Abstract
Capture–mark–recapture (CMR) approaches are the backbone of many studies in population ecology to gain insight on the life cycle, migration, habitat use, and demography of target species. The reliable and repeatable recognition of an individual throughout its lifetime is the basic requirement of a CMR study. Although invasive techniques are available to mark individuals permanently, noninvasive methods for individual recognition mainly rest on photographic identification of external body markings, which are unique at the individual level. The re-identification of an individual based on comparing shape patterns of photographs by eye is commonly used. Automated processes for photographic re-identification have been recently established, but their performance in large datasets (i.e., > 1000 individuals) has rarely been tested thoroughly. Here, we evaluated the performance of the program AMPHIDENT, an automatic algorithm to identify individuals on the basis of ventral spot patterns in the great crested newt (Triturus cristatus) versus the genotypic fingerprint of individuals based on highly polymorphic microsatellite loci using GENECAP. Between 2008 and 2010, we captured, sampled and photographed adult newts and calculated for 1648 samples/photographs recapture rates for both approaches. Recapture rates differed slightly with 8.34% for GENECAP and 9.83% for AMPHIDENT. With an estimated rate of 2% false rejections (FRR) and 0.00% false acceptances (FAR), AMPHIDENT proved to be a highly reliable algorithm for CMR studies of large datasets. We conclude that the application of automatic recognition software of individual photographs can be a rather powerful and reliable tool in noninvasive CMR studies for a large number of individuals. Because the cross-correlation of standardized shape patterns is generally applicable to any pattern that provides enough information, this algorithm is capable of becoming a single application with broad use in CMR studies for many species.
Keywords: GENECAP, noninvasive individual recognition, shape patterns, single-use application, standardized cross-correlation, Wild-ID
Introduction
A species' population ecology is composed of many facets such as life cycle, movement patterns, habitat use, and population size among other important traits and parameters. Capture–mark–recapture (CMR) studies of individuals provide a simple but powerful approach to construct a database to analyze and make inferences on these traits and parameters in natural populations (e.g., Lebreton et al. 1992; Halliday 1995; Phillott et al. 2007) and can therefore be seen as a type of backbone tool in population ecology studies. The basic idea of CMR approaches is the reliable recognition and identification of a specific individual from conspecifics in a population over certain periods or throughout the entire life span. Individuals can therefore exhibit markings that make them unique; markings can either already exist (e.g., individual spot and shape patterns) or must be introduced by invasive methods. Such invasive methods can include toe-clipping (e.g., in amphibians: Clarke 1972; Kenyon et al. 2009; Phillott et al. 2007; Waichman 1992), inert fluorescent polymer (elastomer) subcutaneous marking (Anholt et al. 1998; Schlaepfer 1998), and the use of passive integrated transponder (PIT) tagging (Brown 1997; Gibbons and Andrews 2004). These methods differ in their impact on the individual: The marking process can be disruptive, the markings themselves can influence the behavior (e.g., mating and foraging; see Winandy and Denoel 2011), or the individual survival rate can be affected (Powell and Proulx 2003; McCarthy and Parris 2004). Moreover, the loss of markings over time and the nonreporting of retrieved markings can be a problem and bias results (Lukacs and Burnham 2005; Speed et al. 2007).
Noninvasive or minimally invasive methods do not require that marks must be artificially introduced. The most common noninvasive method is photographic CMR, which uses natural marks of an individual such as color patterns or body spots. This method has become more and more popular to identify individuals because the natural marks are stable and robust over time without substantial changes (Hagstrom 1973; Doody 1995; Bradfield 2004; Bolger et al. 2012). Photographs of the individually unique natural marks are used to create a library for subsequent cross-matchings (Speed et al. 2007). Libraries can be checked manually, that is, by eye, or with the assistance of automatic photo-identification software (e.g., Arzoumanian et al. 2005; Gamble et al. 2008; Kenyon et al. 2009), which can potentially overcome the problems of manual approaches, especially for large datasets (Katonas and Beard 1990; Gamble et al. 2008; Bolger et al. 2012).
With the rise of molecular approaches in population biology, highly variable molecular markers can also be used to distinguish individuals from each other in a population. Genetic sampling for CMR studies has been successfully applied to various species (Woods et al. 1999; Pearse et al. 2001; Eggert et al. 2003), and highly polymorphic molecular markers, such as microsatellite loci or single nucleotide polymorphisms (SNPs), are used to identify individuals by DNA fingerprinting (Lukacs and Burnham 2005). The required DNA samples can be collected using noninvasive methods from hair (Woods et al. 1999), feathers, feces (Puechmaille and Petit 2007), and skin pieces or can be collected less invasively with swabs of the buccal mucosa or by tail-tip clipping (Arntzen et al. 1999). Programs such as GENECAP (Wilberg and Dreher 2004), API-CALC (Ayres and Overall 2004) and GIMLET (Valière 2002) are used to match the identical genotypes of identified individuals across large datasets. As with the methods described above, genetic CMR also has drawbacks, such as comparably high costs and matching errors prone to PCR and genotyping errors.
