Abstract
The acquired immune response against tuberculosis is commonly associated with T-cell responses with little known about the role of B cells or antibodies. There have been suggestions that B cells and humoral immunity can modulate the immune response to Mycobacterium tuberculosis. However, the mechanisms involving B-cell responses in M. tuberculosis are not fully understood, in particular the antibody gene preferences. We hypothesized that a preferential use of V genes can be seen associated with resistance to infection mainly in the IgA isotype, which is of prominent importance for infection by pathogens via the mucosal route. We studied healthy individuals with long-term exposure to tuberculosis, infected (TST+) and uninfected TST−) with M. tuberculosis. From a total of 22 V genes analysed, the TST− population preferred the VH3-23 and Vκ1 genes. The VH3-23 genes were subsequently subjected to 454 amplicon sequencing. The TST− population showed a higher frequency of the D3-10 segment compared with the D3-22 segment for the TST+ population. The J segment usage pattern was similar for both populations with J4 segment being used the most. A preferential pairing of J4 segments to D3-3 was seen for the TST− population. The antibodyome difference between both populations suggests a preference for antibodies with VH3-23, D3-3, JH4 gene usage by the TST− population that could be associated with resistance to infection with M. tuberculosis.
Keywords: 454 sequencing, antibody, tuberculosis, V-D-J gene, VH3-23
Introduction
Tuberculosis (TB) is one of the most important causes of mortality due to infectious diseases. It is estimated that 8·7 million new clinical cases and 1·6 million deaths occur every year associated with TB pandemics, being the major cause of mortality in patients with AIDS.1; – 5 Control of the disease is difficult for many reasons, including low sensitivity of traditional diagnostic methods, insufficient therapeutic coverage, co-infection with HIV, growing appearance of strains resistant to treatment, and non-existence of an efficient vaccine for the prevention of adult pulmonary TB and its transmission.1; – 5 Bacillus Calmette–Guérin, which is currently administered as a TB vaccine, only protects against the severe forms of the disease in children but does not protect against pulmonary TB and transmission, prompting the importance of the development of a vaccine that is effective internationally.1; – 5
Tuberculosis is an infectious lung disease caused by Mycobacterium tuberculosis that is typically spread by airborne droplet nuclei. The adaptive immune response against M. tuberculosis develops within 2–6 weeks of the infection and an influx of lymphocytes and activated macrophages infiltrates the lesion, giving rise to granuloma formation.4,6 Traditionally, T cells are thought to play a significant role in protection against TB with several T-cell receptor studies being reported with little emphasis being given to B cells.4,7; – 9 However, recently B cells have been demonstrated to show protective effects in mouse challenge models with M. tuberculosis, indicating the possible role of B cells in inducing a protective mechanism.10,11 As TB is an airborne disease, secretory IgA should be one of the key immunological elements to interact with M. tuberculosis at the respiratory mucosa to avoid infection.
It has been shown that specific IgA reduces bacterial load in infected lungs of mouse models when administered intranasally.12,13 The potential of using IgA antibodies for the immunotherapy of TB is promising with a report of the successful use of a novel human IgA monoclonal antibody and IgA purified from human colostrum for passive immunotherapy of TB.14; – 16 Even so, not much is understood about the intricacy of the antibody gene maturation process with relation to TB and the IgA isotype.
The minimum prerequisite for the adaptive immune system to recognize an immense assortment of antigens is the ability to boast a diverse repertoire of antibodies. Three main genetic modification processes are attributed to the creation of antibody repertoires. The first is somatic recombination of germline V, D and J segments. This is followed by the addition or deletion of nucleotides at the V-D, D-J and V-J junctions. After antigen stimulation, antibody genes undergo somatic hypermutation to generate an even greater increasing repertoire of unique antibodies.17,18 These processes are the cornerstone of the diverse repertoire of antibodies being generated by the immune system. The assortment of antibodies generated in response to an infection has made antibodies useful therapeutic biological agents.
With the advent of high throughput next-generation sequencing technologies, the characterization and analysis of large antibody repertoires are now possible.19 The patterns of antibody V-D-J rearrangement in the study of V gene usage are principally important for antibody-binding characterization. This is due to the non-random use of certain V gene segments that gives different immune responses towards infections among individuals whereby some individuals exhibit stronger resistance toward certain infections and vice versa. Analyses of the use of immunoglobulin V genes in a number of studies have revealed a preferential antibody V gene repertoire for patients with ankylosing spondylitis,20 systemic lupus erythematosus,21 chronic lymphocytic leukaemia22 and other diseases.
A common challenge in V gene repertoire studies is the identification of a suitable population that is, by definition, to be protected against a disease for the study. In the case of TB, this issue is complicated by the non-existence of a defined correlate of protection.4 There have been documented cases whereby individuals remain unresponsive to Tuberculin Skin Test (TST) despite a high and continued exposure to TB.23,24 The interpretation of this phenomenon has been associated more with an intrinsic resistance to infection rather than an incapability of reacting to this preparation.23,25 It has also been reported that the risk of clinical TB in these individuals is low.23,25 Genetic studies with TST− individuals from highly exposed populations successfully identified a genetic region probably associated with resistance to infection, paving the way for studies focusing on the identification of genetic determinants associated with the resistance to M. tuberculosis infection.25
Taking into consideration that very little is understood about B-cell mechanisms in M. tuberculosis protection, we examined the V gene repertoire of the IgA isotype in healthy TST+ and TST− long-term contacts of M. tuberculosis. A healthy person with long-term exposure but who has remained TST− indicates the possibility of resistance to primary infection, in particular associated with IgA production conferring protection at the portal of infection entry. In this study, we hypothesized that a preferential use of V genes at the IgA level could be associated with protection against infection in TST− individuals. Deep sequencing was carried out to provide an insight into the preferential D-J segment usage and D-J segment pairing.
