Abstract
Na+/I− symporter (NIS)-mediated radioiodide uptake (RAIU) serves as the basis for targeted ablation of thyroid cancer remnants. However, many patients with thyroid cancer have reduced NIS expression/function and hence do not benefit from radioiodine therapy. microRNA (miR) has emerged as a promising therapeutic target in many diseases; yet, the role of miRs in NIS-mediated RAIU has not been investigated. In silico analysis was used to identify miRs that may bind to the 3′UTR of human NIS (hNIS). The top candidate miR-339-5p directly bound to the 3′UTR of hNIS. miR-339-5p overexpression decreased NIS-mediated RAIU in HEK293 cells expressing exogenous hNIS, decreased the levels of NIS mRNA, and RAIU in transretinoic acid/hydrocortisone (tRA/H)-treated MCF-7 human breast cancer cells as well as thyrotropinstimulated PCCl3 rat thyroid cells. Nanostring nCounter rat miR expression assay was conducted to identify miRs deregulated by TGFβ, Akti-1/2, or 17-AAG known to modulate RAIU in PCCl3 cells. Among 38miRs identified, 18 were conserved in humans. One of the 18 miRs, miR-195, was predicted to bind to the 3′ UTR of hNIS and its overexpression decreased RAIU in tRA/H-treated MCF-7 cells. miR-339-5p was modestly increased inmost papillary thyroid carcinomas (PTCs), yet miR-195 was significantly decreased in PTCs. Interestingly, the expression profiles of 18 miRs could be used to distinguish most PTCs from nonmalignant thyroid tissues. This is the first report, to our knowledge, demonstrating that hNIS-mediated RAIU can be modulated by miRs, and that the same miRs may also play roles in the development or maintenance of thyroid malignancy. Accordingly, miRs may serve as emerging targets to halt the progression of thyroid cancer and to enhance the efficacy of radioiodine therapy.
Keywords: microRNA-339-5p, NIS, radioiodide uptake, papillary thyroid carcinoma
Introduction
Na+/I− symporter (NIS)-mediated radioiodide uptake (RAIU) in thyroid cells allows for targeted treatment of thyroid cancer. Many patients with advanced thyroid cancer do not benefit from radioiodine therapy due to reduced/absent expression/function of NIS. microRNA (miR) has emerged as a promising therapeutic target in many diseases (Maqbool & Hussain 2014). However, the role of miRs in NIS-mediated RAIU has not been investigated.
miRs predominantly bind to 3′UTR of target genes, resulting in either degradation of target mRNA or inhibition of translation (Krol et al. 2010). The 3′UTR of human NIS (hNIS) is 1296 bp in length, which is about twice as long as the average length of 3′UTRs of human genes (Pesole et al. 2000). Thus, expression of hNIS is likely to be modulated by miRs. Deregulation of miRs in papillary thyroid carcinomas (PTCs) has been reported by several groups (He et al. 2005, Tetzlaff et al. 2007, Pallante et al. 2010, de la Chapelle & Jazdzewski 2011). As NIS expression is downregulated in most PTCs (Lazar et al. 1999), it is likely that the upregulated miRs in PTCs may play roles in suppression of NIS. In this study, we aim to identify miRs that modulate the expression of NIS as well as NIS-mediated RAIU.
Two different approaches were applied to identify miRs that modulate the expression and function of NIS. In the first approach, in silico analysis was conducted to identify the top candidate miR predicted to bind to the 3′UTR of NIS. The effect of this miR on NIS-mediated RAIU was assessed in MCF-7 human breast cancer cells and PCCl3 rat thyroid cells. Direct binding of this miR to the 3′UTR of hNIS was verified by luciferase-hNIS-3′UTR reporter assay. In the second approach, miRs deregulated by signaling nodes known to modulate NIS-mediated RAIU in PCCl3 cells were identified. Among these deregulated miRs, five miRs were predicted to bind to rat Nis (rNis) 3′UTR and one miR was predicted to bind to the 3′UTR of hNIS. Their effects on NIS-mediated RAIU were investigated in both MCF-7 and PCCl3 cells. Finally, the expression levels of these miRs in PTC tumors vs their adjacent nonmalignant thyroid tissues were examined in two different cohorts.
Materials and methods
In silico miR prediction tools
To predict miRs that may bind to the 3′UTR of hNIS, TargetScan 6.1 (http://www.targetscan.org/), miRanda (http://www.microrna.org/), and miRecords (http://mirecords. biolead.org) were used. To increase robustness, the top predicted miR that had the highest number of binding sites and that was predicted by at least two software packages was selected for further studies. miRanda was used to predict miRs that may bind to the 3′UTR of rNis. TargetScan 6.1 was used to predict the top 100 targets of miR-339-5p and miR-195 in human and rat.
Luciferase reporter assay
Plasmid containing the 3′UTR of hNIS cloned downstream of the firefly luciferase CDS and a Renilla luciferase gene as an internal control was purchased from GeneCopoeia, Inc. (Rockville, MD, USA; pEZX-MT01, ID-HmiT017390-MT01). HeLa cell-line with low endogenous expression of NIS was transfected with this plasmid using Fugene HD (Promega Corporation) for 14 h. The cells were subsequently transfected with 50 µM of synthetic oligo miR-339-5p mimic (Life Technologies; MC12347) or scrambled oligo miR (Negative Control #1) using Lipofectamine 2000 (Life Technologies) for additional 6 h. Luciferase activity was measured in a Glomax-Multi Detection System (Promega Corporation) using a Dual-Luciferase Reporter 1000 Assay System (Promega Corporation). Data are represented as relative fold change of the ratios of firefly luciferase activity normalized to Renilla luciferase activity.
RAIU assay
This assay was performed as described previously (Vadysirisack et al. 2007) with 125I in NaI (80 mCi/mmol). RAIU in all figures represents fold change in NIS-mediated RAIU values acquired after subtracting nonspecific RAIU values from parallel experiments performed in the presence of 100 µM perchlorate.
Cell culture and reagents
Human embryonic kidney 293 cells (HEK293; ATCC, Manassas, VA, USA, CRL-1573) were cultured in 90% high-glucose DMEM and 10% fetal bovine serum (FBS; Life Technologies). HEK293 cells with low endogenous expression of NIS were transfected with either pcDNA3 vector control or full-length (−375)-hNIS-(3247)/pcDNA3 also known as pcDNA3/FLhNIS, including the complete 5′ and 3′ UTRs (Lin 2003) using FuGene 6 transfection reagent (Promega Corporation). Mixed stable clones were selected for 2 weeks using 800 µg/ml G418 (Life Technologies) and then maintained in 400 µg/ml G418. MCF-7 cells (ATCC HTB-22) were cultured in 45% DMEM, 45% F-12, and 10% FBS (Life Technologies). MCF-7 cells were treated with transretinoic acid (tRA; 1 µM)/hydrocortisone (H; 1 µM) (Sigma–Aldrich) for 24 h to induce expression of hNIS. Induction with tRA/H was performed in the presence of 5% charcoal-stripped FBS instead of 10% regular FBS. PCCl3 cells were maintained in 6H media with 5% bovine serum as described by Liu et al. (2012). The experiments were performed under acute thyrotropin (TSH) stimulation, where cells were withdrawn from TSH for 5 days (5H media) and then TSH was added back for 24 h to induce rNIS expression. HEK293TN cells (System Biosciences, Mountainview, CA, USA) used to produce lentiviral particles were kind gifts from Dr Sean Lawler. HEK293TN cells were maintained in DMEM supplemented with 10% heat inactivated FBS and 1% PenStrep (Life Technologies). During production of lentiviral particles, the cells were cultured in DMEM supplemented with 2% heat-inactivated FBS and no antibiotics. The passage number of all cells used was below 25. DMSO vehicle control was purchased from Sigma–Aldrich, Akti-1/2 (AKT inhibitor) and 17-AAG (Hsp90 inhibitor) from EMD Millipore (Billerica, MA, USA) and TGFβ from PeproTech, Inc. (Rockyhill, NJ, USA).
miR delivery methods
Lentiviral expression plasmids (System Biosciences) were kind gifts from Dr Lawler. The pCDH-CMV-MCS-EF1-copGFP control plasmid (pCDH) or pCDH/miR-339-5p was packaged into lentiviral particles using HEK293TN cells according to the manufacturer’s protocol. HEK293/pcDNA3 and HEK293/FLhNIS cells were transduced using lentiviral particles containing pCDH or pCDH/miR-339-5p. GFP-positive cells were enriched using FACS and cells were allowed to recover for 2 weeks before being used for experiments. Synthetic oligonucleotide miR-339-5p mimic, anti-miR-339-5p (MH12347), or scrambled oligonucleotide miR (10 nM) was transfected into MCF-7 or PCCl3 cells using Lipofectamine RNAiMAX reagent (Life Technologies) for 24 h at the same time as tRA/H treatment or TSH stimulation respectively before the cells were subjected to RAIU assay or RNA extraction.