Photographic approaches are a promising method to obtain reliable data in large CMR studies; if consistent and time-effective, re-identification of individuals can be guaranteed, as photographs are minimally invasive and may provide broad coverage of individuals in a population. Such approaches will be especially suited for the monitoring not only of endangered species but also of nonendangered species. As it is obvious that manual approaches soon reach their limit in evaluating large photographic CMR datasets, automatic approaches provide a promising alternative to handle such datasets. Although some of these approaches currently exist, there has been no attempt thus far to test their performance in a large dataset using a completely independent, reliable approach. We therefore chose to test the performance of an automatic photo-identification software, called AMPHIDENT (Matthé et al. 2008), based on individual ventral spot patterns with a genetic fingerprinting approach in a large dataset of more than 1600 great crested newts (Triturus cristatus). On the one hand, the great crested newt is an excellent model species for such a comparison because adults display in general an individual, stable belly pattern; on the other hand, a large set of applicable and highly polymorphic microsatellite loci is available (Drechsler et al. 2013) to allow for individual genotype fingerprinting. We tested whether the AMPHIDENT approach would miss or wrongly assign recaptured individuals who have been detected by the genetic fingerprint approach. We found that recapture rates differed between both approaches and that the cross-correlation of standardized images implemented in AMPHIDENT is a time efficient and highly accurate method for the re-identification of individuals, thus making it a potentially powerful tool that is applicable to a broad range of species.
Materials and Methods
Study species, study site, and CMR study design
The northern or great crested newt Triturus cristatus (Salamandridae) occupies a large geographic distribution area in central and northern Europe and in the western parts of Asia. Triturus cristatus lives in fairly high population densities and can be easily captured during its reproductive period in ponds from March until July (Jehle et al. 2011). From a conservation perspective, the crested newt is a highly protected vertebrate species of the European Union and is listed in Annex II and Annex IV of the Habitats Directive of the European Union (92/43/EEC). Species of Annex II should be protected in species-specific areas of conservation (Natura 2000), and the conservation status of species of Annex II and Annex IV should be kept under surveillance by subsequent and constant monitoring of populations (Maletzky et al. 2008).
For this study, samples of T. cristatus were collected from a population in the Latumer Bruch in Krefeld (North-Rhine Westphalia, Germany, 6°39′17″E, 51°19′5″N) during the reproductive season from March to June in 2008, 2009, and 2010, roughly covering an area of 9 km2 (see Fig.1). We were able to detect crested newts in 20 of the 27 regularly monitored water bodies, including ponds, small lakes, and ditches. We used Ortmann's funnel traps (see Drechsler et al. 2010 for details of design and catchability) to catch the newts. Depending on the size of the monitored water body, 1–34 funnel traps were placed in the water and remained there for 48 h. For each water body, 10 consecutive sampling events were conducted during a first period in early spring, with 10 additional consecutive sampling events during a second period in early summer. All newts that were caught in traps were photographed, and a tissue sample was taken for genetic fingerprint analysis. To standardize image acquisition and optimize image quality, individual crested newts were placed in a transparent box and secured with a light sponge at their dorsal side such that the ventral pattern could be photographed without distortion (see Fig.2A). After sampling, the newts were immediately released in their original aquatic habitat.
Figure 1.

Overview of the study area and study sites of crested newts (Triturus cristatus) near Krefeld in Germany.
Figure 2.

Workflow of pattern extraction with AMPHIDENT. (A) The original, nonprocessed picture. (B) The pattern with superposed grid and the preview to pattern selection in true and false color. (C) The extracted pattern. (D) The extracted pattern and the proposed matches (1–4) in order of likelihood supplemented by the original pictures of the extracted pattern and the selected one.
Automatic photograph-based CMR analysis
Pictures of the ventral side of 1648 trapped newts were entered into the automatic photo-identification software AMPHIDENT. AMPHIDENT is an algorithm based on a modified technique of cross-correlation comparisons to find a specific signal (e.g., a specific spot pattern) in a large sequence of similar signals again. It is based on a two-step approach to recognize individual newts on their belly pattern (see Fig.2). First, it needs to extract the belly pattern of the newt from the original photograph. Second, this pattern is then modified and compared with other belly patterns in the database. To extract the belly pattern from the original photograph, individuals have to be placed in a vertical fashion with the head showing to the top. The belly pattern is then extracted by first marking the area of the belly pattern, that is, defining the region of the belly pattern, by the user. The program then extracts this region from the original photograph into a rectangular, straightened pattern of the newt belly. This picture is then transformed via a gray-scale picture into a black and white picture. It is then further modified by a median filter to remove noise of edges and small spots. A detailed description and illustration of the program workflow is given by Matthé et al. (2008). The obtained pattern is then compared with all existing patterns in the database. For each pairwise comparison, a similarity value is calculated. This value is determined by the number of matching pixels of both patterns after one pattern has been transformed into the other pattern by affine transformation. The higher the number of matching pixels between shapes, that is, the more similar the patterns are, the higher the similarity score. The program offers also the option to partially extract the belly pattern, if some parts of the picture are in poor quality. According to obtained similarity values for a specific pattern (i.e., for an individual), the program offers then the user the best 30 ranked matching patterns in a descending order that could be found in the database. The user needs then to compare by eye whether the pattern is already contained in the database, that is, whether an individual has been recaptured or not (see http://www.amphident.de, Matthé et al. 2008 and Fig.2D for examples).