Materials and methods
Ethics statement
All blood samples were collected at the Respiratory Clinic, Hospital Universiti Sains Malaysia (HUSM), Kubang Kerian, Kelantan, Malaysia. Written informed consent for genetic studies was obtained for this study in agreement with protocols approved by the Universiti Sains Malaysia Human Ethics Committee.
Blood sample collection, sample treatment and PCR methods
Blood was collected from healthy individuals working for more than 10 years at the Respiratory Clinic, Hospital Universiti Sains Malaysia (HUSM), Kubang Kerian, Kelantan, Malaysia (eight TST− and seven TST+) (see Supporting information, Tables SI and S2). All the individuals were of Malay descent with no clinical symptoms of TB or personal TB history, aged between 35 and 53 years, bacillus Calmette–Guérin vaccinated, HIV negative and not receiving immunosuppressive medication. For TST (2 TU), the purified protein derivative RT23 (Statens Serum Institute, Copenhagen, Denmark) was used. The reaction was evaluated 48–72 hr after the injection, and the transverse diameter (in millimetres) of the area of induration was recorded. TST positivity cut-off was considered as ≥ 10 mm for size of induration.
The collected blood from each individual was used for lymphocyte isolation using the density gradient centrifugation method. Total RNA was extracted from the lymphocytes by using QIAamp RNA Blood Mini kit (Qiagen, Hilden, Germany). Five hundred nanograms of RNA was reverse transcribed into cDNA using the SuperScript III Reverse Transcription kit (Invitrogen, Carlsbad, CA).
The IgA VH, Vκ and Vλ genes were amplified from cDNA by PCR for 30 cycles using a set of antibody-specific primers as previously described (Table1).26 The Vκ1, Vκ3, Vκ5, Vκ246 and Vκ2N1 forward primers were used in combination with the κCL Rv reverse primer to cover the Vκ repertoire. The Vλ1, Vλ2, Vλ3(DPL16), Vλ3, Vλ78, Vλ3(38), Vλ1459, Vλ15910 and Vλ6 forward primers were used in combination with equimolar λCL1 and CL2 reverse primer to cover the Vλ genes. The VH1, VH157, VH2, VH3, VH3-23, VH4, VH4(DPL63) and VH6 forward primers were used in combination with the IgA Rv reverse primer to amplify the VH genes. For PCR amplification, 0·4 μl cDNA from the initial 500 ng of RNA was used. Each PCR contained 10 mm dNTPs, 10 μm of forward and reverse primers, 2·5 U/μl Pfu polymerase (Fermentas, Glen Burnie, MD). The conditions used for PCR were an initial denaturation at 95° for 90 seconds followed by 30 cycles of denaturation at 95° for 30 seconds, annealing at 55° or 60° for 30 seconds and extension at 72° for 45 seconds. A final extension was carried out at 72° for 5 min. PCR amplification was carried out for the CD19 gene from each individual. Optimized annealing temperatures for specific primer pairs are listed in the Supporting information (Tables S3 and S4). PCR products of the VH, Vκ and Vλ genes from each of the samples were subjected to agarose gel electrophoresis. Fifteen microlitres of the PCR product was loaded into a 1% agarose gel and stained with ethidium bromide. The gel image was captured using a GENEFlash image analyser (Syngene, Cambridge, UK).
Table 1.
All PCR primers for the amplification of VH, Vκ and Vλ genes
| Primer | Primer sequence |
|---|---|
| Vκ1 | 5′-GAC ATC CRG DTG ACC CAG TCT CC-3′ |
| Vκ3 | 5′-GAA ATT GTR WTG ACR CAG TCT CC-3′ |
| Vκ5 | 5′-GAA ACG ACA CTC ACG CAG TCT C-3′ |
| Vκ246 | 5′-GAT ATT GTG MTG ACB CAG WCT CC-3′ |
| Vκ2N1 | 5′-AGA TGC TGT GTG AMC CAG CCT C-3′ |
| Vλ1 | 5′-CAG TCT GTS BTG ACG CAG CCG CC-3′ |
| Vλ2 | 5′-CAG TCT GYY CTG AYT CAG CCT-3′ |
| Vλ3(DPL16) | 5′-TCC TCT GAG CTG AST CAG GAS CC-3′ |
| Vλ3 | 5′-TCC TAT GWG CTG ACW CAG CCA A-3′ |
| Vλ78 | 5′-CAG DCT GTG GTG ACY CAG GAG CC-3′ |
| Vλ3(38) | 5′-TCC TAT GAG CTG AYR CAG CYA CC-3′ |
| Vλ1459 | 5′-CAG CCT GTG CTG ACT CAR YC-3′ |
| Vλ15910 | 5′-CAG CCW GKG CTG ACT CAG CCM CC-3′ |
| Vλ6 | 5′-AAT TTT ATG CTG ACT CAG CCC C-3′ |
| VH1 | 5′-CAG GTC CAG CTK GTR CAG TCT GG-3′ |
| VH157 | 5′-CAG GTG CAG CTG GTG SAR TCT GG-3′ |
| VH2 | 5′-CAG RTC ACC TTG AAG GAG TCT G-3′ |
| VH3 | 5′-GAG GTG CAG CTG KTG GAG WCY-3′ |
| VH3-23 | 5′-TCA ACA CAA CGG TTC CCA GTT A-3′ |
| VH4 | 5′-CAG GTG CAG CTG CAG GAG TCS G-3′ |
| VH4(DPL63) | 5′-CAG GTG CAG CTA CAG CAG TGG G-3′ |
| VH6 | 5′-CAG GTA CAG CTG CAG CAG TCA-3′ |
| κ CL Rv | 5′-ACA CTC TCC CCT GTT GAA GCT CTT-3′ |
| λ CL1 | 5′-TGA ACA TTC TGT AGG GGC CAC TG-3′ |
| λ CL2 | 5′-TGA ACA TTC CGT AGG GGC AAC GG-3′ |
| IgA Rv | 5′-CAG CGG GAA GAC CTT GGG GCT GGT-3′ |
Bioanalyser and statistical analysis
The amplified VH, Vλ and Vκ region genes were analysed (1 : 2 dilutions) for their size (bp) and DNA concentration by using an Agilent 2100 Bioanalyzer with the DNA 7500 Lab Chip kits (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. All statistical analysis was carried out using SPSS® Statistics version 17 (Chicago, IL). The extent of V gene usage was compared between the TST− and TST+ populations, such as to assess whether the differences between the two population medians were statistically significant by Mann–Whitney U-test. A paired-samples t-test was conducted to compare the frequency in TST− and TST+ across the 144 possible gene types. Comparison between the D and J segment usage for both populations was performed using the Wilcoxon signed rank tests. All statistical analysis was carried out using SPSS Statistics version 17 (SPSS Inc).
