Abstract
Identification of units within species worthy of separate management consideration is an important area within conservation. Mitochondrial DNA (mtDNA) surveys can potentially contribute to this by identifying phylogenetic and population structure below the species level. The American crocodile (Crocodylus acutus) is broadly distributed throughout the Neotropics. Its numbers have been reduced severely with the species threatened throughout much of its distribution. In Colombia, the release of individuals from commercial captive populations has emerged as a possible conservation strategy that could contribute to species recovery. However, no studies have addressed levels of genetic differentiation or diversity within C. acutus in Colombia, thus complicating conservation and management decisions. Here, sequence variation was studied in mtDNA cytochrome b and cytochrome oxidase I gene sequences in three Colombian captive populations of C. acutus. Two distinct lineages were identified: C. acutus-I, corresponding to haplotypes from Colombia and closely related Central American haplotypes; and C. acutus-II, corresponding to all remaining haplotypes from Colombia. Comparison with findings from other studies indicates the presence of a single “northern” lineage (corresponding to C. acutus-I) distributed from North America (southern Florida), through Central America and into northern South America. The absence of C. acutus-II haplotypes from North and Central America indicates that the C. acutus-II lineage probably represents a separate South American lineage. There appears to be sufficient divergence between lineages to suggest that they could represent two distinct evolutionary units. We suggest that this differentiation needs to be recognized for conservation purposes because it clearly contributes to the overall genetic diversity of the species. All Colombian captive populations included in this study contained a mixture of representatives of both lineages. As such, we recommend against the use of captive-bred individuals for conservation strategies until further genetic information is available.
Keywords: Captive population, conservation, crocodile, genetic diversity, management, mitochondrial DNA
Introduction
The application of mitochondrial DNA (mtDNA) analyses has provided unique insights into population differentiation and causes of genetic diversity below the level of the species (Avise 2000). Such analyses are also useful in species conservation, where they can potentially contribute to the identification of phylogenetic, population or other evolutionary significant units (ESUs; Moritz 1994b; Crandall et al. 2000; Ryder 1986; Vogler and DeSalle 1994) worthy of separate management consideration. Although the specific criteria for delimiting ESUs are widely debated (for a review, see Fraser and Bernatchez 2001), the basic principle is to identify units that, due to their reproductive or historical isolation from other populations, contribute substantially to the overall evolutionary history of the species (Moritz 2002; Avise 2005). Identification of genetic diversity relevant to species conservation is particularly necessary for developing conservation programs for threatened and managed species (Moritz 1999; Storfer 1999). Knowledge of genetic diversity and structure can help identify the most closely related populations or individuals for use in translocation and augmentation programs. Furthermore, examination of the genetic composition of existing captive breeding populations can help guide management practices for maintaining the genetic integrity of distinct genetic groups in captivity as well as assessing the suitability of captive individuals for population recovery and conservation strategies (e.g., Ruokonen et al. 2000; Burns et al. 2003; Gaur et al. 2006; Ramirez et al. 2006; Russello et al. 2007; Beauclerc et al. 2010; McGreevy et al. 2011; Roldán et al. 2011; Benavides et al. 2012; Meraner et al. 2014).
The American crocodile, Crocodylus acutus Cuvier, 1807, is a widely distributed species, being found from North America (southern Florida) to northern South America, as well as the Caribbean islands (Thorbjarnarson 2010). The species has suffered severe population declines throughout much of its distribution (Thorbjarnarson et al. 2006) and as a result is listed as vulnerable by the International Union for Conservation of Nature and Natural Resources (IUCN) and included in Appendix I of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) (except for the population of Cuba, which is included in Appendix II). While national and international trade restrictions have allowed population recovery in some parts of the species' distribution, in other parts, populations remain small and threatened (e.g., Colombia, Ecuador, Jamaica) (Thorbjarnarson et al. 2006). Population recovery has been aided by the establishment of regulated commercial exploitation (registered under CITES) of closely managed commercial farm populations (Thorbjarnarson et al. 2006). As populations of the American crocodile continue to be threatened in the wild, there is increasing interest in the use of captive-bred individuals for reintroduction and reinforcement programs in order to prevent local population extinction.
In Colombia, commercial hunting during the first half of the last century reduced numbers of C. acutus greatly (Medem 1981). In spite of a ban on commercial hunting in 1968, recovery has been limited resulting in the survival of population relicts throughout its former distribution (Castaño-Mora 2002). Subsequent censuses and monitoring in 1994 and 1997 indicated the precarious state of this species in Colombia and the need for continued action to prevent local extinction (Castaño-Mora 2002 and references therein). Despite its continued threatened status in Colombia, there exist many thousands of individuals in captivity on commercial crocodile farms within the country. The founder base of these captive populations includes animals caught in Colombia under collecting permits authorized by the corresponding national authority and animals of unknown geographical origin confiscated by local authorities in Colombian territory (records Ministerio de Ambiente y Desarrollo Sostenible, Colombia). Descendants of these animals now represent the major part of the captive breeding population in Colombia. National legislation requires that a percentage of the descendants of these captive populations be returned to the wild as compensation. However, uncertainty regarding genetic relationships in C. acutus raises the possibility that management plans may not adequately take into account actual patterns of genetic diversity within the species.