Biometric performance assessment of AMPHIDENT
To assess the performance of AMPHIDENT, we used metrics that are similar to false-negative and false-positive error and are widely used for biometric assessment performance (Jain 2007). Here, we are using a similar approach as the study of Bolger et al. (2012) in the context of computer-assisted photographic recapture of Masai giraffes. Accordingly, we estimated the rate of false rejections (FRR) and the rate of false acceptances (FAR). FRR describes the probability that two images of the same individual are not considered as matching samples, that is, they are false rejections. FAR, on the other hand, describes the frequency of matching two images of different individuals, that is, being false acceptances. We estimated FRR with a test set of 100 known images matching the underlying database and FAR with 50 belly images from newts, which were not represented in the database. Pictures of both test sets were randomly chosen from the database. As a test database, we used the full image database with 1648 images for the study. Thus, AMPHDIENT had 100 possibilities to make false rejection errors and 50 opportunities to make false acceptances. The images were processed with AMPHDIENT as described above and checked whether each trial image was within the 10 highest ranking images as suggested by AMPHIDENT. We calculated FRR as the number of false rejections divided by the number of true matches (Eq. 1). Accordingly, each picture that AMPHIDENT failed to place within the 10 highest ranking images or that we as the observer failed to recognize within these 10 highest ranking images was defined as a “false” rejection.
| 1 |
The FAR was calculated as the number of false acceptances divided by the number of potentially false acceptances, that is, by 50.
| 2 |
Genetic CMR analysis
To test the performance of AMPHIDENT for a large dataset with a completely independent approach, we used genetic CMR analysis in the course of the genetic characterization of the crested newt population in Krefeld. Accordingly, each newt that was recorded by a photograph was also sampled with permission of the local environmental authorities using a small piece of tissue from the tail fin, which represents a highly regenerative type of tissue during the aquatic phase of newts. Tissue samples, stored in 80% ethanol, were used to extract total genomic DNA using the sodium dodecyl sulfate (SDS)-proteinase K/phenol–chloroform extraction method. Genomic DNA was stored in Tris-EDTA buffer (10 mmol/L Tris-HCl, 0.1 mmol/L EDTA, pH 8.0) and used for all subsequent reactions. A 10 μL Type-it multiplex PCR reaction (Qiagen, Hilden, Germany) containing 1 μL of DNA was performed and 17 microsatellite loci were amplified in three multiplex mixes of primer combinations as described in detail in Drechsler et al. (2013). Applied PCR parameters were as follows: (1) an initial Taq polymerase activation step of 5 min at 95°C, (2) 30 sec at 94°C, (3) 90 sec at an annealing temperature of 60°C, and (4) 60 sec extension at 72°C; steps 2–4 were repeated for 30 cycles, (5) a final extension phase of 30 min at 60°C completed the PCR. Obtained PCR products were diluted with 200 μL of water, and 19 μL of Genescan 500-LIZ size standard (Applied Biosystems, Carlsbad, US) was added to 1 μL of each multiplex reaction before analysis on an ABI 3730 96-capillary automated DNA sequencer. Genotyping of alleles was performed using GENEMARKER software (SoftGenetics version 1.95, State College, PA).
GENECAP 1-4 (http://wilberglab.cbl.umces.edu/downloads.html), a Microsoft EXCEL macro, was used to identify individual samples on the basis of matching microsatellite loci genotypes. As not all individuals could be successfully genotyped for the complete set of 17 loci, we tried to find the optimal number of microsatellite loci that would result in a high exclusion probability and a maximum number of individuals genotyped successfully for this set of loci. Accordingly, to perform this selection, we ran GENECAP with different numbers of loci and chose the loci with the highest resolution, in terms of exclusion probability, and total number of successfully genotyped individuals. The exclusion probability (PE) of different sets of loci was calculated on the basis of the average nonexclusion probability (identity) (P) of each single locus computed by CERVUS (version 3.0.3, Field Genetics Ltd.; see http://www.fieldgenetics.com). PE was calculated for different sets of loci by the multiplication of P for the corresponding single loci and by calculating the complementary probability of the combined value of P (i.e., PE = 1-Pcombined). Nine microsatellite loci matched the desired criteria and were used for the genetic CMR analysis. Using GENECAP, the matching probability, that is, the probability that two individuals sharing the same genotype assuming that both individuals are siblings (Psib) was set to 0.05, and the chosen method was 'sib'. GENECAP showed samples with the same genotype and those that differed in one or two alleles. The detailed program functions are described in Wilberg and Dreher (2004).