454 amplicon sequencing, sequences filtration and analysis
The VH3-23 genes from the TST− and TST+ populations were subjected to the 454 Roche GS-FLX amplicon sequencing that was carried out by Macrogen (Seoul, South Korea). PCR amplicons of VH3-23 genes from the TST− and TST+ populations were amplified using the PCR protocol described earlier. All PCR amplicons were subjected to Bioanalyzer analysis to determine the quality before sequencing. The sequences for the VH3-23 gene from the TST− and TST+ populations had been submitted to GenBank under the accession number SUB 116490. The obtained raw 454 sequence data were filtered using the Perl length filtration script (see Supporting information, Supplementary method S1). All scripts were run in the Windows OS (Microsoft, Redmond, WA) command prompt program with the installation of the Perl package. The first data filtration was to retain sequences with lengths > 300 nucleotides. The length and the file to be filtered can be customized through user-input values. The Output fasta file contains the filtered VH3-23 genes with desired size and was generated upon the completion of the script. The filtered sequences were submitted to VBASE2 for D and J segment alignment and CDR analysis.27 The results obtained were classified to D and J segments alignment assessments by using the Perl analysis script (see Supporting information, Supplementary method S2). The output report text file was generated upon the completion of the Perl analysis script.
Circos plot
All Circos plots were generated using the Circos software (http:http://www.circos.ca/software).
Results
Germline V gene usage between TST+ and TST− population
In this study, relatively stronger bands of VH3-23 gene amplification were observed for TST− population in comparison with the TST+ population using agarose gel electrophoresis (Fig.1a, b). Given this distinct variation, a semi-quantitative analysis for all the PCR amplified V genes of both TST− and TST+ populations were carried out by using the Agilent 2100 Bioanalyzer (Fig.1c, d). By comparing the mean DNA concentration of the amplified DNA between the TST− and TST+ populations, most of the VH genes were found to have higher average DNA concentration in the TST+ population except for the VH3-23 (labelled as VH3N), VH4 and VH6 genes. This indicates that most of the VH genes were more frequently expressed in the TST+ population.
Figure 1.
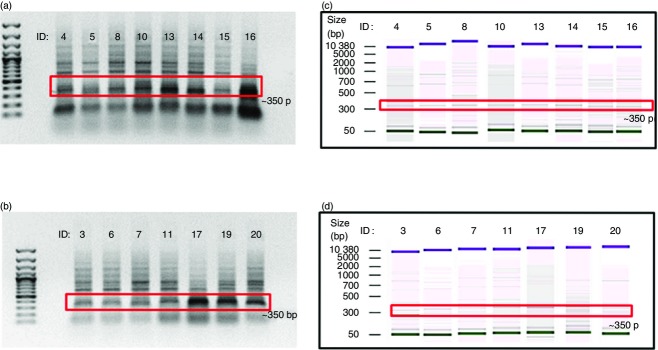
Agarose gel electrophoresis and Bioanalyzer analysis for VH3N. (a) Agarose gel analysis of TST− population; (b) agarose gel analysis of TST+ population; (c) Bioanalyzer analysis of TST− population; (d) Bioanalyzer analysis of TST+ population. The red box indicates the desired bands of approximately 350 bp in size of the VH3N (VH3-23) genes. DNA marker used is a 100 bp plus DNA Ladder from Fermentas. + control is the amplification of a known V gene and the − control is a non-templated DNA sample.
We also studied the V gene usage of the light chains although no direct pairing analysis was carried out with the IgA isotype. Among the 11 Vλ genes analysed, five Vλ genes were expressed more in the TST+ population and four of the Vλ genes were highly expressed in the TST− population. For the Vκ gene segments usage comparison, all the Vκ genes were shown to be expressed in higher amounts for the TST− population (see Supporting information, Tables S5 and S6). The extent of use for each V gene was compared between the TST− and TST+ groups, such that the differences between the two population medians were statistically significant by Mann–Whitney U-test. Mann–Whitney U-test revealed that of the 22 V genes, only VH3-23 (U = 7·5, P < 0·05) and Vκ1(U = 0·00, P < 0·01) registered significantly higher frequencies for TST− compared with TST+. This indicates that both VH3-23 and Vκ1 genes were significantly higher in the TST− than the TST+ population.
PCR amplification and Bioanalyzer analysis of CD19 gene
The concentration of CD19 amplification products was analysed using the Bioanalyzer system (see Supporting information, Table S7). The concentration of CD19 from each sample was analysed statistically. The Mann–Whitney U-test revealed that there was no significant difference between TST− and TST+ in the CD19 gene amplification (U = 27·5, P = n.s).
