Abstract
The calsyntenins are atypical members of the cadherin superfamily that have been implicated in learning in C. elegans and memory formation in humans. As members of the cadherin superfamily, they could mediate cell-cell adhesion, although their adhesive properties have not been investigated. As an initial step in characterizing the calsyntenins, we have cloned clstn1, clstn2 and clstn3 from the zebrafish and determined their expression in the developing zebrafish nervous system. The three genes each have broad, yet distinct, expression patterns in the zebrafish brain. Each of the ectodomains mediates homophilic interactions through two, aminoterminal cadherin repeats. In bead sorting assays, the calsyntenin ectodomains do not exhibit hompphilic preferences. These data support the idea that calsyntenins could either act as adhesion molecules or as diffusible, homophilic or heterophilic ligands in the vertebrate nervous system.
Keywords: calsyntenin, zebrafish, adhesion, cadherin
Introduction
Differential cell-cell adhesion is required to regulate a wide range of cellular processes at every stage of development. Cell surface proteins belonging to large, multigene families mediate the majority of these adhesive interactions. The cadherin superfamily is a large and diverse collection of proteins related by the presence of extracellular repeats of a ~110 residue cadherin motif (Nollet et al., 2000, Redies, 2000). While the classical cadherins were originally identified on the basis of their calcium-dependent adhesive activity and are known to mediate predominantly homophilic interactions (Takeichi, 1988, 1990), much less is known about the adhesive activity of other superfamily members.
Recent results suggest that other cadherin subfamilies exhibit a range of adhesive properties and mechanisms. In contrast to classical cadherins, some superfamily members participate primarily in heterophilic interactions. Protocadherin-15 and Cadherin-23 interact heterophilically to form the tip-links found in stereocilia and mediate adhesion through a "handshake" mechanism involving the first two cadherin repeats of each protein (Kazmierczak et al., 2007, Sotomayor et al., 2012). Similarly, the atypical cadherins Fat and Dachsous have been shown to mediate heterophilic interactions (Ishiuchi et al., 2009). Moreover, an atomic model for Drosophlia N-cadherin (DN-cadherin) and sequence analysis also provide evidence for diversity in cadherin structure and adhesive interactions (Jin et al., 2012). Protocadherins comprise the largest group of molecules within the cadherin superfamily. Although their domain structure is similar to classical cadherins, direct testing of protocadherin ectodomains has generally failed to detect significant adhesive activity for a number of family members (Chen and Gumbiner, 2006, Morishita et al., 2006, Blevins et al., 2011). Recently, Protocadherin-24 and Mucin-like protocadherin were shown to interact heterophilically during assembly of the intestinal brush border (Crawley et al., 2014). In addition, some evidence suggests that protocadherins are components of multiprotein assemblies, which may be required for their adhesive activity (Emond et al., 2011). Thus, each group within the cadherin superfamily may exhibit very distinct adhesive properties and mechanisms.
The Calsyntenins (Clstns) are cadherin superfamily members, each consisting of two cadherin motifs near the amino terminus, a membrane proximal LGN domain, a single-pass transmembrane domain and a highly acidic intracellular domain (Hintsch et al., 2002). The intracellular domain binds calcium (Vogt et al., 2001) and also interacts with Kinesin Light Chain to act as a vesicular cargo adapter (Konecna et al., 2006, Araki et al., 2007). They appear to play important roles in intracellular transport and have been implicated in endosomal trafficking (Ponomareva et al., 2014), NMDA Receptor trafficking (Ster et al., 2014) and APP trafficking (Araki et al., 2003, Araki et al., 2004, Suzuki et al., 2006, Mizumaru et al., 2009). Calsyntenins have been implicated in learning and memory in both C. elegans (Ikeda et al., 2008, Ohno et al., 2014) and humans (Jacobsen et al., 2009, Zhang et al., 2009, Preuschhof et al., 2010). Like many type-I transmembrane proteins, Calsyntenins undergo proteolytic processing and ectodomain shedding (Vogt et al., 2001, Araki et al., 2004, Maruta et al., 2012). While the specific function of this cleavage is not known, the ectodomain alone is sufficient to rescue learning defects in C. elegans (Ikeda et al., 2008).