Individuals which were identified by GENECAP as recaptures were compared manually, that is, by comparing the corresponding belly patterns of the two samples by eye.
Results
During the reproductive periods of crested newts from 2008 to 2010, 1648 newts were captured and photographed in 20 of the 27 monitored sampling sites in the study area of the Latumer Bruch in Krefeld. Of these, 1618 genetic samples were obtained for subsequent microsatellite loci analysis.
Photograph-based recapture analysis of AMPHIDENT
AMPHIDENT could not handle 42 of the 1648 (2.55%) pictures due to poor quality. These pictures showed either bright light spots that impaired photo-identification or the pictures were blurred. Altogether, AMPHIDENT identified 162 of the 1648 (9.83%) pictures as recaptures (Fig.3). Of these, 101 individuals were recaptured once, 11 two times, 6 three times, 4 four times, and 1 individual five times. Figure4 shows a crested newt captured five times between April 2008 and May 2009. Despite the belly pattern has been changing significantly between recapture events, AMPHIDENT was still able to recognize it as the same individual; identification of this individual was also confirmed manually by eye as well as by GENECAP. Even in two cases where individuals were captured first as juveniles, AMPHIDENT was able to recognize them later as adults. Only in two cases was AMPHIDENT not able to detect a recaptured individual, which was detected by GENECAP.
Figure 3.
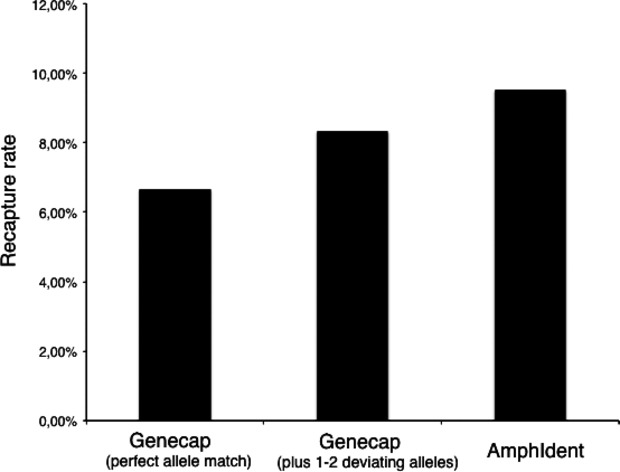
Recapture rates of the genetic fingerprinting method GENECAP (with perfect allele match and with two deviating alleles) and of AMHPIDENT.
Figure 4.

The same adult crested newt and its extracted belly pattern at five distinct capture events (from April 2008 to May 2009). Although ventral spot pattern obviously changes, AMPHIDENT was able to assign the photographs to a single individual.
The FRR rate of AMPHIDENT was rather low, as only two images were not ranked under the 10 highest ranking images (FRR = 2/100 = 2%), whereas the remaining 98 images were ranked in first place. In addition, no test images were wrongly assigned to an individual in the database (FAR = 0/50 = 0.00%).
Genetic recapture analysis
The optimal balance between total number of successfully genotyped individuals and maximum exclusion probability was found for 9 of the 17 available microsatellite loci when using GENECAP. When more than nine microsatellite loci were used, the number of correctly recaptured individuals did not increase, and when less than nine microsatellite loci were applied, the number of incorrectly recognized animals increased. Accordingly, we used the following nine microsatellite loci from Drechsler et al. (2013) and Krupa et al. (2002): Loc13, Loc27, Loc36, Loc35, Loc29, Loc46, Tc68b, Tc50, and Tc74. The calculated exclusion probability for this set of loci based on allele frequencies observed across the entire population in Krefeld was estimated as 99.99%.
GENECAP analysis identified 108 of the 1618 crested newts as recaptured individuals who perfectly matched in all analyzed alleles; an additional 20 matches could be identified that differed in one allele and additional 48 matches that differed in two alleles. All 108 perfectly matching individuals could also be verified manually by comparison of photographs made of these individuals at different capture events. For the 20 individuals differing in one allele, nine matches and 18 matches of the 48 individuals differing in two alleles, respectively, could be verified manually as recaptures. Altogether, we were able to identify 108 individuals (6.67%) unambiguously as recaptures without further manual photographic comparison and 135 individuals (8.34%) with additional manual photographic comparison. According to GENECAP, 89 of the 135 recaptured individuals were recaptured once, 11 twice, 4 three times, and 3 four times.
Comparison of genetic and automatic photograph identification CMR approaches
A total of 206 (12.50%) individuals of the 1648 crested newts could be identified as recaptures by the combined use of GENECAP and AMPHIDENT. Of these, 91 individuals were identified by both methods as recaptures. GENECAP detected 44 caught individuals as recaptures that were not detected by AMPHIDENT, as the picture quality was too poor (42 pictures), or because AMPHIDENT could not handle them (2 pictures, see chapter Photograph-based recapture analysis). By contrast, AMPHIDENT recognized 71 individuals as recaptures that were not detected by GENECAP. PCR and allele calling errors with more than 2 nonreplicated alleles per individual led to missing matches in GENECAP. As a result, AMPHIDENT was able to identify 162 of the 206 total recaptures (78.64%). With a recapture rate of 9.83%, the matching success of AMPHIDENT was higher than the recapture rate of GENECAP with perfect allele matching (6.67%) or with the additional help of manual comparison for individuals with 1 or 2 deviating alleles (8.34%; see Fig.3).