D and J segment usage frequency
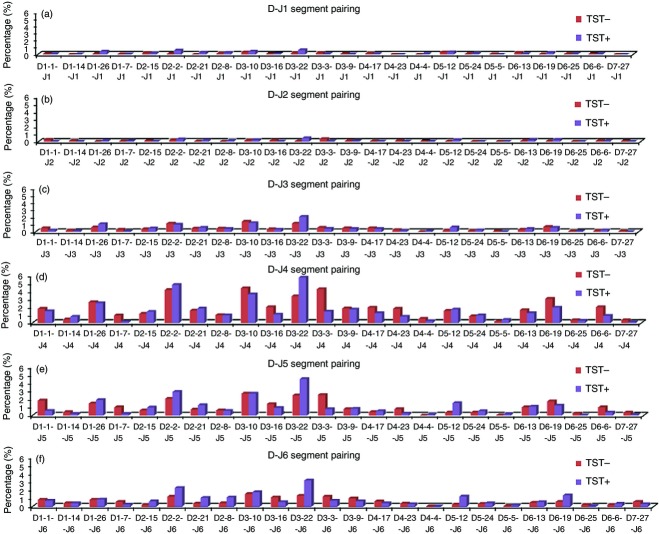
A total of 69 065 reads and 55 414 reads of the VH3-23 gene segment were obtained from the TST− and TST+ populations, respectively. The 454 sequence data set was sieved for quality whereby only sequences with lengths more than 300 nucleotides were retained by performing a Perl script filtering analysis. A total of 27 673 reads for the TST− population and 20 889 reads for the TST+ population with a length exceeding 300 nucleotides were retained for VBASE2 analysis. The individual gene segment frequency analysis shows that for the TST+ population, gene segment D3-22 (16·60%) was the most frequently used segment followed by D2-2 (11·95%) and D3-10 (9·94%). However, in the TST− population, gene segment D3-10 (10·51%) was more frequent, followed by D3-3 (9·19%) and D2-2 (8·93%) (Fig.2a). Segments D-4-4, D5-5, D6-25 and D7-27 were used at very low frequencies for both populations (Fig.2a). A preferential usage of D3-22 is seen for TST+ whereas D3-10 was preferred for TST−.
Figure 2.

D and J segments usage analysis of VH3-23 gene for TST− and TST+ populations. (a) Analysis of D segment usage; (b) analysis of J segment usage. The red and blue bars represent the TST− and TST+ populations, respectively.
The J segment usage pattern was similar for both populations with JH4 segment being used the most (43·54% for TST− population; 37·38% for TST+ population), followed by JH5 segment (25·57% for TST− population; 24·76% for TST+ population), and JH6 segment (14·99% for TST− population; 19·39% for TST+ population) (Fig.2b). Wilcoxon Signed tests showed that the TST− population differed significantly from TST+ for the J segments (Z = −2·201, P < 0·05) but not the D segments (Z = −1·314, P = n.s).
D and J segment pairing frequency analysis
For the TST+ population, the D3-22-J4 pairing showed the highest percentage of pairing (5·76%), followed by D2-2-J4 (4·82%), D3-22-J5 (4·48%), D3-10-J4 (3·63%) and D3-22-J6 (3·16%) in comparison with the estimated frequency of 0·76% for unbiased V-D segment pairings based on the 132 D-J combinations found from the total number of 2344 reads obtained (Fig.3). Hence, D3-22-J4 pairing is about 7·6-fold (5·76/0·76 = 7·6) more than the expected pairing frequency. The TST− population also exhibited preferential pairing whereby D3-10-J4 scored 4·39%, followed by D3-3-J4 (4·28%), D2-2-J4 (4·18%), D3-22-J4 (3·38%) and D6-19-J4 (3·07%) in a total number of 1891 reads obtained (Fig.3).
Figure 3.
D and J segments pairing frequency percentage analysis for both TST− and TST+ populations. (a) Pairing frequency of J1; (b) pairing frequency of J2; (c) pairing frequency of J3; (d) pairing frequency of J4; (e) pairing frequency of J5; (f) pairing frequency of J6. All J segments are with the 23 D segments under the given total number of re-combination events.
A paired-samples t-test was conducted to compare the frequency in TST− and TST+ across all 144 possible gene combinations. There was a significantly higher frequency count for TST+ (M = 16·28, SD = 22·058) compared with TST− (M = 13·13, SD = 16·675); t(143) = 2·914, P = 0·00).
D-J segment Circos analysis
The comparison between the Circos plots shows a higher recombination of D3-3 to J4, J5 and J6 by the TST− than the TST+ population (Fig.4a, b). For the TST+ population, a distinct pattern of usage was the D5-12 gene segment. Here the J6, J5 and J4 segments are paired with D5-12 in relatively higher percentages than in TST− population (Fig.4a). The comparison between the pairing patterns for both populations would mean that D3-3 pairing could potentially be of more interest than D3-22 and D3-10, as these segments are equally prominent in both populations.
Figure 4.
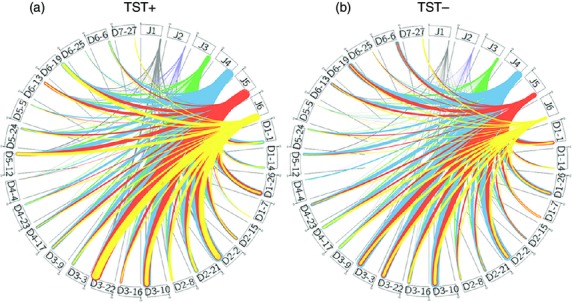
Circos diagram for TST+ and TST− populations. The thickness of the connection line between the J1–J6 segments with the D segments correlates the number of occurrence of the particular pairing in (a) TST+ and (b) TST− populations. The grey, purple, green, blue, orange and yellow colours of the line represent J1–J6 segments, respectively.
