced by the binding of clumps of free PEI molecules to cell plasma membrane, which results in membrane destabilization and a loss of membrane integrity. PEI-mediated phase II cytotoxicity is damage to mitochondria, which can be evaluated by the loss of mitochondrial membrane potential (MMP). The significant damage to mitochondria produced by vector/pDNA complexes will lead to the release of cytochrome c and subsequent induction of executioner caspases (cysteinyl aspartate-specific proteases) and thus induce apoptosis.37 For our NP vector, by observing the ultrathin sections of cells under a TEM, we found that no aggregation of pNPs exists on cell plasma membrane during the transfection process, which may be attributed to the fact that LPSA can spontaneously self-assemble into NPs with good colloid stability, and NPs can be quickly endocytozed by receptor-mediated Cho uptake pathway, avoiding the phase I cytotoxicity.23 Figure 6 shows that, in our studied range, pNPs do not cause severe loss of MMP, that the MMP of MSCs decreases with the increase of N/P ratio, and that the pNP groups at any N/P ratio have higher MMP compared with the PEI 25k group, which loses about 20% of MMP. At N/P of 60, the optimal for in vitro transfection, the MMP of pNP group is close to that of the untreated group (N/P=0) and the Lipofectamine 2000 group, suggesting that pNPs in cells cause low damage to mitochondria of MSCs and thus avoiding the mitochondrially mediated apoptosis induced by pNP transfection. Combined with the results in cell toxicity and zebrafish survival,23,38 we conclude that pNPs are of low toxicity and are promising for clinical applications.
Figure 6.
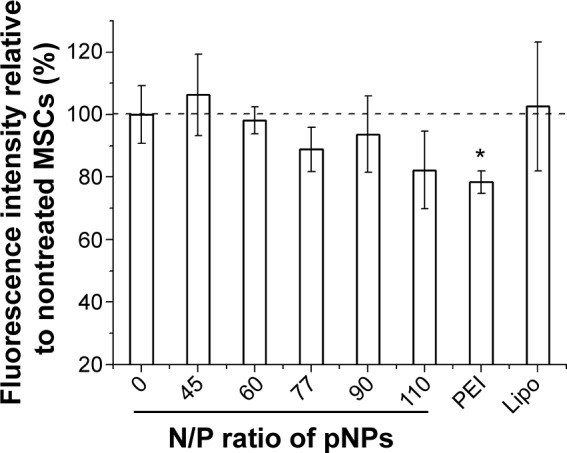
Damage of plasmid deoxyribonucleic acid (pDNA)-loaded lipopolysaccharide-amine nanopolymersomes (pNPs) to the mitochondria of mesenchymal stem cells after serum-free transfection for 48 hours, which is evaluated by the changes in mitochondrial membrane potential quantified by flow cytometer. *P<0.05 versus untreated group (N/P=0).
Conclusion
By monitoring the changes of NPs and pNPs in size, zeta potential, morphology, structure, pDNA-retarding capacity, and transfection ability, a systematic study on their stability under various analogous physiological environments and storage conditions has been carried out. It was found that NPs and pNPs have acceptable stability against dilution, pH, heparin, salts, and serum, which may exactly meet the needs of extracellular and intracellular gene delivery, and thus pNPs have obtained high transfection efficacy in vitro (higher than 95% transfection efficiency in MSCs) and in vivo (the induced significant angiogenesis in zebrafish). Additionally, NPs/pNPs have high storage stability in the form of aqueous solution at 4°C or lyophilized powder (for NPs) at room temperature. Moreover, pNPs do only slight damage to mitochondria of MSCs, avoiding the mitochondrially mediated apoptosis induced by pNP transfection. All of these results highlight their huge potential in clinical gene therapy. We believe that all distinguished properties of NPs are due to the synergy of three functional blocks of polysaccharide, Cho, and low-molecular-weight PEI in LPSA. Our work will provide valuable insights into the design of gene delivery vectors meeting clinical demands and meaningful references for performing systematic stability evaluation for gene vectors, considering the few studies in this field.