In spite of its large geographical distribution and threatened status, limited genetic data from throughout the species' range have been published. While most genetic studies of C. acutus have been limited to local population-level descriptions of genetic diversity and structure as well as interspecific hybridization (e.g., Ray et al. 2004; Cedeño-Vázquez et al. 2008; Porras Murillo et al. 2008; Rodriguez et al. 2008, 2011; Weaver et al. 2008), few have addressed patterns of phylogenetic structure throughout the species' distributional range. Milián-García et al. (2011) analysed mtDNA sequence (complete cyt b and partial COI) and microsatellite DNA in individuals of C. acutus from localities from the Caribbean islands (including Cuba) and Central America (Costa Rica and Panama). Taxonomically, analyses supported the designation of Central American C. acutus but not Cuban C. acutus, the latter exhibiting greater genetic similarity to Crocodylus rhombifer from Cuba. Moreover, these findings supported the recognition of a single mtDNA lineage in C. acutus. Rodriguez et al. (2011) compared partial mtDNA control region sequence of C. acutus from localities across Florida with sequences published for individuals from Costa Rica, Mexico, Belize and Jamaica. Like the study of Milián-García et al. (2011), analyses supported the existence of a single C. acutus mtDNA lineage Oaks (2011), however, in a study of phylogenetic relationships within the genus based on substantially more sequence data, provided evidence of two mtDNA lineages in C. acutus. Unfortunately, the samples of C. acutus used in Oaks (2011) came from individuals of unknown geographical origin from different captive collections in North America (LSU Museum of Natural Science: C. Austin, pers. comm.). The lack of sufficient genetic data from throughout the distributional range of C. acutus and/or uncertainty regarding the geographical origin of samples included means that the actual extent of genetic diversity and structure in this species is still difficult to assess.
This study was primarily motivated by the need to establish patterns of genetic diversity within captive populations of C. acutus in Colombia to underpin conservation and management decisions at a national level. The current study of genetic diversity aimed to: (1) examine genetic diversity within commercial captive breeding populations in Colombia and (2) determine the phylogenetic relationships of C. acutus haplotypes sampled in this study in relation to other populations.
Materials and Methods
Samples and DNA isolation
Samples (blood or tissue) were obtained from 40 individuals of C. acutus (Fig.1) held on three different commercial crocodile farms (or holding facilities) located in the departments of Atlántico and Bolívar in the Caribbean coastal region of Colombia. Three specimens corresponded to wild-caught, captive-held individuals (taken previously from the wild), and 37 corresponded to captive-bred individuals (descendants of the original founder population). The founder base of the three captive populations included animals caught under collecting permits in different localities in the adjoining departments of Magdalena and Atlántico in the Caribbean coastal region of Colombia as well as animals (previously held at Barranquilla Zoo, department of Atlántico) of unknown geographical origin confiscated by local authorities within Colombian territory. Tissue or blood samples were stored in absolute ethanol until further analysis. Total genomic DNA was isolated using proteinase K digestion and silica/guanidinium thiocyanate extraction. Full details of DNA extraction are contained Appendix S1.
Figure 1.

American crocodile (Crocodylus acutus).
DNA amplification and sequencing
Sequence variation was examined in complete mtDNA cytochrome b (cyt b) and cytochrome oxidase I (COI) gene sequences. The cyt b gene was amplified using primers Croc_GluL2 (5′-AAT TCC CAT TAT TCT CAC TTG G-3′) and Croc_ThrH2 (5′-TTG GGA AGG TGT GTG TAT TCC-3′), and the COI gene amplified using primers Croc_CysL1 (5′-CGA GTT TGC AGT TCG TCG TG-3′) and Croc_SerH1 (5′-AGC ATG TCG TAT TGC GGT TG-3′). All primers were designed for this study. Full details of primer design are contained in Appendix S1. Polymerase chain reactions (PCRs) were carried out in a reaction volume of 30 μL containing 1 × PCR buffer [75 mmol/L Tris-HCl, 20 mmol/L (NH4)2SO4, 0.01% (v/v) Tween 20; Fermentas], 2.0 mmol/L MgCl2, 0.1 mmol/L of each dNTP, 0.75 U Taq polymerase (Fermentas), and 0.1 μmol/L of each primer. After an initial denaturation step of 2 min at 94°C, 34 cycles of 30 sec at 94°C, 30 sec at 55°C, and 2 min at 72°C were followed by a final extension of 72°C for 10 min. Removal of unincorporated primer and dNTPs was achieved using ethanol precipitation. Purified PCR products were sequenced using Big-Dye (Applied Biosystems, Foster City, CA 94404, USA) cycle sequencing reactions with the same primers used for PCR amplification and the resultant reaction products run on an ABI 3500 Genetic Analyzer automated sequencer (Applied Biosytems). Resulting sequence traces were assembled and edited using the program CodonCode Aligner ver. 4.2 (CodonCode Corporation; http://www.codoncode.com). This resulted in 1200 bp of complete cyt b gene sequence and 1557 bp of complete COI gene sequence for all individuals. Sequences have been added to GenBank under accession numbers KF273842–KF273849 for cyt b and KF273834–KF273841 for COI.
Data analyses
Complete gene sequences were aligned using “Clustal W” (Thompson et al. 1994) as implemented within the program BioEdit ver. 7.0 (Hall 1999) and translated into amino acid sequences using the program MEGA ver. 5 (Tamura et al. 2011) to check for the presence of premature stop codons. There were no premature stop codons, insertions, or deletions, and therefore pseudogenes were not suspected. The two gene sequences were combined for subsequent analyses.