Discussion
Tuberculosis is a complex disease characterized by an intricate relationship between the microorganism and its host.4 Up to now, attempts to correlate protection mechanisms for the disease have been unsuccessful.28; – 31 After contact with M. tuberculosis there are usually three potential outcomes: non-infection, latent infection, or clinical TB.4 From the epidemiological point of view in relation with M. tuberculosis infection, individuals without clinical TB are classified as either non-infected or latently infected depending on the results of TST and/or interferon-gamma release assay tests.4,24,25 The existence of TST− individuals despite a high and sustained exposure to M. tuberculosis has been documented whereby these individuals are regarded as resistant to infection with this microorganism.24,25,32; – 36 To clarify the mechanisms of resistance to M. tuberculosis infection in healthy TST− individuals with long exposure to M. tuberculosis, several groups have carried out studies focusing on the genetic and immunological traits.25,34,36
The genetic studies proposed a suggestive linkage with two chromosomal regions in TST− individuals that could be attributed to resistance to infection.34 A report in 2009 showed the presence of a major locus that is associated with resistance to infection with M. tuberculosis in TST− individuals.25 An immune response study involving adults in Uganda reported no differences in innate immune responses between TST− and TST+ individuals exposed to M. tuberculosis but found differences in T-cell responses to M. tuberculosis antigens between the groups.36 The existence of healthy individuals despite high exposure to M. tuberculosis for long periods of time argues in favour of resistance to TB. The presence of TST negativity in these individuals could be associated with resistance to infection, and the TST+ individuals could represent subjects susceptible to infection but with resistance for progression to clinical TB, with both populations exhibiting different mechanisms of protection.23; – 25,32,34; – 36
The respiratory mucosa is the port of entry of M. tuberculosis, so, many different mechanisms (innate and acquired) could be involved in the blockage of the infection at this level.37 One of the most important players at the mucosal surfaces is IgA, which by different mechanisms (including inhibition of adhesion, biological effects of interaction with CD89 receptor), could function to block the entrance of different pathogens, including M. tuberculosis.14,37 Several studies using IgA monoclonal antibodies and human secretory IgA, by mucosal route, were able to exhibit protective effects in challenge experiments with M. tuberculosis in mice.12,14,15,38 Based on these antecedents and with the aim of contributing to the study of potential factors involved in protection against M. tuberculosis infection, the main objective of the present work was to explore the IgA V gene usage in healthy individuals (TST− and TST+) with long-term exposure to M. tuberculosis, in order to determine if there are differences in this aspect between the groups. The antibody responses against M. tuberculosis antigens have been shown to be heterogeneous with no clear association between antigen recognition and protection. Therefore, in this study, our focus is on the global V gene usage of the IgA isotype antibodies to identify differences in V gene usage frequencies that could be associated with protection against M. tuberculosis infection.
Common samples used for V gene usage studies revolve around the use of peripheral B cells as they are easily harvested. Although our focus is on V gene usage of the IgA isotype at the point of entry, peripheral B cells have been demonstrated in pneumonia to be suitable alternatives to reflect the conditions and repertoire of B cells in the lungs.39 Therefore it would be conceivable to apply peripheral B cells focusing on IgA-specific V genes for analysis to understand the repertoire for V gene preferences in the lungs of TB-exposed individuals. The extracted pool of antibody genes from the IgA-producing B cells in the peripheral blood that was used for PCR amplification represents the in vivo collection of the encoded antibody genes for each population. If there is no bias in the gene usage for the antibody generation, the resulting PCR profile comparison of the V gene segments between the populations by using identical and controlled amounts of the antibody genes as the template with similar PCR conditions will not show significant differences considering that they contain similar representations of V gene usage. In both populations, a stronger intensity for each V gene amplification band is attributed to the varying degrees of gene expression levels.
The results indicate that VH3-23 of IgA isotype and Vκ1 genes were higher in the TST− population than TST+ population. It has been reported that in IgM memory cells from healthy naive individuals, VH3-23 is also the most commonly used variable gene.40; – 42 Previous studies observed that for many diseases, different individuals are able to produce antibodies with the same V-D-J combinations for effective clearance of infection.43 This observation was seen in antibody responses to bacterial polysaccharide antigens, which are the targets of protective and vaccine responses against infections by some of the life-threatening bacteria, such as Haemophilus influenzae and Streptococcus pneumoniae.40 These protective neutralizing antibodies used V segment VH3-23, either J segment J4 or J6, and a short D segment in response to the Haemophilus influenzae type B vaccine.44 Notably, use of the VH3-23 segment is not only found in Haemophilus influenzae infections,45 but also in lymphocytic leukaemia,46 and in HIV47 and Streptococcus pneumoniae infections.43 The similar outline of preferred VH3-23 gene segment together with bias unique D and J4 segments in the TST− population could therefore indicate a possible induced response by the humoral immune system to M. tuberculosis exposure.
As conventional agarose gel electrophoresis would not be able to provide an accurate estimate of the band intensity and DNA concentration, the Bioanalyzer system was used in this study to provide a more precise estimation of band intensity and DNA concentration. Previous studies have used the Agilent 2100 Bioanalyzer with the DNA Lab-Chip kits for accurate quantification of PCR products for the RT-PCR.48 To ensure that the cDNA used was the best representation of the actual repertoire, the amount of RNA used for each cDNA synthesis was kept constant. PCR primer annealing conditions were optimized to ensure the best possible amplification with customized conditions for each primer pair. The CD19 PCR amplification is indicative of the amount of B lymphocytes harvested during total RNA extraction of peripheral mononuclear cells. Consistency of CD19 band intensity from the amplification suggests that no bias occurred in B-cell numbers during RNA extraction and cDNA amplification.