RNA extraction and quantitative real-time PCR
Total RNA including small RNAs was extracted using the RNeasy Kit (Qiagen) and contaminating DNA was removed by on-column DNase I digestion according to manufacturer’s protocol. Quantitative real-time PCR (RT-qPCR) of NIS and GAPDH was performed as described previously (Liu et al. 2012). Representative data are presented as relative fold changes in NIS mRNA compared with the GAPDH control. For preparation of cDNA and qRT-PCR of miR, TaqMan MicroRNA Assays (Applied Biosystems, Life Technologies) were performed according to the manufacturer’s instructions. Representative data are presented as relative fold changes in miR-339-5p levels compared with the U6 snRNA control.
miR expression profiling
The nCounter rat miR expression assay from NanoString Technologies, Inc. (Seattle, WA, USA; Geiss et al. 2008, Wyman et al. 2011) was used for miR expression profiling at the Ohio State University Comprehensive Cancer Center (OSUCCC) Nucleic Acid Facility. Total RNA (100 ng) was used from two biological replicates for each treatment group, i.e., chronic TSH-induced PCC3 cells treated with DMSO vehicle control, Akti-1/2, 17-AAG, or TGFβ for 24 h. The data were first technically normalized using positive controls and then a Quantile normalization method was performed. miRs with expression levels below the level of noise, defined by negative controls, in 80% of the experimental groups, i.e. in five out of six experimental groups, were excluded from statistical analysis. This resulted in identification of about 100 miRs out of the 420 detectors in each comparison. Pairwise comparisons between each treatment and DMSO vehicle control were performed to determine fold-changes and P values. P values of less than 0.05 were considered significant. miRs significantly upregulated above 1.4-fold or significantly downregulated below 1.8-fold are shortlisted in Table 1.
Table 1.
Fold change of miRs deregulated in PCCl3 rat thyroid cells treated with TGF03B2;, Akti-1/2, or 17-AAG compared with DMSO vehicle control
| Rat miRs | TGFβ/DMSO | Akti-1/2/ DMSO |
17-AAG/DMSO |
|---|---|---|---|
| rno-miR-3578 | −1.6 | −2.1 | −1.2 |
| rno-miR-465 | −1.6 | −1.9 | 1.0 |
| rno-miR-3571 | 1.1 | −2.2 | −1.4 |
| rno-miR-344a-3p | 1.1 | −1.9 | −1.2 |
| rno-miR-3589 | 1.0 | −1.9 | −1.1 |
| rno-miR-449c-3p | −2.1 | −2.0 | −1.4 |
| rno-miR-3594-3p | −1.3 | −1.8 | −1.4 |
| rno-miR-455 | −1.1 | −1.8 | −1.1 |
| rno-miR-532-5p | −1.2 | −1.8 | −1.4 |
| rno-miR-325-3p | 1.0 | −1.8 | −1.3 |
| rno-miR-301a | −1.2 | 1.5 | 1.4 |
| rno-miR-29a | 1.0 | 1.4 | 1.2 |
| rno-miR-29c | −1.2 | 1.6 | 1.6 |
| rno-miR-3585-5p | 1.0 | 1.4 | 1.7 |
| rno-miR-218a | 1.3 | 1.6 | 1.4 |
| rno-miR-29b | −1.1 | 2.0 | 1.9 |
| rno-miR-331 | −1.3 | −1.8 | −2.9 |
| rno-miR-872 | −1.2 | −1.4 | −2.0 |
| rno-miR-106b | 1.4 | −1.1 | 1.4 |
| rno-miR-425 | 1.3 | 1.2 | 1.4 |
| rno-miR-30c | 1.0 | 1.1 | 1.4 |
| rno-miR-96 | 1.0 | 1.6 | 1.7 |
| rno-miR-30b-5p | −1.3 | 2.3 | 2.7 |
| rno-miR-190b | −1.1 | −1.3 | 1.5 |
| rno-miR-3559-5p | −1.4 | 1.1 | 1.6 |
| rno-miR-1949 | −2.1 | −1.7 | −1.3 |
| rno-miR-3581 | −2.0 | −1.6 | −1.4 |
| rno-miR-374 | −1.8 | −1.5 | −1.8 |
| rno-miR-544 | −1.8 | −1.9 | −1.4 |
| rno-miR-3561-3p | 1.4 | −1.5 | 1.2 |
| rno-miR-206 | 1.4 | 1.1 | 1.1 |
| rno-miR-27b | 1.4 | −1.0 | 1.1 |
| rno-miR-322 | 1.4 | 1.2 | 1.1 |
| rno-miR-539 | 1.7 | 1.0 | −1.1 |
| rno-miR-148b-3p | 1.4 | 1.5 | 1.6 |
| rno-miR-15b | 1.4 | 1.5 | 1.9 |
| rno-miR-195 | 1.4 | 1.3 | 1.3 |
| rno-miR-23b | 1.7 | 1.3 | 1.2 |
Fold changes highlighted in bold are significantly upregulated more than 1.4-fold or significantly downregulated more than 1.8-fold. miRs high-lighted in italics are conserved between rats and humans. miRs underlined are predicted to bind to the 3′UTR of rNis. miRs highlighted in bold are predicted to bind to the 3′UTR of hNIS.
Next generation sequencing
Small RNA samples from PTC tumors (PTC-T, n=19), nonmalignant tissue adjacent to but not infiltrated by tumor from the same patient (PTC-N, n=19), and thyroid tissues from normal individuals (NN, n=14) were analyzed using a Solid 5500 sequencing platform. The comparison of miRs deregulated between the PTC-T and PTC-N groups was performed using the paired Welch t-test as described previously (Swierniak et al. 2013). TCGA data (http://cancergenome.nih.gov/) were analyzed using the Wilcoxon signed-rank test. P values of less than 0.05 were considered statistically significant.
Heat map generation
The shortlisted rat miRs from Nanostring analysis were checked for exact sequence matches with human miRs. Expression levels of miRs with exact sequence matches were checked using next generation sequencing (NGS) data from 19 PTC-T/PTC-N pairs and 14 NN samples followed by creation of a heat map using unsupervised clustering. Hierarchical clustering of samples based on expression profiles of selected miRs was performed using Ward’s agglomeration method operated on Euclidean distance measures.