Though Clstns have two amino-terminal cadherin motifs, their role in cell adhesion is not well understood. The two repeats lack conserved cadherin sequence signatures and have an extended linker not present in other cadherin superfamily members (Hintsch et al., 2002). To investigate the adhesive properties of Clstns, we cloned zebrafish Clstn1, 2 and 3, with each exhibiting a distinct expression pattern in the developing nervous system. We used fusions of the ectodomains to the Fc (fragment, crystallizable) region of human IgG to determine whether Clstns mediate homophilic adhesion. We show that Clstn ectodomains (ECs) can mediate adhesion, and that the atypical cadherin repeats are both necessary and sufficient for this adhesive activity. We observe substantial heterophilic interactions among Clstns, suggesting that there is no homophilic specificity among these proteins. As the Clstn ectodomains are constitutitvely shed due to proteolytic cleavage (Vogt et al., 2001, Araki et al., 2004, Ikeda et al., 2008), it is not clear whether Clstns function as adhesion molecules, or whether their ectodomains act as secreted signaling molecules. These results provide the first characterization of the adhesive properties of this family of neural molecules with a demonstrated role in learning.
Materials and Methods
Fish Maintenance
Adult zebrafish (Danio rerio) and embryos of the Tübingen longfin and AB strains were maintained at ~28.5°C and staged according to Westerfield (1995). Embryos were raised in E3 embryo medium (Westerfield, 1995) with 0.003% phenylthiourea (Sigma-Aldrich, St. Louis, MO, USA) to inhibit pigment formation.
Identification of zebrafish calsyntenins
Human Calsyntenin protein sequences were used to BLAST the NCBI zebrafish genomic database, using tblastn. The zebrafish sequences identified from the BLAST search were used to design PCR primers corresponding to the predicted open reading frames of the zebrafish clstns:
Clstn1
Forward primer, 5'- GCGCTCGAGGCCACCATGCGCATTCGCGGGGTCAAACCTTTCGCT-3';
EC Reverse primer, 5'- CGGATCCCCGCCTCCGCCGTAGGTGAGAGTGGAGTCGTCCCACTC-3';
T7, 5'- GTAATACGACTCACTATAGGGCGAAAGCTCCACAGTGCCAGAGCTGGCACC-3'
Clstn2
Forward primer, 5'- GCGCTCGAGGCCACCATGAAGATGCGCGCGATAACAGCGATG-3';
EC Reverse primer, 5'- CGGATCCCCTCCGCCTCCATAGGGTAACGTAGACGGGTCCCACTC-3';
T7, 5'- GTAATACGACTCACTATAGGGCGACAGGTTCTGTGGCACAACCCAATCTGG-3'
Clstn3
Forward primer, 5'- GCGCTCGAGGGCACCATGGCCAGAATGAGTTTCCTATCCTTTCTC-3';
EC Reverse primer, 5'-CGGATCCCCTCCGCCTCCGTAGCGCCTAGGGGCGGAGTCTCTTCC-3';
T7, 5'-GTAATACGACTCACTATAGGGCGATGAAGCCACTGAATCGGGCACTACAGC-3'
Total RNA was extracted from 72 hours postfertilization (hpf) embryos using Trizol (Invitrogen, Carlsbad, CA, USA). Oligo-dT was used to prime the synthesis of cDNA from 1 µg of total RNA. First-strand cDNA synthesis was performed using Superscript III RT (Invitrogen) for 2 hours at 37°C. The coding sequences for the EC domains of each calsyntenin were then obtained by RT-PCR and TOPO-cloning (Invitrogen), and were sequenced.