The ability of next-generation sequencing to sequence a million sequences with high levels of sensitivity helps to provide a proper representation of the abundance of antibodies and their genes that occur in the immune system. Further characterization of the sequencing results would require the reference of online antibody analysis softwares and databases such as VBASE2. VBASE2 is a database of mouse and human immunoglobulin variable genes that offers details of the V genes such as nucleotide and amino acid sequences, V, D, J segment alignment, and CDR analysis.27
A comparison based on frequency for each D and J gene segment would allow a reflection of a numerical percentage of the frequency that a particular gene segment is used in each population. The D3-22 gene segment was most used by the TST+ and D3-10 by the TST− individuals. Antibodies against HIV, hepatitis C and Plasmodium falciparum antigens and in leukaemia, have used both gene segments.49; – 52 Analysis also shows low use of gene segment D4-4 for both populations. Antibody gene usage of D4-4 has been reported in a VH4-34/D4-4/JH6 arrangement for multiple myeloma antibodies with low autoreactivity.53
During V-D-J recombination, initially D and J segments recombine first and this is followed by V-D-J recombination. If the D and J segments are paired at different frequencies, the resulting antibody repertoire will contain highly skewed representation of D-J combinations, which will be further diversified by other mechanisms such as V-D-J pairing, somatic hypermutation and affinity maturation. In this study, the D-J segment pairing frequencies are consistent with the D and J segment usage for both TST populations. For both the TST+ and TST− population, the D3-22-J4 segment pairing was highest. The higher J4 usage would be the result of the memory from long-term exposure to TB. This is in agreement with previous comparison of V and J gene segment usage in naive and memory subsets where the J4 segment was preferred for memory populations with a decrease of J6 gene segments.42
The Circos plots were able to show the relative prominence of each D-J pairing within the VH3-23 repertoire. These plots are useful to reveal trends that are only apparent when the repertoire is analysed in the context of a complete D-J recombination.41 These plots revealed prominent patterns that are not necessarily significant in terms of frequency. The higher frequency pairings, although significant in both populations, reflect a similar pattern whereby D3-22-J4 pairings are abundant. This could be a reflection of the immune response towards TB exposure as a whole but does not clearly echo the difference in B-cell response seen between the two populations. A closer analysis shows that D3-3 pairings to J4 and J5 are unique in TST− populations. The preference for D3-3 segments has only been reported in chronic lymphocytic leukaemia but with a preferential combination with VH1-69 and J6.54
A limitation of this study involves the inability to correlate the phenotypic nature with the antibody genotype. It remains to be seen if the preferences observed would actually correlate with the generation of protective antibodies for TB. However, this report provides the first insight into the antibody genetics of B cells in response to TB exposure. Pairing analysis using the Circos plots showed a distinctive usage of the J4, J5 and J6 segments with D3-3 segment for TST− individuals. These findings could be indicative of a possible M. tuberculosis antigen component with the capacity to elicit, in resistant individuals, protective IgA immune responses with antibodies encoded by this combination that probably have special affinity for critical antigenic epitopes promoting the blockade of the entrance of M. tuberculosis. Further analysis is still required as there are still many questions that remain unanswered to fully understand how antibodies can be coupled to elevate immunity against M. tuberculosis. However, the antibodyome study between both populations suggests that a subset of V-D-J gene pairing, mainly VH3-23-D3-3-J4, could be of interest in populations protected from infection with M. tuberculosis.
Acknowledgments
This study was supported by the Higher Institution Centre of Excellence (HICoE) Grant (Grant No. 311/CIPPM/44001005) and Long term Research Grant Scheme (Grant No. 203.PSK.6722001), Ministry of Education, Malaysia. We thank all donors for participating in the study. We would like also thank Mr Hajar Fauzan Bin Ahmad and nurses at HUSM for their kind assistance throughout the sampling process.
Disclosure
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1Size of induration for TST− population.
Table S2. Size of induration for TST+ population.
Table S3. Primer annealing temperature at 55°.
Table S4. Primer annealing temperature at 60°.
Table S5. Bioanalyzer analysis of the VH, Vκ and Vλ genes concentration (ng/µl) for TST− population.
Table S6. Bioanalyzer analysis of the VH, Vκ and Vλ genes of the TST+ population.
Table S7. Bioanalyzer analysis of the CD19 gene concentration for TST− and TST+ populations.
Method S1. Perl length filtration script.
Method S2. Perl antibody analysis script.