Statistical analysis
All experiments had at least two independent trials with three replicates for each experimental group within each trial. For RAIU assay, all the data values were log10 transformed to reduce variance and skewness, and then linear mixed effects models were used to take account of the correlations among observations from the same trial. For RT-qPCR data, linear mixed effects models were used for analysis along with the ΔΔCT method. From the model, all the pre-specified comparisons for each experiment were obtained and adjusted for multiple comparisons using the sequentially rejective Holm’s method (Holm 1979) to control for type 1 error at 0.05. For luciferase assays, the two sample t-test was performed for comparisons after taking the average of three replicates, and P values of less than 0.05 were considered to be significant. For TCGA data, Pearson’s correlation method was used to test the association between the expression of NIS and the selected miRs from patients’ tumor samples. Expression of selected miRs from TCGA data were compared between normal and tumor tissues using paired t-tests. SAS v9.2 Software was used for all the analysis (SAS Institute, Inc., Cary, NC, USA).
Results
miR-339-5p targets the 3′UTR of hNIS and reduces hNIS-mediated RAIU in HEK293 cells expressing exogenous hNIS
In silico analysis predicted that miR-339-5p has the highest binding score and the highest number of binding sites in the 3′UTR of hNIS, i.e. two binding sites at nucleotides 291–297 (TargetScan context score: −0.25 and miRanda mirSVR score:−0.43) and nucleotides 458–464 (TargetScan context score:−0.22 and miRanda mirSVR score:−0.58). As shown in Fig. 1A, overexpression of miR-339-5p resulted in a significant decrease (21%; P=0.006) in luciferase activity of the luciferase-hNIS-3′UTR reporter. Consistent with the luciferase assay, overexpression of miR-339-5p resulted in a significant decrease (36%; P=0.002) in hNIS-mediated RAIU activity in HEK293 cells stably expressing exogenous hNIS (Fig. 1B). On the basis of these findings, it is concluded that miR-339-5p directly binds to the 3′UTR of hNIS and decreases hNIS-mediated RAIU activity.
Figure 1.
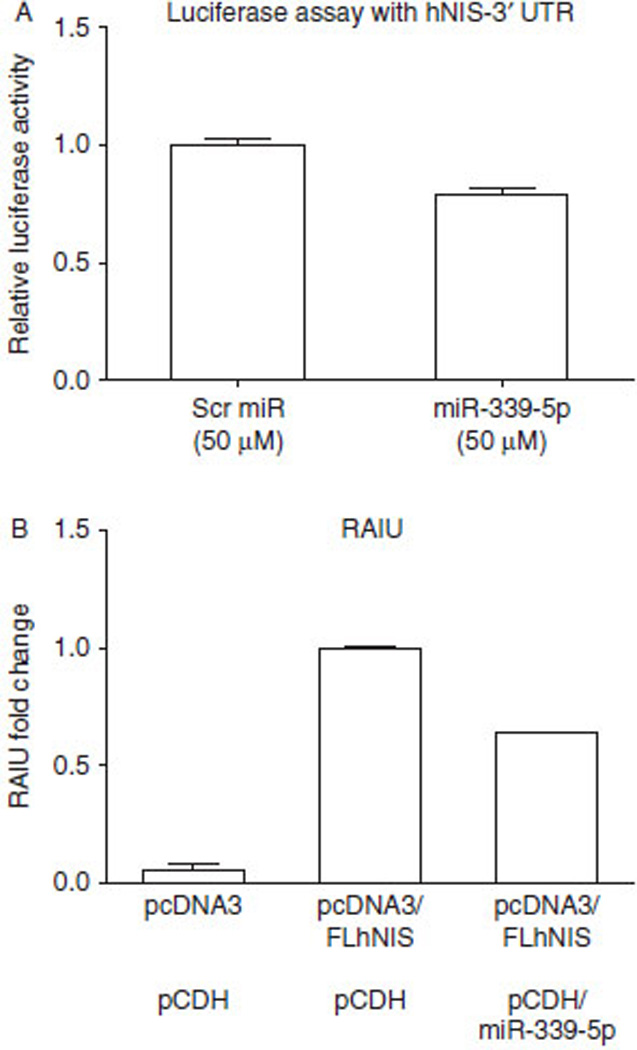
miR-339-5p targets the 3′UTR of hNIS and reduces hNIS-mediated RAIU in HEK293 cells expressing exogenous hNIS. (A) The results of the luciferase assay indicated that overexpression of miR-339-5p resulted in a significant decrease (21‰; P=0.006) in luciferase activity from the luciferase-hNIS-3′UTR reporter. After seeding for 24 h, HeLa cells were transfected with plasmid containing firefly luciferase-hNIS-3′UTR for 14 h and subsequently transfected with scrambled (Scr) miR ormiR-339-5p for 6 h before being subjected to luciferase assay. (B) Overexpression of miR-339-5p resulted in a significant decrease (36‰; P=0.002) in hNIS-mediated RAIU activity in HEK293 cells stably expressing exogenous hNIS. HEK293 cells were stably transfected with either pcDNA3 vector control or pcDNA3/FLhNIS including 5′ and 3′ UTRs. Stable overexpression of miR-339-5p was mediated by transduction of lentiviral particles containing either the pCDH control plasmid or pCDH/miR-339-5p. Relative fold-changes in RAIU activity are shown. (A and B) The results are representative of at least two independent experiments performed in triplicates and the mean±s.d. for each group are shown.
miR-339-5p reduces the level of tRA/H-induced hNIS mRNA and hNIS-mediated RAIU in MCF-7 human breast cancer cells
There is no immortalized human thyroid cell-line that consistently expresses endogenous hNIS conferring evident RAIU activity. We choose tRA/H-treated MCF-7 cells to investigate the effect of overexpression of miR-339-5p on the levels of endogenous hNIS mRNA and hNIS-mediated RAIU, as it is well established that tRA/H significantly induces the expression and function of hNIS in MCF-7 cells (Kogai et al. 2000, Beyer et al. 2011). As shown in Fig. 2A and B, miR-339-5p overexpression significantly decreased tRA/H-induced hNIS mRNA levels (26%; P<0.0001) as well as hNIS-mediated RAIU activity (30%; P<0.0001). Note that anti-miR-339-5p counteracted the effects of overexpression of miR-339-5p on the expression/function of hNIS, albeit anti-miR-339-5p alone had little effect. As shown in Fig. 2C, miR-339-5p was overexpressed by approximately 1000-fold and this was reduced to approximately 100-foldbyanti-miR-339-5p. This is consistent with the notion that anti-miR counteracts the effect of miR most probably by both miR degradation and functional inhibition. Note that the level of endogenous miR-339-5p was not affected by tRA/H treatment, indicating that hNIS expression/function of hNIS induced by tRA/H in MCF-7 cells was not mediated by miR-339-5p. On the basis of these results, it is concluded that expression and function of hNIS was decreased by overexpression of miR-339-5p.
Figure 2.