Discussion
Automatic recognition software approaches based on pattern recognition hold the promise to become useful for monitoring efforts and CMR studies for a wide range of organisms that differ individually in observable pattern traits and for which a noninvasive marking method is desired or even requested (e.g., for threatened species). Although various promising automatic approaches have been developed, there have been few attempts to rigorously test their performance across large datasets. However, performance tests in large datasets are crucial if automatic approaches aim to be competitive and challenge the predominant manual approaches still used in many CMR studies. This study evaluated the performance of an automatic photo-identification approach (AMPHIDENT) based on a cross-correlation algorithm of standardized images in the context of a large CMR study in the great crested newt. We were able to test this fully noninvasive automatic pattern recognition approach applying a completely independent approach, that is, genetic fingerprinting of individuals using highly polymorphic microsatellite loci. Our results clearly demonstrate that the automatic recognition algorithm of AMPHIDENT is able to reliably identify individuals as recaptures within large datasets (>1600 individuals) with a high accuracy and even outperforms the applied genetic re-identification of individuals. In the following sections, we will discuss the performance of this algorithm in comparison with existing approaches, as well as its potential to become a single application platform for a broad range of species. As no perfect approach to estimate the real number of recaptures exist, we set the total number of recaptures to 206 as informed by combined results of AMPHIDENT and GENECAP, assuming that this conservative estimate is very close to the real number of recaptured individuals.
Performance of AMPHIDENT in large datasets
In recent years, different automatic pattern recognition algorithms have been developed (e.g., Speed et al. 2007; Van Tienhoven et al. 2007; Sacchi et al. 2010), but only few approaches have been tested in large datasets (e.g., Gamble et al. 2008; Bolger et al. 2012). The program AMPHIDENT has been developed on the basis of a cross-correlation comparison technique to identify individuals based on unique coloration patterns in crested newts and fire-belly toads (genus Bombina). Although this program was initially described in 2008, its performance has not been rigorously tested in large datasets until now. Therefore, we applied this automatic recognition algorithm to identify recapture rates of individual crested newts across 27 different potential breeding sites on the basis of their unique belly patterns. To our knowledge, only the study of Bendik et al. (2013) has tested the performance of an automatic photo-identification approach using a completely independent method, that is, colored visible implant elastomers, whereas other studies (e.g., Gamble et al. 2008; Bolger et al. 2012) did not independently test the performance of their algorithms.
Genetic fingerprinting was performed based on the variation of 17 mainly tetranucleotide microsatellite loci, which have been developed and tested extensively for an excellent performance in our target species and underlying study population in Krefeld (see Drechsler et al. 2013 for details). If allele frequencies are known in the natural population, which is the case in our study, highly confident exclusion probabilities can be calculated providing a probability that another individual with the exact genotype would exist in the population. As some of the individuals had missing data for some microsatellite loci, we sought to reduce the number of loci to include as many individuals as possible, while maintaining a high exclusion probability, that is, >0.99. GENECAP is an efficient and time-saving method to identify recaptures out of a large dataset of individuals genotyped for a large number of microsatellite loci. In our study, GENECAP was able to identify 135 recaptures based on a maximum of two deviating alleles—a threshold recommended by Paetkau (2003)—and missed 71 recaptures as informed by combined AMPHIDENT/GENECAP results. Thus, overall error rate due to allele misidentification was 4.3% and is, compared to other studies using a comparable number of polymorphic loci, quite low (see McKelvey and Schwartz 2004). As applied loci have been developed de novo for our study species, errors due to null alleles or allele dropout should be rare or even absent. One common approach to estimate occurrence and rate of genotypic errors is to test for deviations from Hardy–Weinberg equilibrium (see Gomes et al. 1999). As shown in Drechsler et al. (2013), none of the loci used here showed a significant deviation (P ≥ 0.05) from Hardy–Weinberg equilibrium when tested for a large number of individuals in our study population, thus suggesting that typical allele errors should be negligible.
Although no false matches could be observed for the 108 perfectly allele matching recaptured individuals identified by GENECAP, the necessary manual control of matches that differed in one or two alleles revealed a relatively large number of 27 overlooked recaptured individuals using this approach. Thus, an obvious drawback of the genetic fingerprint approach is the additional source of information needed to increase the detection probability of recaptures. In general, the observed recapture results are strongly influenced by the proper selection of loci as well as their adequate number and observed exclusion probability in the target population (Paetkau 2004). Altogether, 36 individuals could not be identified as recaptures by GENECAP due to PCR errors caused by poor DNA quality as well as allele shifting artifacts caused by the allele calling process with GENEMARKER.