References
- Falzon D, Raviglione MC. TB control: Achievements, obstacles and the role of vaccines. In: Norazmi MN, Acosta A, Sarmiento ME, editors. The Art &Science of Tuberculosis Vaccine Development. 2nd edn. Malaysia: Oxford University Press; 2014. pp. 13–29. [Google Scholar]
- Zumla A, Raviglione M, Hafner R, von Reyn CF. Tuberculosis. N Engl J Med. 2013;368:745–55. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- Zumla A, George A, Sharma V, Herbert N Baroness Masham of IIton. WHO's 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–7. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- Kaufmann SH, Lange C, Rao M, Balaji KN, Lotze M, Schito M, Zumla AI, Maeurer M. Progress in tuberculosis vaccine development and host-directed therapies – a state of the art review. Lancet Respir Med. 2014;2:301–20. doi: 10.1016/S2213-2600(14)70033-5. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, Sprenger M. Drug-resistance – a challenge for tuberculosis control in the European Union and European Economic Area. Euro Surveill. 2014;19:1–3. doi: 10.2807/1560-7917.es2014.19.11.20737. [DOI] [PubMed] [Google Scholar]
- Summers L. Tuberculosis a persistent health care problem. J Nurse-Midwifery. 1987;32:68–78. doi: 10.1016/0091-2182(87)90002-4. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Flynn J. CD8+ T cells in tuberculosis. Am J Respir Crit Care Med. 2002;166:1116–21. doi: 10.1164/rccm.2204027. [DOI] [PubMed] [Google Scholar]
- Mahon RN, Sande OJ, Rojas RE, Levine AD, Harding CV, Boom WH. Mycobacterium tuberculosis ManLAM inhibits T-cell-receptor signaling by interference with ZAP-70, Lck and LAT phosphorylation. Cell Immunol. 2012;275:98–105. doi: 10.1016/j.cellimm.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindestam Arlehamn CS, Sette A. Definition of CD4 immunosignatures associated with MTB. Front Immunol. 2014;5:124. doi: 10.3389/fimmu.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglione PJ, Chan J. How B cells shape the immune response against Mycobacterium tuberculosis. Eur J Immunol. 2009;39:676–86. doi: 10.1002/eji.200839148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta A, Lopez Y, Norazmi MN, Pando RH, Alvarez N, Sarmiento ME, Glatman-Freedman A. In: Tuberculosis-Current Issues in Diagnosis and Management. Mahboub BH, Vats MG, editors. 2013. The role of antibodies in the defence against tuberculosis, Croatia: InTech: doi: 10.5772/53950. [Google Scholar]
- Lopez Y, Yero D, Falero-Diaz G. Induction of a protective response with an IgA monoclonal antibody against Mycobacterium tuberculosis 16 kDa protein in a model of progressive pulmonary infection. Int J Med Microbiol. 2009;299:447–52. doi: 10.1016/j.ijmm.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Reljic R, Williams A, Ivanyi J. Mucosal immunotherapy of tuberculosis: is there a value in passive IgA? Tuberculosis. 2006;86:179–90. doi: 10.1016/j.tube.2006.01.011. [DOI] [PubMed] [Google Scholar]
- Balu S, Reljic R, Lewis MJ. A novel human IgA monoclonal antibody protects against tuberculosis. J Immunol. 2011;186:3113–9. doi: 10.4049/jimmunol.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Reljic R, Naylor I. Passive protection with immunoglobulin A antibodies against tuberculous early infection of the lungs. Immunology. 2004;111:328–33. doi: 10.1111/j.1365-2567.2004.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez N, Otero O, Camacho F. Passive administration of purified secretory IgA from human colostrum induces protection against Mycobacterium tuberculosis in a murine model of progressive pulmonary infection. BMC Immunol. 2013;14:S3. doi: 10.1186/1471-2172-14-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Market E, Papavasiliou FN. VDJ Recombination and the evolution of the adaptive immune system. PLoS Biol. 2003;1:e16. doi: 10.1371/journal.pbio.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JA, Jiang N, White RA, Fisher DS, Quake SR. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–10. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Kim N, Lee M-K, Choi H-J, Baek HJ, Nam C-H. Overexpression and unique rearrangement of VH2 transcripts in immunoglobulin variable heavy chain genes in ankylosing spondylitis patients. Exp Mol Med. 2010;42:319–26. doi: 10.3858/emm.2010.42.5.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Chung J, Shin GT, Lee SY, Jang Y-J. Variable region genes of human monoclonal autoantibodies to histones H2A and H2B from a systemic lupus erythematosus patient. Mol Immunol. 2005;42:311–7. doi: 10.1016/j.molimm.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Pritsch O, Troussard X, Magnac C. VH gene usage by family members affected with chronic lymphocytic leukaemia. Br J Haematol. 1999;107:616–24. doi: 10.1046/j.1365-2141.1999.01757.x. [DOI] [PubMed] [Google Scholar]
- Rose DN, Schechter CB, Adler JJ. Interpretation of the tuberculin skin test. J Gen Intern Med. 1995;10:635–42. doi: 10.1007/BF02602749. [DOI] [PubMed] [Google Scholar]
- Rieder HL. Epidemiologic Basis of Tuberculosis Control. 1st edn. Paris, France: International Union Against Tuberculosis and Lung Disease; 1999. [Google Scholar]
- Cobat A, Gallant CJ, Simkin L. Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med. 2009;206:2583–91. doi: 10.1084/jem.20090892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim TS, Mollova S, Rubelt F, Sievert V, Dübel S, Lehrach H, Konthur Z. V-gene amplification revisited – an optimised procedure for amplification of rearranged human antibody genes of different isotypes. New Biotechnol. 2010;27:108–17. doi: 10.1016/j.nbt.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Retter I, Althaus HH, Münch R, Müller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33:D671–4. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Alves C, Booty MG, Carpenter SM, Jayaraman P, Rothchild AC, Behar SM. In search of a new paradigm for protective immunity to TB. Nat Rev Microbiol. 2014;12:289–99. doi: 10.1038/nrmicro3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley AM, McShane H. Tuberculosis vaccines: progress and challenges. Trends Pharmacol Sci. 2011;32:601–6. doi: 10.1016/j.tips.