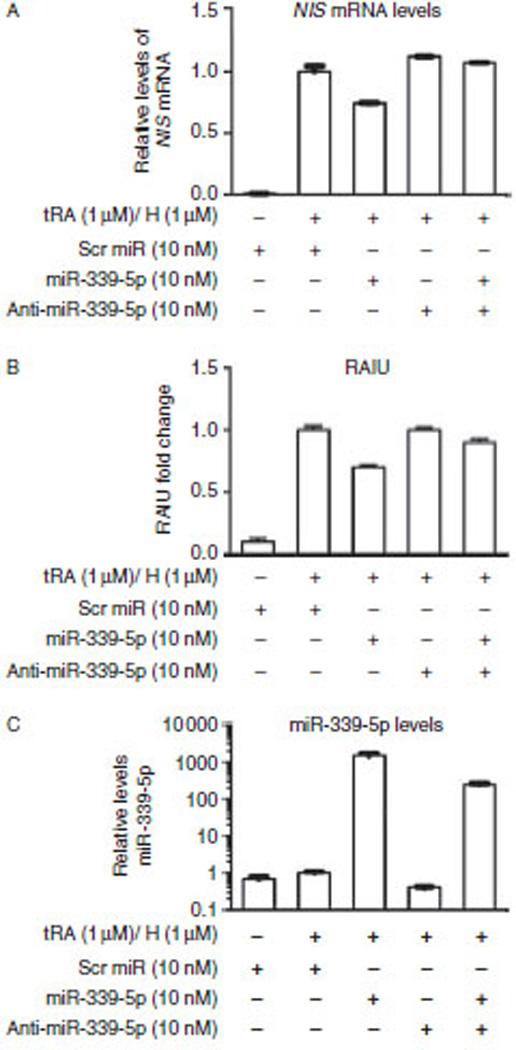
miR-339-5p reduces the levels of tRA/H-induced hNIS mRNA and hNIS-mediated RAIU in MCF-7 human breast cancer cells. (A) The results of RT-qPCR for hNIS indicated that overexpression of miR-339-5p resulted in a significant decrease in the levels of tRA/H-induced hNIS mRNA (26‰; P<0.0001). Relative fold changes in levels of hNIS mRNA normalized to levels of GAPDH mRNA are shown. (B) overexpression of miR-339-5p resulted in a significant decrease in tRA/H-induced hNIS-mediated RAIU activity (30‰; P<0.0001). Relative fold changes in RAIU activity are shown. (C) The results from RT-qPCR indicated that tRA/H had little effect on the levels of endogenous miR-339-5p and that the level of miR-339-5p increased by approximately 1000-fold upon transfection with oligo miR-339-5p. The increase in the level of miR-339-5p was reduced to approximately 100-fold by anti-miR-339-5p. Relative fold changes in the levels of miR-339-5p normalized to the levels of U6 snRNA are shown. (A, B and C) Scrambled oligo miRs or oligo miR-339-5p mimic were transfected into MCF-7 cells for 24 h at the same time as tRA/H treatment, then the cells were subjected to RNA extraction or RAIU. The results are representative of two independent experiments performed in triplicates and the mean±s.d. for each set of results is shown.
miR-339-5p reduces the levels of TSH-induced rNis mRNA and rNIS-mediated RAIU in PCCl3 rat thyroid cells
As miR-339-5p is 100% conserved between human and rat, we examined the effect of overexpression of miR-339-5p on levels of endogenous rNis mRNA and rNIS-mediated RAIU in PCCl3 rat thyroid cells that express functional rNIS upon stimulation with TSH. The 3′UTR of hNIS and the 3′UTR of rNis share only 35.2% nucleotide sequence identity and miRanda predicted that miR-339-5p has only one binding site in the 3′UTR of rNis on nucleotides 686–691 with a very low score (mirSVR score: −0.02). As shown in Fig. 3A and B, miR-339-5p overexpression resulted in a significant decrease in the levels of TSH-induced rNis mRNA (30%; P=0.0016) as well as TSH-induced rNIS-mediated RAIU activity (30%; P <0.0001). Note that anti-miR-339-5p counteracted the effects of overexpression of miR-339-5p on the expression/function of rNIS. As shown in Fig. 3C, miR-339-5p was overexpressed by approximately 200-fold and was reduced to approximately 20-fold by anti-miR-339-5p. TSH had little effect on levels of endogenous miR-339-5p, which is consistent with other findings (Leone et al. 2011, Akama et al. 2012) that the expression of miR-339-5p is not modulated by TSH, the major regulator of the expression and function of NIS. On the basis of these results, it is concluded that the expression and function of rNIS was significantly decreased by overexpression of miR-339-5p.
Figure 3.
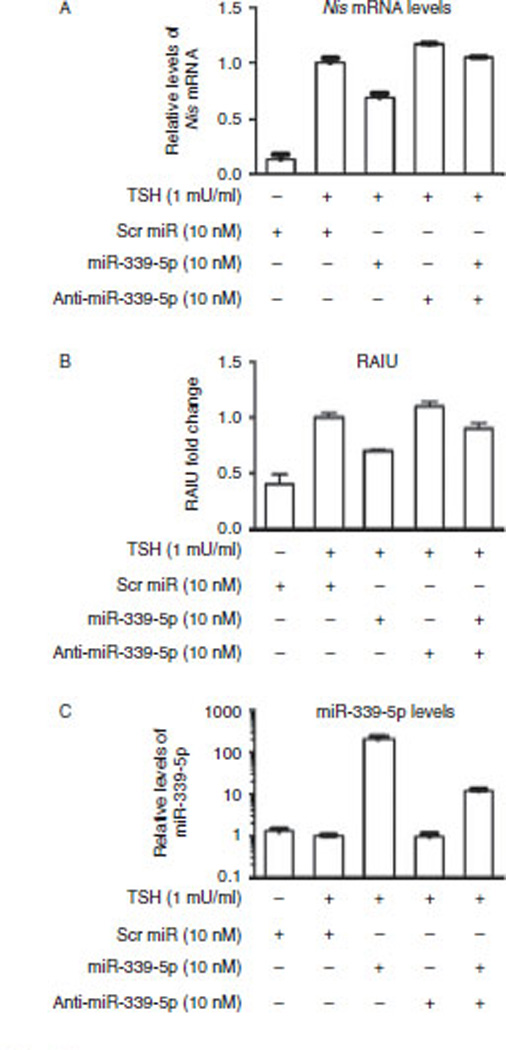
miR-339-5p reduces the levels of TSH-induced rNis mRNA and rNIS-mediated RAIU inPCCl3 rat thyroid cells. (A)The results of RT-qPCR for rNis indicated that overexpression of miR-339-5p resulted in a significant decrease in TSH-induced levels of rNis mRNA (30‰; P=0.0016). Relative fold change in the levels of rNis mRNA normalized to the levels of Gapdh mRNA are shown. (B)Overexpression of miR-339-5p resulted in a significant decrease in TSH-induced rNIS-mediated RAIU activity (30‰; P<0.0001). Relative fold changes in RAIU activity are shown. (C) The results of RT-qPCR indicated that TSH had little effect on the levels of endogenous miR-339-5p and that the levels of miR-339-5p increased by approximately 200-fold upon transfection with oligo miR-339-5p. The increase in the level of miR-339-5p was reduced to approximately 20-fold by anti-miR-339-5p. Relative fold changes in the levels of miR-339-5p normalized to the levels of U6 snRNA are shown. (A, Band C) Scrambled oligo miRs or oligo miR-339-5p mimic were transfected into PCCl3 cells for 24 h at the same time as TSH stimulation, then the cells were subjected to RNA extraction or RAIU. The results are representative of two independent experiments performed in triplicates and the mean±s.d. for each set of results is shown.
Several miRs deregulated by signaling nodes that modulate rNIS-mediated RAIU in PCCl3 cells are predicted to bind to the 3′UTR of Nis
TSH-stimulated RAIU in rat thyroid cells can be modulated by TGFβ (Pekary & Hershman 1998, Nicolussi et al. 2003, Costamagna et al. 2004), AKT (Kogai et al. 2008, Liu et al. 2012), and HSP90 (Marsee et al. 2004) by modulating the expression of rNIS, the function of rNIS, and iodide efflux respectively. To uncover candidate miRs that modulate rNIS-mediated RAIU in rat thyroid cells, miRs deregulated by TGFβ, Akti-1/2, or 17-AAG in PCCl3 cells were identified (Table 1). Among 38 miRs identified, miR-218a, miR-425, miR-96, miR-27b, and miR-539 were predicted to bind to the 3′UTR of rNis (mirSVR score range: −0.38 to −0.01). Among these five miRs, two miRs were significantly upregulated by TGFβ (1.4-and 1.7-fold) indicating their possible roles in the mediation of repression of rNIS by TGFβ. As Akti-1/2 and 17-AAG do not modulate expression of rNis, it is puzzling that one out of five miRs predicted to bind to the 3′UTR of rNis was upregulated by both 17-AAG and Akti-1/2 (1.4- and 1.6-fold respectively), and three out of five miRs by 17-AAG (1.4- to 1.7-fold). Nevertheless, we examined the direct effects of these five miRs on TSH-stimulated rNIS-mediated RAIU in PCCl3 cells. As shown in Fig. 4A, unlike miR-339-5p, overexpression of these five miRs did not result in a significant decrease in RAIU in PCCl3 cells. miR-339-5p was not included in the list of 38 miRs due to its low expression level in PCCl3 cells, which did not meet the cut-off value of Nanostring analysis. Interestingly, despite its low levels, miR-339-5p was upregulated by TGFβ (1.3-fold), indicating that miR-339-5p may mediate the effect of TGFβ on rNIS expression.