Acknowledgments
This work was supported by the National Science Foundation of China (81101153, 81371665) and the Science and Technology Plan Funds of Guangdong (2013B051000017).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chandrakasan S, Malik P. Gene therapy for hemoglobinopathies: the state of the field and the future. Hematol Oncol Clin North Am. 2014;28:199–216. doi: 10.1016/j.hoc.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Reilly M, Federoff H, Fong Y, et al. Gene therapy: charting a future course – summary of a National Institutes of Health Workshop. Hum Gene Ther. 2014;25:488–497. doi: 10.1089/hum.2014.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka M, Taketomi K, Yonemitsu Y. Therapeutic angiogenesis: recent and future prospects of gene therapy in peripheral artery disease. Curr Gene Ther. 2014;14:300–308. doi: 10.2174/156652321404140902124838. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Li W, Ma N, et al. Non-viral gene delivery methods. Curr Pharm Biotechnol. 2013;14:46–60. [PubMed] [Google Scholar]
- 5.Jones C, Chen C, Ravikrishnan A, et al. Overcoming nonviral gene delivery barriers: perspective and future. Mol Pharm. 2013;10:4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vercauteren D, Rejman J, Martens TF, et al. On the cellular processing of non-viral nanomedicines for nucleic acid delivery: mechanisms and methods. J Control Release. 2012;161:566–581. doi: 10.1016/j.jconrel.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Zhang Z, Zhou C, et al. Hydrophobic modifications of cationic polymers for gene delivery. Prog Polym Sci. 2010;35:1144–1162. [Google Scholar]
- 8.Logie J, Owen SC, McLaughlin CK, et al. PEG-graft density controls polymeric nanoparticle micelle stability. Chem Mater. 2014;26:2847–2855. [Google Scholar]
- 9.Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41:2562–2574. doi: 10.1039/c1cs15258k. [DOI] [PubMed] [Google Scholar]
- 10.Mishra S, Webster P, Davis M. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 11.Oe Y, Christie RJ, Naito M, et al. Actively-targeted polyion complex micelles stabilized by cholesterol and disulfide cross-linking for systemic delivery of siRNA to solid tumors. Biomaterials. 2014;35:7887–7895. doi: 10.1016/j.biomaterials.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, Anchordoquy TJ. Effect of cholesterol nanodomains on the targeting of lipid-based gene delivery in cultured cells. Mol Pharm. 2010;7:1311–1317. doi: 10.1021/mp100097b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang DA, Narang AS, Kotb M, et al. Novel branched poly(ethylenimine)-cholesterol water-soluble lipopolymers for gene delivery. Biomacromolecules. 2002;3:1197–1207. doi: 10.1021/bm025563c. [DOI] [PubMed] [Google Scholar]
- 14.Han S, Mahato RI, Kim SW. Water-soluble lipopolymer for gene delivery. Bioconjug Chem. 2001;12:337–345. doi: 10.1021/bc000120w. [DOI] [PubMed] [Google Scholar]
- 15.Semple SC, Chonn A, Cullis PR. Influence of cholesterol on the association of plasma proteins with liposomes. Biochemistry. 1996;35:2521–2525. doi: 10.1021/bi950414i. [DOI] [PubMed] [Google Scholar]
- 16.Bajaj A, Kondaiah P, Bhattacharya S. Synthesis and gene transfection efficacies of PEI-cholesterol-based lipopolymers. Bioconjugate Chem. 2008;19:1640–1651. doi: 10.1021/bc700381v. [DOI] [PubMed] [Google Scholar]
- 17.Furgeson DY, Chan WS, Yockman JW, et al. Modified linear polyethylenimine–cholesterol conjugates for DNA complexation. Bioconjugate Chem. 2003;14:840–847. doi: 10.1021/bc0340565. [DOI] [PubMed] [Google Scholar]
- 18.Pawar SN, Edgar KJ. Alginate derivatization: a review of chemistry, properties and applications. Biomaterials. 2012;33:3279–3305. doi: 10.1016/j.biomaterials.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Patnaik S, Arif M, Pathak A, et al. PEI-alginate nanocomposites: efficient non-viral vectors for nucleic acids. Int J Pharm. 2010;385:194–202. doi: 10.1016/j.ijpharm.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 20.Jones RA, Cheung CY, Black FE, et al. Poly(2-alkylacrylic acid) polymers deliver molecules to the cytosol by pH-sensitive disruption of endosomal vesicles. Biochem J. 2003;372:65–75. doi: 10.1042/BJ20021945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju HK, Kim SY, Kim SJ, et al. pH/temperature-responsive semi-IPN hydrogels composed of alginate and poly(N-isopropylacrylamide) J Appl Polym Sci. 2002;83:1128–1139. [Google Scholar]