We were rather surprised by the high performance quality of the program AMPHIDENT. The false rejection rate (FRR) was 2%, and no false acceptances (FAR) were observed. Of the 206 total recaptures identified by both approaches, AMPHIDENT was able to correctly identify 78.86%. In comparison, Bendik et al. (2013) found a FRR of 0.76% for Jollyville Plateau salamanders (Eurycea tonkawae) using high-quality images and 15.9% when using lower quality images, respectively; the FFR of the visible implant elastomer approach used in the same study was estimated at 1.90%. In addition, compared with other computer-assisted photographic identification approaches, error rates of AMPHIDENT belong to the best values found so far (see Bolger et al. 2012), thus stating the high accuracy of the program in large datasets.
The obtained false rejection rate of 2% in our study for AMPHIDENT is in line with an error rate estimated manually (i.e., by eye comparison) based on 319 photographs taken from sites with a known high recapture rate of crested newts (Matthé et al. 2008). Here, 105 pictures could be assigned manually to an existing pattern, whereas AMPHIDENT identified 103 recaptures, resulting in an error rate of less than 2%. The high performance of AMPHIDENT may be due to several reasons: (1) The shape pattern is transferred to a rectangular figure, so that the original body shape and potential body bending do not impact further analysis; (2) a median filter eliminates intervening image noise; (3) AMPHIDENT can handle changes in patterns of an individual over time (see Fig.4), even from juvenile to adult stages, whereas photograph matching scores of an individual were negatively correlated with time in the approach of Bendik et al. (2013); and (4) unusual areas of a shape pattern that makes the individual special and are less frequently found in other individuals can be marked as a striking character for identification.
Normally, we quickly adjusted to working with AMPHIDENT after a short period of training, and the overall handling time of an image from the pattern extraction to the final database comparison took on average only 90 sec and remained constant even when the underlying database increased.
AMPHIDENT – an overlooked approach with high potential for other species
Until now, most of the computer-based individual pattern recognition approaches have been developed for a single or a group of target species and have limited use for accurate identification in other species. However, the program module application WILD-ID, based on the scale-invariant feature transform (SIFT) algorithm (Lowe 2004), has been developed by Bolger et al. (2012) as a pattern matching software that has the potential for application to other species. This approach has been successfully extended and applied to work for re-identification of salamander individuals in a large population (Bendik et al. 2013) and therefore demonstrates its potential to be broadly applicable. AMPHIDENT has thus far been developed for shape pattern discrimination in a limited number of amphibian species. Even in Germany, where the program was published in a national journal several years ago, the approach has been vastly overlooked and only rarely applied. Our results suggest that the underlying algorithm of AMPHIDENT provides an outstanding recognition precision for individuals and performs at least as accurately as the WILD-ID software in large datasets. Because the underlying cross-correlation of standardized pictures should be easily transferable to other species with highly variable and discriminative patterns, this approach has the potential to also be applicable for a wide range of taxa (see Fig.5 for possible examples). At the moment, AMPHIDENT is operating as a single species application program, where the different modules have been developed for distinct species. A reasonable future development of the existing approach would be therefore a single application program for a broad range of species. Here, the user could simply enter the shape pattern of the target species under investigation, and after a short training period, the program should be able to discriminate individuals on the basis of such a species-specific pattern.
Figure 5.
Possible other candidate species providing an individual recognition pattern as shown for (I) dorsal side of sand lizards (Lacerta agilis); (II) ventral chest pattern of Galápagos marine iguanas (Amblyrhynchus cristatus); (III) dorsal skull plate of the adder (Vipera berus); (IV) dorsal side of the Near East fire salamander (Salamandra infraimmaculata); (V) ventral side of the Pyrenean mountain brook newt (Calotriton asper).
Conclusion
Currently, successfully working algorithms such as WILD-ID and AMPHIDENT have the potential to become software applications with broad species applicability. Therefore, it seems necessary to shift the focus away from developing new single species applications toward the improvement of existing high performance algorithms into one broadly applicable software tool. The first crucial and easy step to do would be the systematic implementation and adoption of existing promising species-specific CMR datasets into one of the existing algorithms. Such an implementation and extension would be a powerful tool making many monitoring and CMR studies feasible on a larger scale and would be highly appreciated by many conservation biologists and researchers involved in monitoring efforts.
Acknowledgments
We thank Andrea Funke (Untere Landschaftsbehörde Krefeld, Germany) for providing collection permits and Barbara Caspers (University of Bielefeld), Björn Stockhausen, and Benedikt Schmidt (Zürich) for useful comments on the manuscript. Eliane Küpfer and Dirk Schmeller provided the images of Salamandra infraimmaculata and Calotriton asper, respectively. This research has been funded by the Deutsche Bundesstiftung Umwelt (DBU) through a Ph.D. fellowship to AD and by a research grant of the German Science Foundation (DFG) to Sebastian Steinfartz (STE 1130/7-1).
Conflict of Interest
None declared.