Phylogenetic relationships among sequences were estimated using Bayesian phylogenetic analysis (BY) and maximum parsimony (MP) to provide an alternative approach (see Simmons and Miya 2004; Lewis et al. 2005). For the BY analysis, the aligned full-length gene sequence dataset obtained from C. acutus in this study (hereinafter referred to as Colombia-only dataset) was partitioned into three partitioning schemes: (1) combined gene sequences, (2) gene sequences separated, and (3) separate partitions for codon positions 1 and 2 (cp 1 + 2) and codon position 3 (cp3). The most appropriate model of sequence evolution was selected for each partitioning scheme using the Bayesian Information Criterion (BIC) as performed in jModelTest 2 (Darriba et al. 2012). BY analyses (parameters unlinked) were performed in MrBayes ver. 3.2.1 (Ronquist et al. 2012). Two MCMC samplers were run in parallel (four chains each, temperature parameter set at 0.5) starting from a random tree, using 2.0 × 106 generations (samples recorded every 100 generations) with the first 200,000 generations of each run discarded as burn-in. Convergence was established using the program Tracer ver. 1.4 (Rambaut and Drummond 2007). MP analyses (unweighted) were performed in PAUP* ver. 4.0b10 (Swofford 1998) with 1000 bootstrap samples from the sequence data (heuristic search, 10 random addition replicates). Analyses included Crocodylus moreletii as an outgroup (GenBank accession number: HQ585889; Meganathan et al. 2011). Relationships among sequences were also estimated by an unrooted parsimony network based on a statistical parsimony procedure (Templeton et al. 1992) using the 95% probability of parsimony criterion to connect sequences using the program TCS ver. 1.21 (Clement et al. 2000). This procedure is based on the probability that only a single mutation occurs where there is a nucleotide difference between sequences (as opposed to multiple mutations at the site).
To understand better the phylogenetic relationships of C. acutus haplotypes sampled in this study in relation to other populations, analyses were performed that included complete cyt b and partial COI sequence data from C. acutus from Central America (Panama and Costa Rica) and Cuba as well as sequences from C. rombifer from Cuba (hereinafter referred to as C. acutus-extended dataset) reported by Milián-García et al. (2011) (Appendix S2). Two of the sequences included correspond to individuals reported to be hybrids of C. acutus/C. rombifer. Finally, a third phylogenetic analysis was performed that included partial cyt b sequence data from Oaks (2011) corresponding to individuals of C. acutus from different captive collections in North America (St. Augustine Alligator Farm Zoological Park, Bronx Zoo, Atlanta Zoo and Silver Springs State Park; LSU Museum of Natural Science: C. Austin, pers. comm.).
Results
Sequence variation and genetic diversity
The Colombia-only dataset contained ten variable sites (nine parsimony informative) in the cyt b sequence and eight variable sites (eight parsimony informative) in the COI sequence. The combined gene sequences resulted in eight distinct haplotypes within C. acutus sampled in this study (Table1). Uncorrected sequence divergence among sequences was 0.0–0.8% for cyt b 0.0–0.4% for COI.
Table 1.
Variable sites for the eight mitochondrial DNA haplotypes found within Colombian captive Crocodylus acutus samples based on the full-length alignment (2757 bp) of complete cytochrome b (cyt b) and cytochrome oxidase I (COI) gene sequences
| Variable Position number |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COI |
cyt b |
|||||||||||||||||||
| 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||||||||
| 2 | 4 | 4 | 5 | 5 | 6 | 0 | 7 | 9 | 9 | 0 | 0 | 1 | 6 | 6 | 6 | 6 | ||||
| 1 | 6 | 0 | 7 | 5 | 9 | 1 | 6 | 2 | 6 | 7 | 2 | 5 | 1 | 3 | 5 | 5 | 9 | |||
| Haplotype | GenBank Accession (COI, cyt b) | 2 | 1 | 8 | 4 | 8 | 7 | 2 | 9 | 8 | 5 | 7 | 2 | 1 | 3 | 0 | 3 | 8 | 4 | n |
| Cac01 | KF273834, KF273842 | C | T | G | C | G | A | A | G | A | G | A | G | T | C | C | T | A | G | 21 |
| Cac02 | KF273835, KF273843 | . | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | 1 |
| Cac03 | KF273836, KF273844 | . | C | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | 1 |
| Cac04 | KF273837, KF273845 | . | C | . | . | . | G | . | . | . | . | . | . | C | . | . | . | . | . | 2 |
| Cac05 | KF273838, KF273846 | . | C | A | . | . | G | . | . | . | . | . | . | C | A | . | . | . | . | 3 |
| Cac06 | KF273839, KF273847 | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | 1 |
| Cac07 | KF273840, KF273848 | T | . | A | . | A | G | G | . | G | A | . | A | . | . | T | C | G | A | 2 |
| Cac08 | KF273841, KF273849 | T | . | A | T | A | G | G | A | G | A | . | A | . | . | T | C | G | A | 9 |
The C. acutus-extended dataset contained 79 variable sites (78 parsimony informative) in the cyt b sequence and 29 variable sites (28 parsimony informative) in the partial COI gene sequence (not shown), excluding outgroup taxa. The combined gene sequences resulted in seven distinct haplotypes within C. acutus sampled in this study and four within C. acutus included from Central America. None of the haplotypes detected in C. acutus sampled in this study were shared with sequences included from Central America. Uncorrected sequence divergence among C. acutus haplotypes sampled in this study was 0.0–0.8% for cyt b and 0.0–0.1% for COI. Uncorrected sequence divergence among Central America/Colombia C. acutus haplotypes was relatively low: 0.0–0.8% sequence divergence in cyt b and 0.0–0.4% divergence in COI. Divergence between C. acutus haplotypes from Central America/Colombia and Cuba was much greater (uncorrected cyt b: 5.4–5.7%, COI: 4.6–4.8%) and similar to that between C. acutus haplotypes from Central America/Colombia and C. rombifer (uncorrected cyt b: 5.2–5.7%, COI: 4.9–5.1%). Divergence between C. rombifer haplotypes and C. acutus haplotypes from Cuba was relatively low (0.5–0.9% sequence divergence in cyt b and 0.4–0.4% divergence in COI) and similar to that among C. acutus from Central America and Colombia.