Generation of Clstn1 deletion mutants
To make the Clstn1EC deletions, we performed overlapping PCR reactions, using Clstn1EC-Fc as the template DNA, an upstream forward primer in the vector sequence and a downstream reverse primer, 5'- GGTCTCCAACATGATGCTGGC-3'. The resulting fragments were then cut with AfeI, subcloned back into the Clstn1EC-Fc plasmid and screened for orientation. The Clstn1EC1–2 fragment was amplified and subcloned using XhoI and BamHI. The primers used were:
Clstn1ΔEC1
Forward primer, 5'- CTGGACCCTCCTCTGATAAAAGAGAAGTCCTACAAA-3';
Reverse primer, 5'- TTTGTAGGACTTCTCTTTTATCAGAGGAGGGTCCAG-3';
Clstn1ΔEC2
Forward primer, 5'- GACAGCATCATGAAGGTGCAAGGATTTAACAAAAGG-3';
Reverse primer, 5'- CCTTTTGTTAAATCCTTGCACCTTCATGATGCTGTC-3';
Clstn1Δ(EC1–2)
Forward primer, 5'- CCTTTTGTTAAATCCTTGCACCTTCATGATGCTGTC-3';
Reverse primer, 5'- CCTTTTGTTAAATCCTTGTATCAGAGGAGGGTCCAG-3';
Clstn1EC1–2
Forward primer, 5'- GCGCTCGAGGCCACCATGCGGATCCGCGGGGTCAAACTT-3';
Reverse primer, 5'- CGCGGATCCCCTCCACCTCCCAGATGCATGCTGGGAAACAGCGC-3';
Bead aggregation and bead sorting assays
Bead aggregations were performed essentially as described previously (Sivasankar et al., 2009, Biswas et al., 2010). Plasmids encoding the Fc-fusion proteins were independently transfected into HEK293 cells using FuGene 6 (Roche Applied Science, Indianapolis, IN, USA) according to the manufacturer’s instructions. After 24 hours, the growth medium was replaced with serum-free medium. After an additional 48 hours, the culture medium containing the secreted Fc-fusion proteins was collected. The media was concentrated using Amicon Ultra-15 centrifugal filters (EMD Millipore, Billerica, MA, USA) and incubated with 1.5 µL Protein A-Dynabeads (Invitrogen) for 2 hours with gentle agitation at 4°C. The beads were washed extensively in binding buffer (50 mM Tris, 100 mM NaCl, 10 mM KCl, 0.2% BSA, pH 7.4). The beads were split into two tubes and either 2 mM EDTA or 2 mM CaCl2 (final concentrations) was added to each tube, the beads were mixed thoroughly, and then transferred to glass depression slides. Beads were allowed to aggregate with gentle rocking for 1 hour. To document bead aggregation, images were collected on an Axiostar microscope (Zeiss, Thornwood, NY, USA) using a 10× objective. Bead sorting assays were performed essentially the same as bead aggregation assays, except that the Protein A-magnetic beads were fluorescent (green: screenMAG/G-Protein A or red: screenMAG/OP-Protein A; Chemicell, Berlin, Germany).
Whole mount in situ hybridization