References
- Anholt BR, Negovetic S. Som C. Methods for anaesthetizing and marking larval anurans. Herpetol. Rev. 1998;29:153–154. [Google Scholar]
- Arntzen JW, Oldham RS. Smithson A. Marking and tissue sampling effects on growth and survival in the newt Triturus cristatus. J. Herpetol. 1999;33:567–576. [Google Scholar]
- Arzoumanian Z, Holmberg J. Norman B. An astronomical pattern-matching algorithm for computer-aided identification of whale sharks Rhincodon typus. J. Appl. Ecol. 2005;42:999–1011. [Google Scholar]
- Ayres KL. Overall ADJ. API-CALC 1.0: a computer program for calculating the average probability of identity allowing for substructure, inbreeding and the presence of close relatives. Mol. Ecol. Notes. 2004;4:315–318. [Google Scholar]
- Bendik NF, Morrison TA, Gluesenkamp AG, Sanders MS. O'Donnell LJ. Computer-assisted photo identification outperforms visible implant elastomers in an endangered salamander, Eurycea tonkawae. PLoS One. 2013;8:e59424. doi: 10.1371/journal.pone.0059424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger DT, Morrison TA, Vance B, Lee D. Farid H. A computer-assisted system for photographic mark–recapture analysis. Methods Ecol. Evol. 2012;3:813–822. [Google Scholar]
- Bradfield KS. Photographic identification of individual Archey's Frogs, Leiopelma archeyi, from natural markings. DOC Sci. Int. Series. 2004;191:36. , Department of Conservation, Wellington. [Google Scholar]
- Brown LJ. An evaluation of some marking and trapping techniques currently used in the study of anuran population dynamics. J. Herpetol. 1997;31:410–419. [Google Scholar]
- Clarke RD. The effect of toe clipping on survival in Fowler's Toad (Bufo woodhousei fowleri. Copeia. 1972;1972:182–185. [Google Scholar]
- Doody JS. A photographic mark-recapture method for patterned amphibians. Herpetol. Rev. 1995;26:19–21. [Google Scholar]
- Drechsler A, Bock D, Ortmann D. Steinfartz S. Ortmann's funnel trap – a highly efficient tool for monitoring amphibian species. Herpetol. Notes. 2010;3:13–21. [Google Scholar]
- Drechsler A, Geller D, Freund K, Schmeller DS, Künzel S, Rupp O, et al. What remains from a 454 run: estimation of success rates of microsatellite loci development in selected newt species (Calotriton asper, Lissotriton helveticus, and Triturus cristatus) and comparison with Illumina based approaches. Mol. Ecol. Resour. 2013;3:3947–3957. doi: 10.1002/ece3.764. doi: 10.1002/ece3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert LS, Eggert JA. Woodruff DS. Estimating population sizes for elusive animals: the forest elephants of Kakum National Park, Ghana. Mol. Ecol. 2003;12:1389–1402. doi: 10.1046/j.1365-294x.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- Gamble L, Ravela S. McGarigal K. Multi-scale features for identifying individuals in large biological databases: an application of pattern recognition technology to the marbled salamander Ambystoma opacum. J. Appl. Ecol. 2008;45:170–180. [Google Scholar]
- Gibbons JW. Andrews KA. PIT tagging: simple technology at its best. Bioscience. 2004;54:447–454. [Google Scholar]
- Gomes I, Collins A, Lonjou C, Thomas NS, Wilkinson J, Watson M. Morton N. Hardy-Weinberg quality control. Annals of Human Genetics, 1999;63:535–538. doi: 10.1017/S0003480099007824. doi: 10.1046/j.1469-1809.1999.6360535.x. [DOI] [PubMed] [Google Scholar]
- Hagstrom T. Identification of newt specimens (Urodela, Triturus) by recording the belly pattern and a description of photographic equipment for such registrations. Brit. J. Herpetol. 1973;4:321–326. [Google Scholar]
- Halliday T. More on toe-clipping. Froglog. 1995;12:3. [Google Scholar]
- Jain AK. Biometric recognition. Nature. 2007;449:38–40. doi: 10.1038/449038a. [DOI] [PubMed] [Google Scholar]
- Jehle R, Thiesmeier B. Foster J. The crested newt: a dwindling pond dweller. Bielefeld, Germany: Laurenti-Verlag; 2011. [Google Scholar]
- Katonas K. Beard JA. Population size, migrations and feeding aggregations of the humpback whale (Megaptera novaenangliae) in the Western North Atlantic Ocean. Reports Int. Whaling Comm. 1990;12:295–305. [Google Scholar]
- Kenyon N, Phillott AD. Alford RA. Evaluation of the photographic identification method (PIM) as a tool to identify adult Litoria genimaculata (Anura: Hylidae) Herpetol. Cons. Biol. 2009;4:403–410. [Google Scholar]