Phylogenetic analyses
For the Colombia-only dataset, jModelTest 2 indicated that the best model of DNA substitution was the HKY model for the combined mtDNA gene sequences (partition scheme [1]) and for each mtDNA gene sequence (partition scheme [2]). The best models of DNA substitution for codon positions (partition scheme [3]) were HKY for cp1 + 2 and GTR + G for cp3. The BY and MP trees recovered two well-supported groupings (hereinafter referred to as haplogroups I and II) within Colombian captive C. acutus (Fig.2). The unrooted parsimony network (not shown) revealed that at least 11 mutational steps separate haplogroups I and II. Differentiation is generally low among the remaining haplotypes (two mutational steps between neighboring haplotypes).
Figure 2.
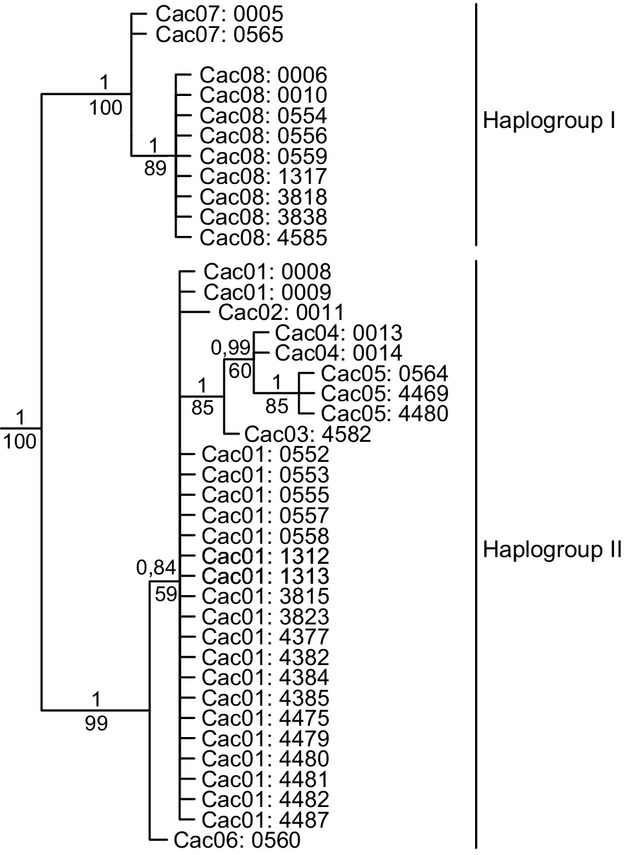
Bayesian phylogram based on the full-length alignment (2757 bp) of complete cytochrome b and cytochrome oxidase I gene sequences for the 40 captive Crocodylus acutus individuals from Colombia (outgroup removed). Internal posterior probabilities (above) and bootstrap support values (below) are provided for all nodes. A number identifying the haplotype, followed by a number specific to the individual, designates samples from Colombia. Haplogroup designations correspond to Fig.3 and Fig.4.
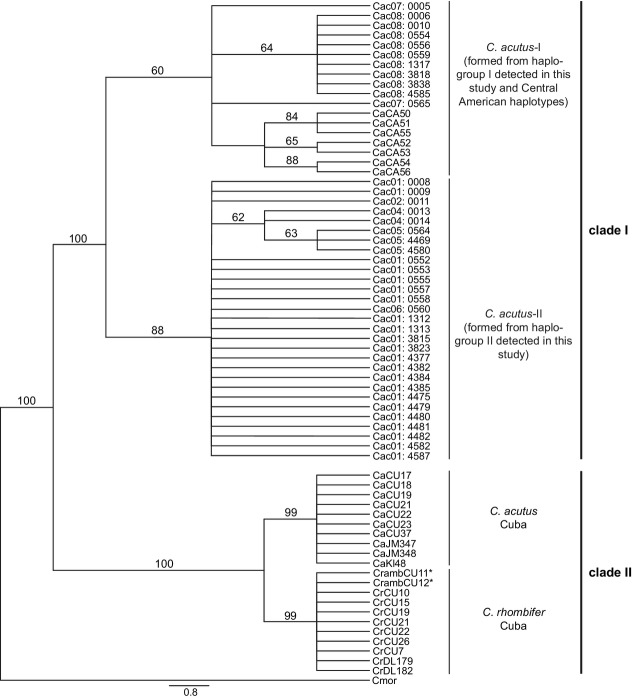
For the C. acutus-extended dataset, jModelTest 2 indicated that the best model of DNA substitution was the HKY + I model for all partitioning schemes, except for the cyt b gene sequence in partition scheme (2) where jModelTest 2 indicated that the best model of DNA substitution was HKY + G. The three different partitioning schemes did not affect the topologies considerably. The BY and MP trees both recovered two main clades: clade I, corresponding to C. acutus from Central America and Colombian captive populations; and clade II, corresponding to C. acutus from Cuba and C. rombifer (Fig.3). Individuals of C. acutus from Cuba are clearly divergent from C. acutus from Central America and Colombian captive populations, being more closely related to C. rombifer. Depending on the analysis (BY or MP), Central American haplotypes grouped either with haplogroup I or haplogroup II to form two separate mtDNA lineages (C. acutus-I and C. acutus-II; Fig 3). As can be seen in the unrooted parsimony network constructed among C. acutus from Colombian captive populations and Central America (Fig.4), two alternatively parsimonious pathways connect the two haplogroups resulting in a closed loop. The unrooted parsimony network reveals that at least three mutational steps separate haplogroup I from Central American haplotypes while haplogroup II is separated from Central American haplotypes by at least six mutational steps. These differences clearly support the phylogenetic grouping of Central American haplotypes with haplogroup I.
Figure 3.