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Minassian AM, Ronan EO, Poyntz H, Hill AV, McShane H. Preclinical development of an in vivo BCG challenge model for testing candidate TB vaccine efficacy. PLoS One. 2011;6:e19840. doi: 10.1371/journal.pone.0019840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagina BMN, Abel B, Scriba TJ. Specific T cell frequency and cytokine expression profile do not correlate with protection against tuberculosis after bacillus Calmette–Guérin vaccination of newborns. Am J Respir Crit Care Med. 2010;182:1073–9. doi: 10.1164/rccm.201003-0334OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrall AJ, Netea MG, Alisjahbana B, Hill PC, van Crevel R. Early clearance of Mycobacterium tuberculosis: a new frontier in prevention. Immunology. 2014;141:506–13. doi: 10.1111/imm.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Zalwango S, Malone LL. Clinical and epidemiological characteristics of individuals resistant to M. tuberculosis infection in a longitudinal TB household contact study in Kampala, Uganda. BMC Infect Dis. 2014;14:352. doi: 10.1186/1471-2334-14-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Zalwango S, Malone LL. Genome scan of M. tuberculosis infection and disease in Ugandans. PLoS One. 2008;3:e4094. doi: 10.1371/journal.pone.0004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel L, El-Baghdadi J, Bousfiha AA, Casanova JL, Schurr E. Human genetics of tuberculosis: a long and winding road. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130428. doi: 10.1098/rstb.2013.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan CS, Zalwango S, Thiel BA. Innate and adaptive immune responses during acute M. tuberculosis infection in adult household contacts in Kampala, Uganda. Am J Trop Med Hyg. 2012;86:690–7. doi: 10.4269/ajtmh.2012.11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olafsdottir T, Harandi AM. Mucosal immunity and vaccination with special emphasis on respiratory tract. In: Norazmi MNAA, Sarmiento ME, editors. The Art & Science of Tuberculosis Vaccine Development. 2nd edn. Malaysia: Oxford University Press; 2014. pp. 985–97. [Google Scholar]
- Alvarez N, Otero O, Camacho F. Passive administration of purified secretory IgA from human colostrum induces protection against Mycobacterium tuberculosis in a murine model of progressive pulmonary infection. BMC Immunol. 2013;14(Suppl 1):S3. doi: 10.1186/1471-2172-14-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HM, He Y, Sangster MY. Broad dispersion and lung localization of virus-specific memory B cells induced by influenza pneumonia. Proc Natl Acad Sci. 2008;105:3485–90. doi: 10.1073/pnas.0800003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R, Lee W, Cahill P. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney BS, Willis JR, McKinney BA, Crowe JE. High-throughput antibody sequencing reveals genetic evidence of global regulation of the naive and memory repertoires that extends across individuals. Genes Immun. 2012;13:469–73. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- Wu Y-C, Kipling D, Leong HS, Martin V, Ademokun AA, Dunn-Walters DK. High-throughput immunoglobulin repertoire analysis distinguishes between human IgM memory and switched memory B-cell populations. Blood. 2010;116:1070–8. doi: 10.1182/blood-2010-03-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lottenbach KR, Barenkamp SJ, Lucas AH, Reason DC. Recurrent variable region gene usage and somatic mutation in the human antibody response to the capsular polysaccharide of Streptococcus pneumoniae type 23F. Infect Immun. 2002;70:4083–91. doi: 10.1128/IAI.70.8.4083-4091.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas AH, Reason DC. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol Rev. 1999;171:89–104. doi: 10.1111/j.1600-065x.1999.tb01343.x. [DOI] [PubMed] [Google Scholar]
- Silverman GJ, Lucas AH. Variable region diversity in human circulating antibodies specific for the capsular polysaccharide of Haemophilus influenzae type b. Preferential usage of two types of VH3 heavy chains. J Clin Invest. 1991;88:911–20. doi: 10.1172/JCI115394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fält S, Merup M, Tobin G, Thunberg U, Gahrton G, Rosenquist R, Wennborg A. Distinctive gene expression pattern in VH3-21 utilizing B-cell chronic lymphocytic leukemia. Blood. 2005;106:681–9. doi: 10.1182/blood-2004-10-4073. [DOI] [PubMed] [Google Scholar]
- Scamurra RW, Miller DJ, Dahl L, Abrahamsen M, Kapur V, Wahl SM, Milner EC, Janoff EN. Impact of HIV-1 infection on VH3 gene repertoire of naive human B cells. J Immunol. 2000;164:5482–91. doi: 10.4049/jimmunol.164.10.5482. [DOI] [PubMed] [Google Scholar]
- Gottwald E, Müller O, Polten A. Semiquantitative reverse transcription-polymerase chain reaction with the Agilent 2100 Bioanalyzer. Electrophoresis. 2001;22:4016–22. doi: 10.1002/1522-2683(200110)22:18<4016::AID-ELPS4016>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Kunert R, Ruker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res Hum Retroviruses. 1998;14:1115–28. doi: 10.1089/aid.1998.14.1115. [DOI] [PubMed] [Google Scholar]
- Chan CH, Hadlock KG, Foung SK, Levy S. VH1-69 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97:1023–6. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- Widhopf GF, 2nd, Kipps TJ. Normal B cells express 51p1-encoded Ig heavy chains that are distinct from those expressed by chronic lymphocytic leukemia B cells. J Immunol. 2001;166:95–102. doi: 10.4049/jimmunol.166.1.95. [DOI] [PubMed] [Google Scholar]
- Cheng X-J, Hayasaka H, Watanabe K. Production of high-affinity human monoclonal antibody fab fragments to the 19-kilodalton C-terminal merozoite surface protein 1 of Plasmodium falciparum. Infect Immun. 2007;75:3614–20. doi: 10.1128/IAI.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froyland M, Thompson KM, Thorpe SJ, Sahota SS, Gedde-Dahl T, Bogen B. A VH4-34+ myeloma protein with weak autoreactivity. Haematologica. 2007;92:690–3. doi: 10.3324/haematol.10850. [DOI] [PubMed] [Google Scholar]
- Tobin G, Thunberg U, Karlsson K. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104:2879–85. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1Size of induration for TST− population.
Table S2. Size of induration for TST+ population.
Table S3. Primer annealing temperature at 55°.
Table S4. Primer annealing temperature at 60°.
Table S5. Bioanalyzer analysis of the VH, Vκ and Vλ genes concentration (ng/µl) for TST− population.
Table S6. Bioanalyzer analysis of the VH, Vκ and Vλ genes of the TST+ population.
Table S7. Bioanalyzer analysis of the CD19 gene concentration for TST− and TST+ populations.
Method S1. Perl length filtration script.
Method S2. Perl antibody analysis script.