Figure 4.
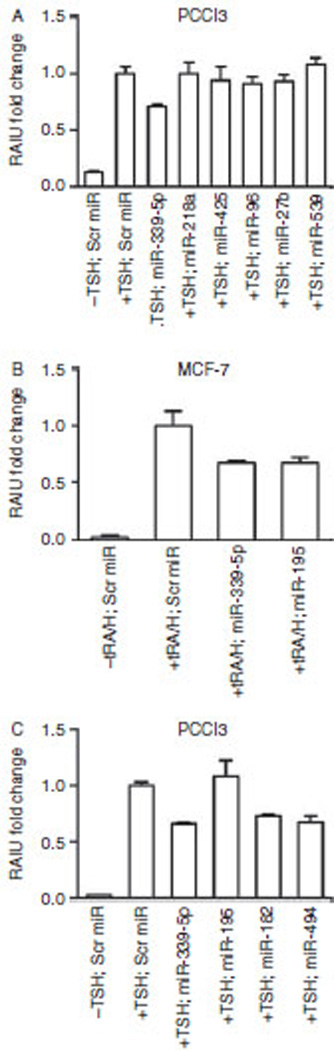
Several miRs deregulated by signaling nodes that modulate rNIS-mediated RAIU in PCCl3 cells are predicted to bind to the 3′UTR of Nis. (A) Overexpression of miR-218a, miR-425, miR-96, miR-27b, or miR-539 did not result in a significant decrease (P=0.6529, 0.1834, 0.1777, 0.2638, and 0.9686 respectively) in TSH-induced rNIS-mediated RAIU activity in PCCl3 cells. (B) Overexpression of miR-195 resulted in a significant decrease in tRA/H-induced hNIS-mediated RAIU (30‰; P<0.0001) in MCF-7 cells. (C) Overexpression of miR-195 did not significantly decrease (P=0.2059) but overexpression of miR-182 and miR-494 did significantly decrease TSH-induced rNIS-mediated RAIU (27‰; P<0.0001 and 33‰; P<0.0001 respectively) in PCCl3 cells. (A, B and C) Oligo miRs were transfected into PCCl3 cells or MCF-7 cells for 24 h at the same time as TSH or tRA/H treatment respectively and then the cells were subjected to RAIU. The results are representative of two independent experiments performed in triplicates and the mean±s.d. for each set of results is shown.
Among 38 rat miRs deregulated by TGFβ, Akti-1/2, or 17-AAG in PCCl3 cells, 18 of them have exact sequence matches between human and rat, and miR-195 is predicted to bind the 3′UTR of hNIS (mirSVR score: −0.01). Overexpression of miR-195 significantly decreased RAIU by 30% (P<0.0001) which was similar to the effect of miR-339-5p in tRA/H-treated MCF-7 cells (Fig. 4B). However, miR-195 is not predicted to bind to the 3′UTR of rNIS and its overexpression did not significantly decrease (P=0.2059) rNIS-mediated RAIU in PCCl3 cells (Fig. 4C). In comparison, overexpression of rno-miR-182 and rno-miR-494, which are predicted to bind to the 3′UTR of rNIS (mirSVR score: −0.77 and −0.16 respectively), did significantly decrease rNIS-mediated RAIU in PCCl3 cells (27%; P<0.0001 and 33%; P<0.0001 respectively). On the basis of these results, it is concluded that miR-339-5p modulates the expression of NIS in both human and rat cells, yet miR-195 appears to modulate the expression of NIS in human but not in rat cells, as indicated by its effects on NIS-mediated RAIU activity.
Expression profiles of 18 hsa-miRs distinguish most PTCs from nonmalignant thyroid tissues
Almost all PTCs have reduced NIS-mediated RAIU activity. Accordingly, many signaling pathways driving thyroid tumorigenesis are also known to reduce NIS-mediated RAIU in thyroid. We therefore investigated the expression profiles of the 18 miRs deregulated by TGFβ, Akti-1/2, or 17-AAG in 19 PTC-T/PTC-N pairs and 14 NN. As shown in Fig. 5, the expression profile of these 18 miRs could be used to distinguish most PTC-T samples from PTC-N and NN samples. The fold changes of these 18 miRs in PTC-T compared with PTC-N were examined in the cohort from Medical University of Warsaw (n=19) as well as from thyroid cancer TCGA database (n=59). As shown in Table 2, hsa-miR-96 and hsa-miR-27b were significantly upregulated in PTC-T compared with PTC-N in both cohorts. In contrast, hsa-miR-455 and hsa-miR-195 were significantly downregulated in PTC-T compared with PTC-N in both cohorts. As hsa-miR-195 was predicted to bind to the 3′UTR of hNIS and its overexpression decreased NIS-mediated RAIU activity, it is surprising that hsa-miR-195 was downregulated rather than upregulated in PTC-T versus PTC-N. Accordingly, miR that plays a role in the development or maintenance of thyroid malignancy may also modulate NIS-mediated RAIU, yet the underlying mechanisms could be distinct and complex in nature.
Figure 5.
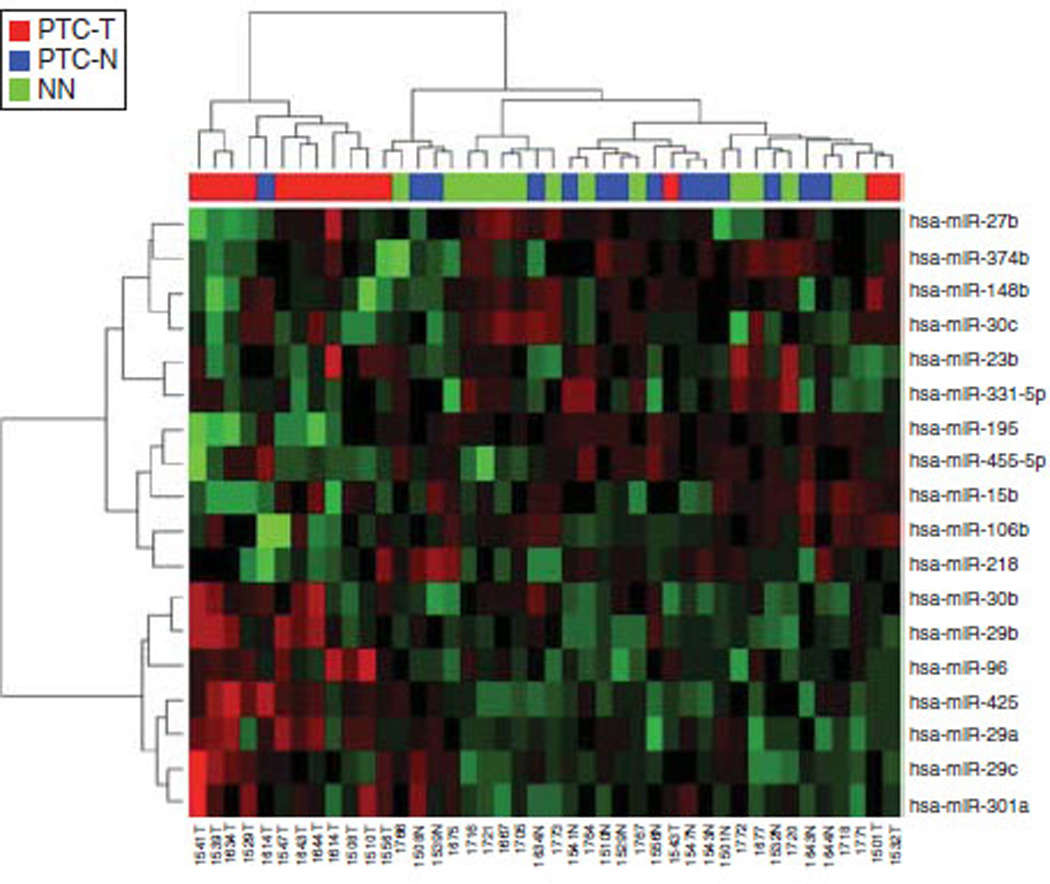
Expression profiles of 18 hsa-miRs distinguish most papillary thyroid carcinomas from nonmalignant thyroid tissues. Among the 38 rat miRs deregulated by TGFβ, Akti-1/2, or 17-AAG in PCCl3 cells, 18 of them were conserved in humans. The expression levels of the 18 hsa-miRs were extracted from the NGS data from 19 PTC-T/PTC-N pairs and 14 NN. A heat map created by unsupervised clustering using the expression profiles of the 18 hsa-miRs as shown could be used to distinguish most PTC tumors from nonmalignant thyroid tissues. in triplicates and the mean±s.d. for each set of results is shown.