Maximum Parsimony cladogram based on the shorter alignment (1746 bp) of complete cytochrome b and partial cytochrome oxidase I gene sequences for the 40 Crocodylus acutus from Colombia and homologous sequences available in GenBank for C. acutus and C. rombifer (Milián-García et al. 2011). Internal bootstrap support values are provided. Asterisks identify C. acutus/C. rombifer hybrid individuals. A number identifying the haplotype, followed by a number specific to the individual, designates samples from Colombia. Haplogroup designations correspond to Fig.2 and Fig.4. C. acutus-I and C. acutus-II identify the two distinct mtDNA lineages detected within samples of C. acutus from Colombian captive populations and Central America.
Figure 4.
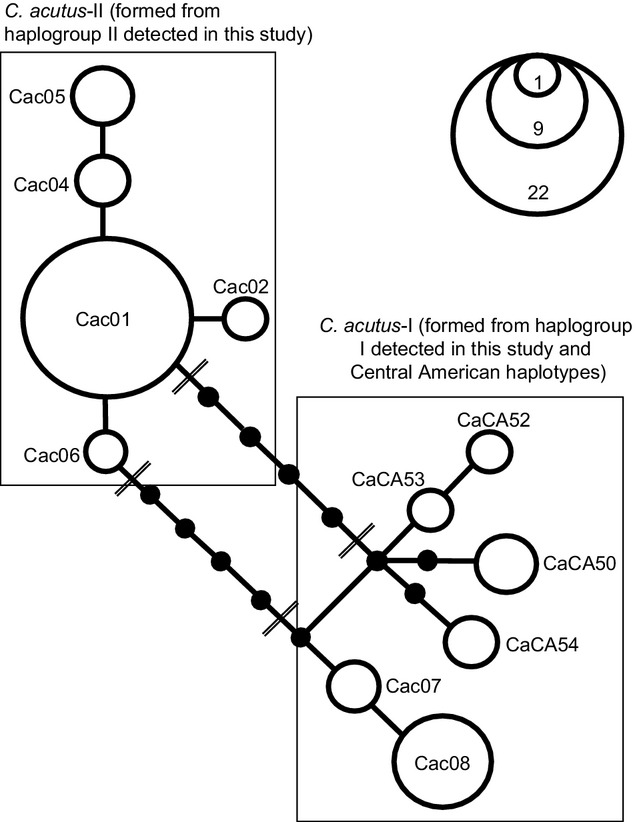
Unrooted cladogram based on the 95% probability of parsimony procedure (Templeton et al. 1992) to show relationships among Crocodylus acutus haplotypes from Colombia and Central America based on the shorter alignment (1746 bp) of complete cytochrome b and partial cytochrome oxidase I gene sequences. All branches are of unit length (one mutational step). Open circles represent observed haplotypes; areas of circles are proportional to the number observed for each haplotype. Filled circles indicate inferred haplotypes not found among sampled individuals. Double lines indicate the most feasible resolutions for the ambiguity in the network. Haplotypes identified in Colombian captive populations and Central America are designated by Cac and CaCA, respectively.
When analyses included published C. acutus cyt b sequence data from Oaks (2011), sequences either formed part of the C. acutus-I lineage or C. acutus-II lineage identified in this study. Only two of the haplotypes (Cac04 and Cac05) found in Colombian captive populations were identical to haplotypes reported by Oaks (2011).
Discussion
The inferences made here are based on mtDNA sequences alone and so may not accurately describe the complete evolutionary history of the species. However, the present analyses strongly indicate that C. acutus from Cuba lies outside the clade containing C. acutus from Central America and this study, forming together with C. rombifer a sister clade relative to the Central America/Colombia C. acutus clade. This relationship is in agreement with that previously suggested by Milián-García et al. (2011) [see Milián-García et al. (2011) for a detailed discussion of possible explanations for this phylogenetic pattern]. Whatever the cause, the closer genetic similarity of individuals assigned to C. acutus from Cuba to individuals assigned to C. rhombifer than to C. acutus from Central America and Colombia is clearly of interest and raises the question of whether it should be included within C. acutus or not.
Within C. acutus from Central America and Colombian captive populations, two distinct mDNA lineages were identified (C. acutus-I and C. acutus-II): C. acutus-I formed from previously published haplotypes sampled from Central America and closely related haplotypes from Colombian captive populations, while C. acutus-II formed from all remaining haplotypes detected in Colombian captive populations. The relationships within C. acutus from Colombian captive populations and Central America are difficult to resolve, but given the intraspecific network the reason is apparent: both Central American and closely related haplotypes from Colombian captive populations are separated from haplogroup II by a similar number of mutational steps. This similarity may help explain the aforementioned ambiguity with regard to the relationships within C. acutus from Central America and Colombian captive populations. However, independent of which of the two alternative mutational pathways is chosen, haplotypes Cac07 and Cac08 (corresponding to haplogroup I) from Colombian captive populations are clearly divergent from haplotypes from haplogroup II, being more closely related to haplotypes from Central America. The mtDNA lineages C. acutus-I and C. acutus-II detected here clearly correspond to the two evolutionary lineages previously recovered by Oaks (2011) based on a much larger sequence dataset. However, unlike the study of Oaks (2011), this study allows speculation on possible geographical distributions of the two lineages (see below).