- Krupa AP, Jehle R, Dawson DA, Gentle LK, Gibbs M, Arntzen JW, et al. Microsatellite loci in the crested newt (Triturus cristatus) and their utility in other newt taxa. Conserv. Genet. 2002;3:85–87. [Google Scholar]
- Lebreton JD, Burnham KP, Clobert J. Anderson DR. Modelling survival and testing biological hypotheses using marked animals: a unified approach with case studies. Ecol. Monogr. 1992;62:67–118. [Google Scholar]
- Lowe DG. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vision. 2004;60:91–110. [Google Scholar]
- Lukacs PM. Burnham KP. Review of capture–recapture methods applicable to noninvasive genetic sampling. Mol. Ecol. 2005;14:3909–3919. doi: 10.1111/j.1365-294X.2005.02717.x. [DOI] [PubMed] [Google Scholar]
- Maletzky A, Goldschmid A. Kyek M. Crested newt (Triturus cristatus superspecies) populations in Salzburg, Austria, their distribution, size and conservation status. Herpetozoa. 2008;20:145–163. [Google Scholar]
- Matthé M, Schönbrodt T. Berger G. Computergestützte Bildanalyse von Bauchfleckenmustern des Kammmolchs (Triturus cristatus. Zeitschrift für Feldherpetologie. 2008;15:89–94. [Google Scholar]
- McCarthy MA. Parris KM. Clarifying the effect of toe clipping on frogs with Bayesian statistics. J. Appl. Ecol. 2004;41:780–786. [Google Scholar]
- McKelvey KS. Schwartz MK. Genetic errors associated with population estimation using non-invasive molecular tagging: problems and new solutions. J. Wildl. Manage. 2004;68:439–448. [Google Scholar]
- Paetkau D. An empirical exploration of data quality in DNA-based population inventories. Mol. Ecol. 2003;12:1375–1387. doi: 10.1046/j.1365-294x.2003.01820.x. [DOI] [PubMed] [Google Scholar]
- Paetkau D. The optimal number of markers in genetic capture–mark–recapture studies. J. Wildl. Manage. 2004;68:449–452. [Google Scholar]
- Pearse DE, Eckerman CM, Janzen J. Avise JC. A genetic analogue of ‘mark–recapture’ methods for estimating population size: an approach based on molecular parentage assessments. Mol. Ecol. 2001;10:2711–2718. doi: 10.1046/j.0962-1083.2001.01391.x. [DOI] [PubMed] [Google Scholar]
- Phillott AD, Skerratt LF, McDonald K, Lemckert FL, Hines HB, Clark JM, et al. Toe-clipping as an acceptable method of identifying individual anurans in mark recapture studies. Herpetol. Rev. 2007;38:305–308. [Google Scholar]
- Powell RA. Proulx G. Trapping and marking terrestrial mammals for research: integrating ethics, performance criteria, techniques, and common sense. ILAR J. 2003;44:259–276. doi: 10.1093/ilar.44.4.259. [DOI] [PubMed] [Google Scholar]
- Puechmaille SJ. Petit EJ. Empirical evaluation of non-invasive capture–mark– recapture estimation of population size based on a single sampling session. J. Appl. Ecol. 2007;44:843–852. [Google Scholar]
- Sacchi R, Sacli S, Pelliteri-Rosa D, Pupin D, Gentilli A, Tettamanti S, et al. Photographic identification in reptiles: a matter of scales. Amphibia-Reptilia. 2010;31:489–502. [Google Scholar]
- Schlaepfer MA. Use of a fluorescent marking technique on small terrestrial anurans. Herpetol. Rev. 1998;29:25–26. [Google Scholar]
- Speed CW, Meekan MG. Bradshaw CJ. Spot the match – wildlife photo-identification using information theory. Front. Zool. 2007;4:1–11. doi: 10.1186/1742-9994-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valière N. GIMLET: a computer program for analysing genetic individual identification data. Mol. Ecol. Notes. 2002;2:377–379. [Google Scholar]
- Van Tienhoven AM, Den Hartog JE, Reijns RA. Peddemors VM. A computer-aided program for pattern-matching of natural marks on the spotted raggedtooth shark Carcharias taurus. J. Appl. Ecol. 2007;44:273–280. [Google Scholar]
- Waichman AV. An alphanumeric code for toeclipping amphibians and reptiles. Herpetol. Rev. 1992;23:19–21. [Google Scholar]
- Wilberg MJ. Dreher BP. GENECAP: a program for analysis of multilocus genotype data for non-invasive sampling and capture-recapture population estimation. Mol. Ecol. Notes. 2004;4:783–785. [Google Scholar]
- Winandy L. Denoel M. The use of automatized behavioural markers to assess methodologies: a study case on PIT-tagging in the Alpine newt. Behav. Res. Methods. 2011;43:568–576. doi: 10.3758/s13428-011-0058-z. [DOI] [PubMed] [Google Scholar]
- Woods JG, Paetkau D, Lewis D, McLellan BN, Proctor M. Strobeck C. Genetic tagging of free-ranging black and brown bears. Wildl. Soc. Bull. 1999;27:616–627. [Google Scholar]