Table 2.
Fold change of 18 hsa-miRs in papillary thyroid carcinoma (PTC-T) compared with paired adjacent nonmalignant tissue (PTC-N)
| Cohort from Medical University of Warsaw (n = 19) | Cohort from TCGA data (n = 59) | |||||||
|---|---|---|---|---|---|---|---|---|
| Human miRs | Mean PTC-T (RPM) |
Mean PTC-N (RPM) |
Fold T/N | P value | Mean PTC-T (RPM) |
Mean PTC-N (RPM) |
Fold T/N | P value |
| hsa-miR-455 | 30 | 53 | 0.56 | 0.01 | 139 | 240 | 0.58 | 5.94×10−6 |
| hsa-miR-301a | 8 | 5 | 1.65 | 0.03 | 14 | 16 | 0.93 | 2.77×10−6 |
| hsa-miR-29a | 164 | 124 | 1.32 | 0.06 | 21 730 | 21 702 | 1.00 | 6.25×10−2 |
| hsa-miR-29c | 619 | 923 | 0.67 | 0.15 | 5732 | 8610 | 0.67 | 5.16×10−9 |
| hsa-miR-218a | 2127 | 2365 | 0.90 | 0.62 | 136 | 243 | 0.56 | 5.74×10−8 |
| hsa-miR-29b | 25 | 16 | 1.57 | 0.14 | 964 | 801 | 1.20 | 5.43×10−2 |
| hsa-miR-331 | 4 | 9 | 0.49 | 0.05 | 66 | 69 | 0.96 | 4.19×10−1 |
| hsa-miR-106b | 499 | 512 | 0.97 | 0.90 | 406 | 601 | 0.68 | 1.64×10−6 |
| hsa-miR-425 | 1314 | 805 | 1.63 | 0.02 | 109 | 203 | 0.54 | 9.68×10−9 |
| hsa-miR-30c | 10 250 | 11 010 | 0.93 | 0.47 | 1518 | 2236 | 0.68 | 7.88×10−6 |
| hsa-miR-96 | 132 | 66 | 2.01 | 0.02 | 18 | 9 | 2.03 | 1.98×10−6 |
| hsa-miR-30b-5p | 13 784 | 11 000 | 1.25 | 0.03 | 745 | 982 | 0.76 | 4.48×10−4 |
| hsa-miR-374 | 377 | 552 | 0.68 | 0.16 | 92 | 152 | 0.61 | 2.23×10−8 |
| hsa-miR-27b | 64 | 28 | 2.26 | 0.01 | 3099 | 2432 | 1.27 | 5.22×10−5 |
| hsa-miR-148b-3p | 533 | 628 | 0.85 | 0.24 | 308 | 436 | 0.71 | 8.73×10−8 |
| hsa-miR-15b | 1791 | 2008 | 0.89 | 0.47 | 242 | 309 | 0.78 | 4.76×10−3 |
| hsa-miR-195 | 1031 | 2019 | 0.51 | 0.00 | 35 | 80 | 0.44 | 8.05×10−10 |
| hsa-miR-23b | 6 | 5 | 1.26 | 0.38 | 2508 | 1850 | 1.36 | 2.07×10−5 |
RPM, reads per million. Fold T/N highlighted in italics do not correlate between the Cohort from the Medical University of Warsaw and the Cohort from TCGA. Fold T/N highlighted in bold are significantly deregulated in the same direction in both cohorts.
Discussion
In this study, we have identified miR-339-5p that modulates the levels of NIS mRNA and NIS-mediated RAIU. miR-339-5p directly bound to 3′UTR of hNIS and reduced exogenously-expressed-hNIS-mediated RAIU in HEK293 cells. miR-339-5p not only reduced tRA/H-induced hNIS mRNA and hNIS-mediated RAIU inMCF-7 human breast cancer cells but also reduced TSH-induced rNis mRNA and rNIS-mediated RAIU in PCCl3 rat thyroid cells. We have also identified 38 miRs deregulated by reagents known to modulate RAIU in PCCl3 cells, and 18 of them had exact sequence matches between human and rat. miR-195 was predicted to bind to the 3′UTR of hNIS but not to the 3′UTR of rNis, and its overexpression decreased tRA/H-induced hNIS-mediated RAIU in MCF-7 cells but did not decrease TSH-induced rNIS-mediated RAIU in PCCl3 cells. Interestingly, the 18 miRs deregulated by reagents known to modulate RAIU could be used to distinguish between PTCs from nonmalignant tissues. This finding indicates that miRs that play a role in the development or maintenance of thyroid malignancy may also modulate NIS-mediated RAIU, although the underlying mechanisms could be distinct and complex in nature.
miR-339-5p was significantly upregulated in PTC-T by 1.3-fold compared with NN, yet there was no significant difference between PTC-T and PTC-N in the cohort from the Medical University of Warsaw. Similarly, there was no significant difference in the levels of expression of miR-339-5p between PTC-T and PTC-N in the cohort of thyroid cancer from the TCGA database. Pearson’s correlation test on 485 PTCs from the TCGA database showed a non-significant negative correlation between the levels of miR-339-5p and NIS mRNA (correlation coefficient= −0.063 and P=0.163). These findings are consistent with the findings that the effect of a single miR on a specific target could be miniscule (Hausser & Zavolan 2014). In addition, the effect of a given miR on its target can be genetic-context-dependent, such that its effect on a specific target may not reach statistical significance in a cohort of tumors with various genetic contexts. Indeed, miR-339-5p was one of the top ten upregulated miRs (Nikiforova et al. 2008) in conventional follicular adenomas (13.4-fold), oncocytic follicular adenomas (18.2-fold), oncocytic follicular carcinomas (42.5-fold), and poorly differentiated thyroid carcinomas (6.1-fold).