The geographical distributions of C. acutus-I and C. acutus-II lineages are not easy to assess in this study due to a lack of information regarding the geographical origin of all individuals used to found the Colombian captive population and an absence of genetic information from other parts of the species' range. However, we suggest that the current genetic diversity detected in Colombian captive populations of C. acutus most likely reflects phylogeographic structure within the species. Both haplogroup I and Central American haplotypes clearly form part of the same evolutionary unit (designated C. acutus-I in this study). The close phylogenetic relatedness of haplotypes from haplogroup I to Central American haplotypes suggests that the former grouping most likely originates from geographical populations in northwestern Colombia. Further clues exist as to the possible spatial distribution of the C. acutus-I lineage. Rodriguez et al. (2011) compared partial mtDNA control region sequence of C. acutus from localities across Florida with sequences published for individuals from Costa Rica, Mexico, Belize and Jamaica (differing gene fragments meant they were not included in this study). Analyses supported the presence of a single evolutionary unit. Given the findings of this study, this raises the possibility of a single “northern” evolutionary unit (corresponding to C. acutus-I recovered here) that extends from North America (southern Florida), through Central America and into northern South America. The apparent absence of haplotypes from the C. acutus-II lineage in North and Central America suggests that the C. acutus-II lineage could represent a separate evolutionary unit restricted to South America. Owing to the lack of genetic data from other parts of the species' range it is not possible to determine the geographical limits of these evolutionary units or whether they are geographically overlapping or not. Clearly, analysis of populations from throughout the species distribution (particularly from South America) is required if actual genetic relationships among populations and patterns of distribution of genetic diversity within C. acutus are to be understood.
The lack of overall rate heterogeneity within the Colombia/Central America clade makes it possible to obtain broad estimates of their divergence time. A study on related Crocodylus species provided convincing inter-and intraspecific calibrations of a molecular clock (Oaks 2011). We calibrated cyt b sequences [corresponding to the fragment used in Oaks (2011)] by calculating the sequence divergence between C. acutus and Crocodylus intermedius clades reported in Oaks (2011). This suggests that the main cladogenesis event in the Central America/Colombia clade occurred about 0.56–0.62 myr (extreme estimates: 0.56–0.87 myr).
Implications for conservation and management
In many analyses of mtDNA within species, molecular phylogenies can be used to identify phylogenetic and population structure below the species level worthy of separate conservation management (Avise 2005). In this study, results support the presence of at least two mtDNA lineages within C. acutus (C. acutus-I and C. acutus-II) which could form the basis for the designation of separate ESUs. The confirmation of similar differentiation between mtDNA lineages in nuclear markers would provide strong support for ESU status (Ryder 1986; Moritz 1994a,b).
There is no theoretical or empirical standard for setting levels of sequence divergence beyond which phylogenetic units should be recognized as distinct ESUs, although comparisons between levels of divergence within and among related species may provide an empirical guide. A study on the congeneric species C. rombifer would seem to support the idea of separate ESU status for the two mtDNA lineages detected in C. acutus. This species contains two mtDNA lineages [C. rhombifer-α and C. rhombifer-β; sensu Weaver et al. (2008)] with sequence divergence of 0.9% based on cyt b. Microsatellite DNA analyses confirmed differentiation between C. rhombifer-α and C. rhombifer-β, providing strong support for ESU status (Weaver et al. 2008; Milián-García et al. 2011). Similar levels of mtDNA divergence for the same gene (albeit for a longer sequence fragment) between C. acutus-I and C. acutus-II (0.8% cyt b) would seem to raise the possibility of similar differentiation at microsatellite DNA and support the use of ESU status for C. acutus-I and C. acutus-II. However, any consideration of possible ESU status for the two mtDNA lineages within C. acutus would also need to include information on the geographical distributions of the two lineages.
An important finding of this study is the presence of two mtDNA lineages within the Colombian captive populations of C. acutus analysed. This pattern most likely results from historical stocking practices (which may have involved mixtures of individuals from separate geographical localities within Colombia). Although the use of mixed stocks for reintroductions and/or population augmentation is typically discouraged by conservation managers due to concerns of outbreeding depression (Templeton et al. 1986; Edmands 2007), it does have the potential advantage of reversing the adverse effects of inbreeding depression by increasing genetic diversity of inbred populations (Moritz 1999; Tallmon et al. 2004; Edmands 2007; Frankham et al. 2011). The decision to use mixed stocks in conservation strategies should be based on a balance between the concerns of inbreeding depression and outbreeding depression. If inbreeding depression is not of immediate concern, it would seem prudent to avoid the use of genetically mixed stocks for conservation purposes and maintain the genetic integrity of distinct phylogenetic units within the species.
The effect of crossing distinct mtDNA lineages on fitness has never been addressed in crocodiles. However, interspecific hybridization has been reported between numerous Crocodylus species (Fitzsimmons et al. 2002; Cedeño-Vázquez et al. 2008; Rodriguez et al. 2011; Tabora et al. 2012), apparently without negative consequences to hybrid individuals. Although the potential for reduced fitness due to outbreeding depression cannot be excluded completely, the apparent lack of adverse consequences to interspecific hybrids suggests that concerns of outbreeding depression within C. acutus should be minimal. Independent of the possible concerns of outbreeding depression related to the crossing of C. acutus mtDNA lineages, we suggest that this differentiation needs to be taken into account for conservation purposes because it clearly contributes to the overall genetic diversity of the species. Given historical stocking and management practices, the mixed stock structure detected in this study is likely to be a common feature of the commercial captive breeding population in Colombia. As such, it would seem prudent to avoid the use of captive-bred individuals in reintroduction or augmentation programs if conservation planning for C. acutus is to protect distinct evolutionary units, or at least until further and more conclusive genetic information is available.
Although preliminary, results of the current study are important to ongoing conservation and genetic management programs for the species, both locally and throughout the species distribution. This study provides an important step in the description of genetic diversity relevant to conservation efforts and genetic management of this species in Colombia.
Acknowledgments
The funding sources had no involvement in study design, interpretation of data or in the decision to submit the article for publication. Many people contributed to the collection of samples. These include Adriana Restrepo, Gonzalo Jiménez Alonso, Sandra Hernández Rangel, Lina Pedraza, Jorge Augusto Osorio Kuan, and Viviana Hernández Montealegre.