- 22.He W, Guo ZH, Wen YT, et al. Alginate-graft-PEI as a gene delivery vector with high efficiency and low cytotoxicity. J Biomater Sci Polym Ed. 2012;23:315–331. doi: 10.1163/092050610X550359. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Teng W, Liu L, et al. Efficient cytosolic delivery mediated by polymersomes facilely prepared from a degradable, amphiphilic, and amphoteric copolymer. Nanotechnology. 2013;24:265104. doi: 10.1088/0957-4484/24/26/265104. [DOI] [PubMed] [Google Scholar]
- 24.Teng W, Huang Z, Chen Y, et al. pVEGF-loaded lipopolysaccharide-amine nanopolymersomes for therapeutic angiogenesis. Nanotechnology. 2014;25:65702. doi: 10.1088/0957-4484/25/6/065702. [DOI] [PubMed] [Google Scholar]
- 25.Evans C. Gene therapy for the regeneration of bone. Injury. 2011;42:599–604. doi: 10.1016/j.injury.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menger FM, Galloway AL, Chlebowski ME. Surface tension of aqueous amphiphiles. Langmuir. 2005;21:9010–9012. doi: 10.1021/la051363l. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz CC, Molina-Bolivar JA, Aguiar J. Thermodynamic and structural studies of triton X-100 micelles in ethylene glycol-water mixed solvents. Langmuir. 2001;17:6831–6840. [Google Scholar]
- 28.Quignard S, Mosser G, Boissière M, et al. Long-term fate of silica nanoparticles interacting with human dermal fibroblasts. Biomaterials. 2012;33:4431–4442. doi: 10.1016/j.biomaterials.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Hu L, Yu D, et al. Influence of silica particle internalization on adhesion and migration of human dermal fibroblasts. Biomaterials. 2010;31:8465–8474. doi: 10.1016/j.biomaterials.2010.07.060. [DOI] [PubMed] [Google Scholar]
- 30.Nam K, Nam HY, Kim PH, et al. Paclitaxel-conjugated PEG and arginine-grafted bioreducible poly (disulfide amine) micelles for co-delivery of drug and gene. Biomaterials. 2012;33:8122–8130. doi: 10.1016/j.biomaterials.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian H, Deng C, Lin H, et al. Biodegradable cationic PEG-PEI-PBLG hyperbranched block copolymer: synthesis and micelle characterization. Biomaterials. 2005;26:4209–4217. doi: 10.1016/j.biomaterials.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Roesler S, Koch FP, Schmehl T, et al. Amphiphilic, low molecular weight poly(ethylene imine) derivatives with enhanced stability for efficient pulmonary gene delivery. J Gene Med. 2011;13:123–133. doi: 10.1002/jgm.1538. [DOI] [PubMed] [Google Scholar]
- 33.Muthu M, Feng S. Pharmaceutical stability aspects of nanomedicines. Nanomedicine. 2009;4:857–860. doi: 10.2217/nnm.09.75. [DOI] [PubMed] [Google Scholar]
- 34.Mohammadi Z, Dorkoosh FA, Hosseinkhani S, et al. Stability studies of chitosan-DNA-FAP-B nanoparticles for gene delivery to lung epithelial cells. Acta Pharm. 2012;62:83–92. doi: 10.2478/v10007-012-0008-z. [DOI] [PubMed] [Google Scholar]
- 35.Zheng M, Liu Y, Samsonova O, et al. Amphiphilic and biodegradable hy-PEI-g-PCL-b-PEG copolymers efficiently mediate transgene expression depending on their graft density. Int J Pharm. 2012;427:80–87. doi: 10.1016/j.ijpharm.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Glodde M, Sirsi SR, Lutz GJ. Physiochemical properties of low and high molecular weight poly(ethylene glycol)-grafted poly(ethylene imine) copolymers and their complexes with oligonucleotides. Biomacromolecules. 2006;7:347–356. doi: 10.1021/bm050726t. [DOI] [PubMed] [Google Scholar]
- 37.Hunter AC, Moghimi SM. Cationic carriers of genetic material and cell death: a mitochondrial tale. Biochimica et Biophysica Acta. 2010;1797:1203–1209. doi: 10.1016/j.bbabio.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 38.Holmes CA, Tabrizian M. Substrate-mediated gene delivery from glycol-chitosan/hyaluronic acid polyelectrolyte multilayer films. ACS Appl Mater Interfaces. 2013;5:524–531. doi: 10.1021/am303029k. [DOI] [PubMed] [Google Scholar]