Given the fact that each miR has multiple targets and each target could be modulated by multiple miRs (Hausser & Zavolan 2014), miRs may exert both direct and indirect effects. In silico prediction scores may not translate to direct binding of miRs to target 3′UTRs, and the effects of miRs on the expression or function of target gene may be a consequence of indirect effects. For example, the 5 out of 38 miRs predicted to bind to the 3′UTRof rNis had a predicted score range from high to low, yet none of them decreased TSH-stimulated rNIS-mediated RAIU. Conversely, overexpression of miR-339-5p, miR-182, ormiR-494 decreased rNIS-mediated RAIU, yet we were unable to demonstrate their direct binding to the 3′UTR or rNis (data not shown). To investigate possible indirect effects of miR-339-5p in the modulation of NIS-mediated RAIU, we analyzed its top 100 predicted targets from TargetScan in both human and rat. Ingenuity pathway analysis of the top 100 targets in humans indicated that MDM2 and SYT2 are involved in RXR pathway activation. As activated RXR is implicated in tRA-induced expression of NIS (Kogai et al. 2000), overexpression of miR-339-5p may also indirectly suppress tRA-induced NIS by targeting the RXR pathway. Ingenuity pathway analysis of the top 100 targets in rat indicated that BCL2L11, MAGI2, and SOS1 participate in PTEN signaling activation. PTEN inhibition suppresses NIS expression (Dima et al. 2011); thus, miR-339-5p overexpression may indirectly suppress NIS expression via PTEN inhibition in rat thyroid cells.
miR-195 was significantly downregulated in PTC-T compared with PTC-N in both cohorts examined. Pearson’s correlation test on 485 PTCs from TCGA database showed a moderate but significant positive correlation between miR-195 and NIS mRNA levels (coefficient=0.139 and P=0.0021). This unexpected finding suggests that miR-195 may modulate other target(s) that counteract it’s repression of NIS mRNA, and these target(s) are present in thyroid tumors but absent in tRA/H-treated MCF-7 cells. Alternatively, repression of hNIS mRNA by miR-195 could be overridden by other factors unique to thyroid tumors. To investigate possible indirect effects of miR-195 in the modulation of NIS-mediated RAIU, we examined its top 100 targets in humans. Ingenuity pathway analysis of the top 100 targets in humans indicated that AKT3, EIF4G2, INSR, and RPS6KA6 are involved in activation of them TOR pathway and RAB9B and RasGEF1B are involved in activation of the RAS pathway. Activation of the PI3K/AKT/mTOR pathway (Ohashi et al. 2009) as well as the RAS pathway (Alsayed et al. 2001) upregulates tRA-induced expression and function of NIS in MCF-7 cells; thus, overexpression of miR-195 may indirectly suppress the function of NIS via inhibition of the mTOR pathway or the RAS pathway in MCF-7 cells.
The signaling nodes of TGFβ, AKT, or HSP90 are known to play a role in progression of thyroid tumors, thus it is not surprising that 18 out of 38 miRs deregulated by TGFβ, Akti-1/2, or 17-AAG could be used to separate PTC-T from PTC-N. The listed 18 miRs as a group were not previously reported to be important players in thyroid tumorigenesis, possibly due to their relatively low expression levels. miR-195 (T/N ratio=0.51) was recently shown to target ZNF367, an important regulator of cancer progression (Jain et al. 2014). miR-218 and miR-30c (Tetzlaff et al. 2007) as well as miR-148 and miR-15b (Pallante et al. 2006) have been reported to be downregulated in PTCs versus nonmalignant thyroid tissues. miR-29 a/b/c was upregulated in PTC-T versus PTC-N (He et al. 2005). The T/N values of these miRs, except miR-29c, are consistent with results described in previous publications. Finally, it is interesting to note that 11 out of 38 miRs that we identified were among those miRs reported to be downregulated by 24 h TSH stimulation in FRTL-5 cells (Akama et al. 2012). The finding that 6 out of 11 TSH-modulated miRs were upregulated by TGFβ is indicative of a potential role of miRs in the cross talk between TSH and TGFβ signaling.
In conclusion, this is the first report, to our knowledge, demonstrating that expression and function of NIS can be modulated by miRs. miR-339-5p may play a role in decreasing hNIS-mediated RAIU in follicular thyroid tumors but not in papillary thyroid tumors. As miR-195 was not upregulated in papillary thyroid tumors, it does not directly contribute to the reduction of levels of NIS in papillary thyroid tumors. It is of interest to note that both miR-339-5p (Ju et al. 2009, Pizzimenti et al. 2009, Ueda et al. 2009, Wu et al. 2010, Fayyad-Kazan et al. 2013, Ulivi et al. 2013, Li et al. 2014) and miR-195 (Soon et al. 2009, Heneghan et al. 2010, Li et al. 2011, Chabre et al. 2013, Yang et al. 2013, Ouyang et al. 2014) have been implicated in other types of cancer.
Acknowledgements
The authors thank Dr H Alder and Dr G H Ozer for help with performing and analyzing Nanostring miR assay, Dr S Lawler and Dr J Godlewski for providing reagents and technical guidance to produce lentiviral particles and Anna Kubiak for the miR TCGA data analysis. The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Funding
This work was supported by National Institutes of Health P01CA124570 (PI: M Ringel; Project 3 leader: S M Jhiang), Polish National Science Centre grant number 2012/07/D/NZ3/04149, Polish Ministry of Science grant number N401584838 and Foundation for Polish Science, Program TEAM, co-financed by the European Union European Regional Development Fund. The Laboratory of Genomic Medicine of the Medical University of Warsaw participates in Bastion, a Program financed by the European Union (FP7-REGPOT-2012-CT2012-316254-BASTION).
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution statement
A Lakshmanan, A Wojcicka, M Kotlarek, K Jazdzewski, and S M Jhiang conceived and designed the experiments. A Lakshmanan, A Wojcicka, X Zhang, and M Kotlarek performed the experiments. A Lakshmanan, A Wojcicka, M Kotlarek, X Zhang, K Jazdzewski, and S M Jhiang analyzed the data. A Lakshmanan, A Wojcicka, K Jazdzewski, and SM Jhiang wrote the paper.