Conflict of Interest
None declared.
Supporting Information
Appendix S1. DNA extraction and primer design.
Appendix S2. Additional sequences obtained from GenBank (Milián-García et al. 2011) for 17 Crocodylus acutus from Central America (Panama and Costa Rica) and Cuba, 10 Crocodylus rombifer from Cuba and two individuals identified as hybrids (C. acutus/C. rombifer) from Cuba.
References
- Avise JC. Phylogeography: the history and formation of species. Cambridge, Massachusetts: Harvard Univ. Press; 2000. [Google Scholar]
- Avise JC. Phylogenetic units and currencies above and below the species level. In: Purvis A, Gittleman JL, Brooks T, editors. Phylogeny and conservation, vol 8. New York: Published in the United States of America by Cambridge University Press; 2005. pp. 76–100. , eds... Conservation Biology Series. [Google Scholar]
- Beauclerc KB, Johnson B. White BN. Genetic rescue of an inbred captive population of the critically endangered Puerto Rican crested toad (Peltophryne lemur) by mixing lineages. Conserv. Genet. 2010;11:21–32. [Google Scholar]
- Benavides E, Russello M, Boyer D, Wiese RJ, Kajdacsi B, Marquez L, et al. Lineage identification and genealogical relationships among captive Galápagos tortoises. Zoo. Biol. 2012;31:107–120. doi: 10.1002/zoo.20397. [DOI] [PubMed] [Google Scholar]
- Burns CE, Ciofi C, Beheregaray LB, Fritts TH, Gibbs JP, Márquez C, et al. The origin of captive Galápagos tortoises based on DNA analysis: implications for the management of natural populations. Anim. Conserv. 2003;6:329–337. [Google Scholar]
- Castaño-Mora OV. Libro rojo de reptiles de Colombia. Libros rojos de especies amenazadas de Colombia. Colombia: Instituto de Ciencias Naturales-Universidad Nacional de Colombia, Ministerio del Medio Ambiente, Conservación Internacional-Colombia Bogotá; 2002. [Google Scholar]
- Cedeño-Vázquez JR. Rodriguez D. Calmé S. Ross JP. Thorbjarnarson LD., III Densmore JB. Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: I. Evidence from mitochondrial DNA and morphology. J. Exp. Zool. A Ecol. Genet. Physiol. 2008;309A:661–673. doi: 10.1002/jez.473. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D. Crandall KA. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Crandall KA, Bininda-Emonds ORP, Mace GM. Wayne RK. Considering evolutionary processes in conservation biology. Trends Ecol. Evol. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R. Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons NN, Buchan JC, Lam PV, Polet G, Hung TT, Thang NQ, et al. Identification of purebred Crocodylus siamensis for reintroduction in Vietnam. J. Exp. Zool. B Mol. Dev. Evol. 2002;294:373–381. doi: 10.1002/jez.10201. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Eldridge MDB, Lacy RC, Ralls K, Dudash MR, et al. Predicting the probability of outbreeding depression. Conserv. Biol. 2011;25:465–475. doi: 10.1111/j.1523-1739.2011.01662.x. [DOI] [PubMed] [Google Scholar]
- Fraser DJ. Bernatchez L. Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol. Ecol. 2001;10:2741–2752. [PubMed] [Google Scholar]
- Gaur A, Reddy A, Annapoorni S, Satyarebala B. Shivaji S. The origin of Indian Star tortoises (Geochelone elegans) based on nuclear and mitochondrial DNA analysis: a story of rescue and repatriation. Conserv. Genet. 2006;7:231–240. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Lewis PO, Holder MT. Holsinger KE. Polytomies and Bayesian phylogenetic inference. Syst. Biol. 2005;54:241–253. doi: 10.1080/10635150590924208. [DOI] [PubMed] [Google Scholar]
- McGreevy TJ, Dabek L. Husband TP. Genetic evaluation of the Association of Zoos and Aquariums Matschie's tree Kangaroo (Dendrolagus matschiei) captive breeding program. Zoo. Biol. 2011;30:636–646. doi: 10.1002/zoo.20362. [DOI] [PubMed] [Google Scholar]
- Medem F. Los crocodylia de Sur America. I. Colciencias, Bogotá: Los crocodylia de Colombia; 1981. Vol. [Google Scholar]
- Meganathan PR, Dubey B, Batzer MA, Ray DA. Hague I. Complete mitochondrial genome sequences of three Crocodylus species and their comparison within the Order Crocodylia. Gene. 2011;478:35–41. doi: 10.1016/j.gene.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Meraner A, Cornetti L. Gandolfi A. Defining conservation units in a stocking-induced genetic melting pot: unraveling native and multiple exotic genetic imprints of recent and historical secondary contact in Adriatic grayling. Ecol. Evol. 2014;4:1313–1327. doi: 10.1002/ece3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milián-García Y, Venegas-Anaya M, Frias-Soler R, Crawford AJ, Ramos-Targarona R, Rodríguez-Soberon R, et al. Evolutionary history of Cuban crocodiles Crocodylus rhombifer and Crocodylus acutus inferred from multilocus markers. J. Exp. Zool. A Ecol. Genet. Physiol. 2011;315A:358–375. doi: 10.1002/jez.683. [DOI] [PubMed] [Google Scholar]
- Moritz C. Applications of mitochondrial DNA analysis in conservation: a critical review. Mol. Ecol. 1994a;3:401–411. [Google Scholar]
- Moritz C. Defining ‘Evolutionarily Significant Units' for conservation. Trends Ecol. Evol. 1994b;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- Moritz C. Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas. 1999;130:217–228. [Google Scholar]
- Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst. Biol. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- Oaks JR. A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution. 2011;65:3285–3297. doi: 10.1111/j.1558-5646.2011.01373.x. [DOI] [PubMed] [Google Scholar]
- Porras Murillo LP, Bolanos Montero JR. Robert Barr B. Genetic variation and gene flow among populations of Crocodylus acutus (Crocodylia: Crocodylidae) in three rivers of Central Pacific, Costa Rica. Rev. Biol. Trop. 2008;56:1471–1480. [PubMed] [Google Scholar]
- Rambaut A. Drummond AJ. Tracer v1.4. 2007. , and. Available via http://beast.bio.ed.ac.uk/Tracer. [Google Scholar]
- Ramirez O, Altet L, Ensenat C, Vila C, Sanchez A. Ruiz A. Genetic assessment of the Iberian wolf Canis lupus signatus captive breeding program. Conserv. Genet. 2006;7:861–878. [Google Scholar]
- Ray DA, Dever JA, Platt SG, Rainwater TR, Finger AG, McMurry ST, et al. Low levels of nucleotide diversity in Crocodylus moreletii and evidence of hybridization with C. acutus. Conserv. Genet. 2004;5:449–462. [Google Scholar]
- Rodriguez D. Cedeño-Vázquez JR. Forstner MRJ. Densmore LD., III Hybridization between Crocodylus acutus and Crocodylus moreletii in the Yucatan Peninsula: II. Evidence from microsatellites. J. Exp. Zool. A Ecol. Genet. Physiol. 2008;309A:674–686. doi: 10.1002/jez.499. [DOI] [PubMed] [Google Scholar]
- Rodriguez D, Forstner MRJ, Moler PE, Wasilewski JA, Cherkiss MS. Densmore L. Effect of human-mediated migration and hybridization on the recovery of the American crocodile in Florida (USA) Conserv. Genet. 2011;12:449–459. [Google Scholar]
- Roldán VA, Navarro JL, Gardenal CN. Martella MB. May captive populations of Greater Rhea (Rhea americana) act as genetic reservoirs in Argentina? Zoo. Biol. 2011;30:65–70. doi: 10.1002/zoo.20314. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S. Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz S, editors; Bioinformatics Methods and Protocols, vol 132. Totowa, New Jersey: Humana Press Inc; 2000. pp. 365–386. eds., and.. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- Ruokonen M, Kvist L, Tegelström H. Lumme J. Goose hybrids, captive breeding and restocking of the Fennoscandian populations of the Lesser White-fronted goose (Anser erythropus. Conserv. Genet. 2000;1:277–283. [Google Scholar]
- Russello MA, Hyseni C, Gibbs JP, Cruz S, Marquez C, Tapia W, et al. Lineage identification of Galápagos tortoises in captivity worldwide. Anim. Conserv. 2007;10:304–311. [Google Scholar]
- Ryder OA. Species conservation and systematics: the dilemma of subspecies. Trends Ecol. Evol. 1986;1:9–10. [Google Scholar]
- Simmons MP. Miya M. Efficiently resolving the basal clades of a phylogenetic tree using Bayesian and parsimony approaches: a case study using mitogenomic data from 100 higher teleost fishes. Mol. Phylogenet. Evol. 2004;31:351–362. doi: 10.1016/j.ympev.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Storfer A. Gene flow and endangered species translocations: a topic revisited. Biol. Conserv. 1999;87:173–180. [Google Scholar]
- Swofford DL. PAUP*, Phylogenetic analysis using parsimony (*and other methods), Version 4. Sunderland, Massachusetts: Sinauer Associates; 1998. [Google Scholar]
- Tabora JAG, Hinlo MRP, Bailey CA, Lei RH, Pomares CC, Rebong G, et al. Detection of Crocodylus mindorensis x Crocodylus porosus (Crocodylidae) hybrids in a Philippine crocodile systematics analysis. Zootaxa. 2012;3560:1–31. [Google Scholar]
- Tallmon DA, Luikart G. Waples RS. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 2004;19:489–496. doi: 10.1016/j.tree.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M. Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR, Hemmer H, Mace G, Seal US, Shields WM. Woodruff DS. Local adaptation, coadaptation, and population boundaries. Zoo. Biol. 1986;5:115–125. [Google Scholar]
- Templeton AR, Crandall KA. Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG. Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorbjarnarson JB. Crocodiles: status survey and conservation action plan. 3rd edn. Darwin: Crocodile Specialist Group; 2010. American crocodile Crocodylus acutus; pp. 46–53. [Google Scholar]
- Thorbjarnarson J, Mazzotti F, Sanderson E, Buitrago F, Lazcano M, Minkowski K, et al. Regional habitat conservation priorities for the American crocodile. Biol. Conserv. 2006;128:25–36. [Google Scholar]
- Vogler AP. DeSalle R. Diagnosing units of conservation management. Conserv. Biol. 1994;8:354–363. [Google Scholar]
- Weaver JP, Rodriguez D, Venegas-Anaya M, Cedeño-Vázquez JR, Forstner MRJ. Densmore LD., III Genetic characterization of captive Cuban crocodiles (Crocodylus rhombifer) and evidence of hybridization with the American crocodile (Crocodylus acutus. J. Exp. Zool. A Ecol. Genet. Physiol. 2008;309A:649–660. doi: 10.1002/jez.471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. DNA extraction and primer design.
Appendix S2. Additional sequences obtained from GenBank (Milián-García et al. 2011) for 17 Crocodylus acutus from Central America (Panama and Costa Rica) and Cuba, 10 Crocodylus rombifer from Cuba and two individuals identified as hybrids (C. acutus/C. rombifer) from Cuba.