References
- Akama T, Sue M, Kawashima A, Wu H, Tanigawa K, Suzuki S, Hayashi M, Yoshihara A, Ishido Y, Ishii N, et al. Identification of microRNAs that mediate thyroid cell growth induced by TSH. Molecular Endocrinology. 2012;26:493–501. doi: 10.1210/me.2011-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsayed Y, Uddin S, Mahmud N, Lekmine F, Kalvakolanu DV, Minucci S, Bokoch G, Platanias LC. Activation of Rac1 and the p38 mitogenactivated protein kinase pathway in response to all-trans-retinoic acid. Journal of Biological Chemistry. 2001;276:4012–4019. doi: 10.1074/jbc.M007431200. [DOI] [PubMed] [Google Scholar]
- Beyer S, Lakshmanan A, Liu YY, Zhang X, Wapnir I, Smolenski A, Jhiang S. KT5823 differentially modulates sodium iodide symporter expression, activity, and glycosylation between thyroid and breast cancer cells. Endocrinology. 2011;152:782–792. doi: 10.1210/en.2010-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabre O, Libe R, Assie G, Barreau O, Bertherat J, Bertagna X, Feige JJ, Cherradi N. Serum miR-483-5p and miR-195 are predictive of recurrence risk in adrenocortical cancer patients. Endocrine-Related Cancer. 2013;20:579–594. doi: 10.1530/ERC-13-0051. [DOI] [PubMed] [Google Scholar]
- de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. Journal of Clinical Endocrinology and Metabolism. 2011;96:3326–3336. doi: 10.1210/jc.2011-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costamagna E, Garcia B, Santisteban P. The functional interaction between the paired domain transcription factor Pax8 and Smad3 is involved in transforming growth factor-β repression of the sodium/iodide symporter gene. Journal of Biological Chemistry. 2004;279:3439–3446. doi: 10.1074/jbc.M307138200. [DOI] [PubMed] [Google Scholar]
- Dima M, Miller KA, Antico-Arciuch VG, Di Cristofano A. Establishment and characterization of cell lines from a novel mouse model of poorly differentiated thyroid carcinoma: powerful tools for basic and preclinical research. Thyroid. 2011;21:1001–1007. doi: 10.1089/thy.2011.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyad-Kazan H, Bitar N, Najar M, Lewalle P, Fayyad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani R, et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. Journal of Translational Medicine. 2013;11:31. doi: 10.1186/1479-5876-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature Biotechnology. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions – beyond repression of gene expression. Nature Reviews. Genetics. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. PNAS. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ. Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist. 2010;15:673–682. doi: 10.1634/theoncologist.2010-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- Jain M, Zhang L, Boufraqech M, Liu-Chittenden Y, Bussey K, Demeure MJ, Wu X, Su L, Pacak K, Stratakis CA, et al. ZNF367 inhibits cancer progression and is targeted by miR-195. PLoS ONE. 2014;9:e101423. doi: 10.1371/journal.pone.0101423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X, Li D, Shi Q, Hou H, Sun N, Shen B. Differential microRNA expression in childhood B-cell precursor acute lymphoblastic leukemia. Pediatric Hematology and Oncology. 2009;26:1–10. doi: 10.1080/08880010802378338. [DOI] [PubMed] [Google Scholar]
- Kogai T, Schultz JJ, Johnson LS, Huang M, Brent GA. Retinoic acid induces sodium/iodide symporter gene expression and radioiodide uptake in the MCF-7 breast cancer cell line. PNAS. 2000;97:8519–8524. doi: 10.1073/pnas.140217197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogai T, Sajid-Crockett S, Newmarch LS, Liu YY, Brent GA. Phosphoinositide-3-kinase inhibition induces sodium/iodide symporter expression in rat thyroid cells and human papillary thyroid cancer cells. Journal of Endocrinology. 2008;199:243–252. doi: 10.1677/JOE-08-0333. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The wide spread regulation of microRNA biogenesis, function and decay. Nature Reviews. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lazar V, Bidart JM, Caillou B, Mahe C, Lacroix L, Filetti S, Schlumberger M. Expression of the Na+/I− symporter gene in human thyroid tumors: a comparison study with other thyroid-specific genes. Journal of Clinical Endocrinology and Metabolism. 1999;84:3228–3234. doi: 10.1210/jcem.84.9.5996. [DOI] [PubMed] [Google Scholar]
- Leone V, D’Angelo D, Ferraro A, Pallante P, Rubio I, Santoro M, Croce CM, Fusco A. A TSH-CREB1-microRNA loop is required for thyroid cell growth. Molecular Endocrinology. 2011;25:1819–1830. doi: 10.1210/me.2011-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clinical Cancer Research. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhao W, Bao P, Li C, Ma XQ, Li Y, Chen LA. miR-339-5p inhibits cell migration and invasion and may be associated with the tumor-node-metastasis staging and lymph node metastasis of non-small cell lung cancer. Oncology Letters. 2014;8:719–725. doi: 10.3892/ol.2014.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. PhD Thesis. The Ohio State University; 2003. Regulation of sodium iodide symporter expression/function and tissue-targeted gene transfer of sodium iodide symporter. [Google Scholar]
- Liu YY, Zhang X, Ringel MD, Jhiang SM. Modulation of sodium iodide symporter expression and function by LY294002, Akti-1/2 and rapamycin in thyroid cells. Endocrine-Related Cancer. 2012;19:291–304. doi: 10.1530/ERC-11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool R, Hussain MU. MicroRNAs and human diseases: diagnostic and therapeutic potential. Cell and Tissue Research. 2014;358:1–15. doi: 10.1007/s00441-013-1787-3. [DOI] [PubMed] [Google Scholar]
- Marsee DK, Venkateswaran A, Tao H, Vadysirisack D, Zhang Z, Vandre DD, Jhiang SM. Inhibition of heat shock protein 90, a novel RET/PTC1-associated protein, increases radioiodide accumulation in thyroid cells. Journal of Biological Chemistry. 2004;279:43990–43997. doi: 10.1074/jbc.M407503200. [DOI] [PubMed] [Google Scholar]
- Nicolussi A, D’Inzeo S, Santulli M, Colletta G, Coppa A. TGF-β control of rat thyroid follicular cells differentiation. Molecular and Cellular Endocrinology. 2003;207:1–11. doi: 10.1016/s0303-7207(03)00238-7. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. Journal of Clinical Endocrinology and Metabolism. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi E, Kogai T, Kagechika H, Brent GA. Activation of the PI3 kinase pathway by retinoic acid mediates sodium/iodide symporter induction and iodide transport in MCF-7 breast cancer cells. Cancer Research. 2009;69:3443–3450. doi: 10.1158/0008-5472.CAN-08-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, Chang G, Li X, Li Q, Wang S, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE. 2014;9:e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocrine-Related Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- Pallante P, Visone R, Croce CM, Fusco A. Deregulation of microRNA expression in follicular-cell-derived human thyroid carcinomas. Endocrine-Related Cancer. 2006;17:F91–F104. doi: 10.1677/ERC-09-0217. [DOI] [PubMed] [Google Scholar]
- Pekary AE, Hershman JM. Tumor necrosis factor, ceramide, transforming growth factor-β1, and aging reduce Na+/I− symporter messenger ribonucleic acid levels in FRTL-5 cells. Endocrinology. 1998;139:703–712. doi: 10.1210/endo.139.2.5760. [DOI] [PubMed] [Google Scholar]
- Pesole G, Grillo G, Larizza A, Liuni S. The untranslated regions of eukaryotic mRNAs: structure, function, evolution and bioinformatic tools for their analysis. Briefings in Bioinformatics. 2000;1:236–249. doi: 10.1093/bib/1.3.236. [DOI] [PubMed] [Google Scholar]
- Pizzimenti S, Ferracin M, Sabbioni S, Toaldo C, Pettazzoni P, Dianzani MU, Negrini M, Barrera G. MicroRNA expression changes during human leukemic HL-60 cell differentiation induced by 4-hydroxynonenal, a product of lipid peroxidation. Free Radical Biology & Medicine. 2009;46:282–288. doi: 10.1016/j.freeradbiomed.2008.10.035. [DOI] [PubMed] [Google Scholar]
- Soon PS, Tacon LJ, Gill AJ, Bambach CP, Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson BG, et al. miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clinical Cancer Research. 2009;15:7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- Swierniak M, Wojcicka A, Czetwertynska M, Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M, Koperski L, de la Chapelle A, et al. In-depth characterization of the microRNA transcriptome in normal thyroid and papillary thyroid carcinoma. Journal of Clinical Endocrinology and Metabolism. 2013;98:E1401–E1409. doi: 10.1210/jc.2013-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocrine Pathology. 2007;18:163–173. doi: 10.1007/s12022-007-0023-7. [DOI] [PubMed] [Google Scholar]
- Ueda R, Kohanbash G, Sasaki K, Fujita M, Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze MT, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. PNAS. 2009;106:10746–10751. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulivi P, Foschi G, Mengozzi M, Scarpi E, Silvestrini R, Amadori D, Zoli W. Peripheral blood miR-328 expression as a potential biomarker for the early diagnosis of NSCLC. International Journal of Molecular Sciences. 2013;14:10332–10342. doi: 10.3390/ijms140510332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadysirisack DD, Venkateswaran A, Zhang Z, Jhiang SM. MEK signaling modulates sodium iodide symporter at multiple levels and in a paradoxical manner. Endocrine-Related Cancer. 2007;14:421–432. doi: 10.1677/erc.1.01263. [DOI] [PubMed] [Google Scholar]
- Wu ZS, Wu Q, Wang CQ, Wang XN, Wang Y, Zhao JJ, Mao SS, Zhang GH, Zhang N, Xu XC. MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer. 2010;10:542. doi: 10.1186/1471-2407-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Research. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian J, Weng Y. Upregulation of miR-195 increases the sensitivity of breast cancer cells to adriamycin treatment through inhibition of Raf-1. Oncology Reports. 2013;30:877–889. doi: 10.3892/or.2013.2532. [DOI] [PubMed] [Google Scholar]