The ectodomain of each calsyntenin was amplified by PCR. A T7 RNA Polymerase binding site was included in each of the reverse primers, and these PCR products were used as templates for in vitro transcription (Promega, Madison, WI, USA). Antisense riboprobes were labeled with digoxygenin-dUTP (Roche). Whole mount in situ hybridizations were carried out using standard methods (Westerfield, 1995). Briefly, embryos were fixed at 4°C overnight in 4% paraformaldehyde in PBS, dehydrated in a methanol series and stored in 100% methanol overnight at −20°C. They were rehydrated in decreasing concentrations of methanol and embryos 24 hpf and older were permeabilized using Proteinase K (10µg/ml, Roche). Embryos were refixed in 4% paraformaldehyde prior to hybridization. Digoxygenin-dUTP-labeled riboprobe was added to the hybridization buffer at a final concentration of 200 ng/ml and hybridization was carried out at 65°C overnight. Alkaline phosphatase-conjugated antidigoxygenin Fab fragments (Roche) were used at 1:5000 dilution. NBT/BCIP (Roche) was used for the coloration reaction. Images were captured on a MZ16F stereomicroscope (Leica Microsystems, Wetzlar, Germany).
Results
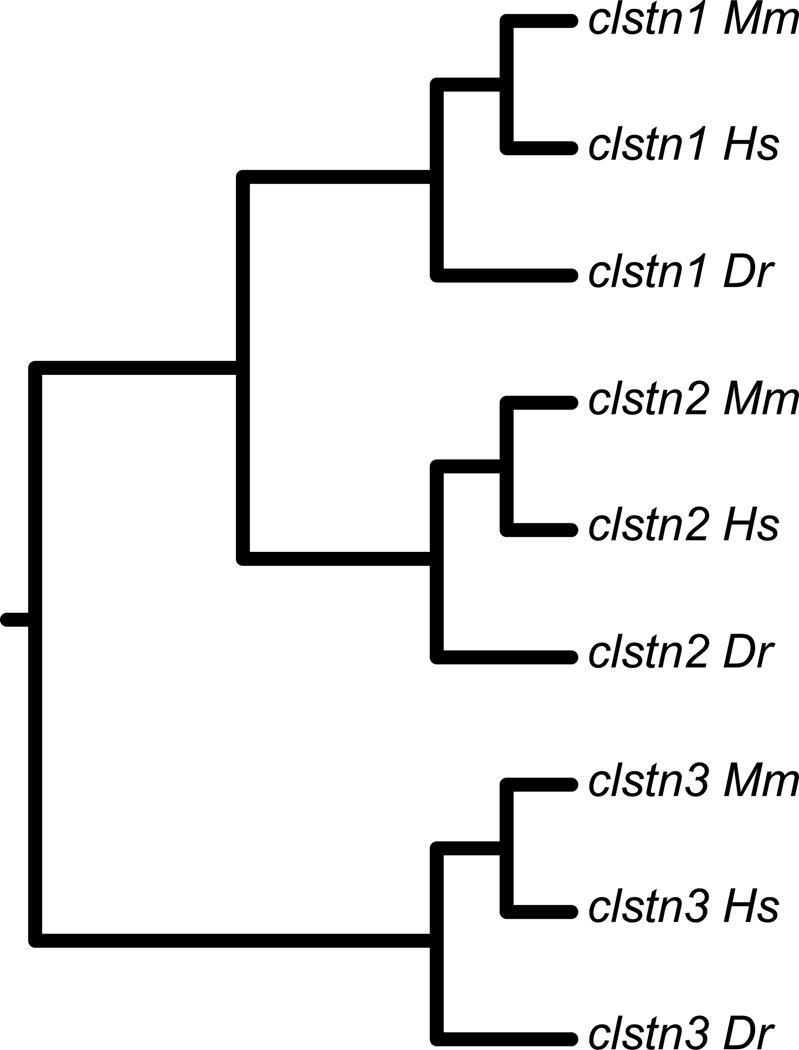
To obtain zebrafish calsyntenins, we first cloned the three zebrafish calsyntenin genes in silico, then used these sequences to design gene specific primers for RT-PCR. We used the protein sequences for the three human calsyntenins to BLAST zebrafish genomic and EST databases. From these sequence data, we assembled full open reading frames for each calsyntenin gene. We then cloned the ectodomains from each of the three genes using RT-PCR (Fig. 1). The calsyntenins each have two, amino-terminal cadherin motifs and a Leu-Gly-Asn repeat-enriched (LGN) domain in the extracellular domain, a single-pass transmembrane segment and a short intracellular region (Fig. 1). The cadherin motifs are separated by an extended linker and lack some of the sequence features conserved in other cadherin subfamilies. Comparsion to sequences from mouse and humans shows that the three zebrafish calsyntenin sequences correspond to clstn1, clstn2 and clstn3 (Fig. 2).
Figure 1. Protein sequences and alignments for the zebrafish calsyntenins.
The three calsyntenin proteins exhibit two, amino-terminal cadherin repeats (orange and purple lines), an LGN domain, (yellow line), a single pass transmembrane segment (green line) and a short cytoplasmic tail.
Figure 2. Phylogenetic analysis of zebrafish Calsyntenins.
The protein sequences of the zebrafish calsyntenins were aligned with those of human and mouse, then analyzed by MAFFT/ClustalX. Each of the zebrafish calsyntenin protein sequences clusters with a human and mouse pair, indicating that the zebrafish genes encode homologs for Clstn1, Clstn2 and Clstn3.
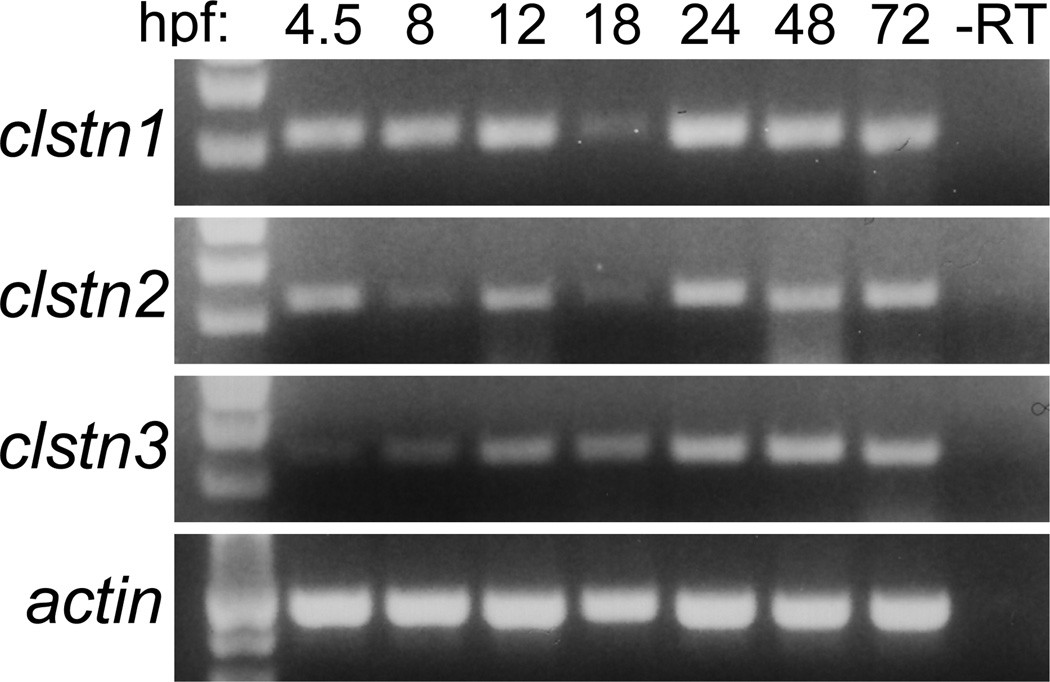
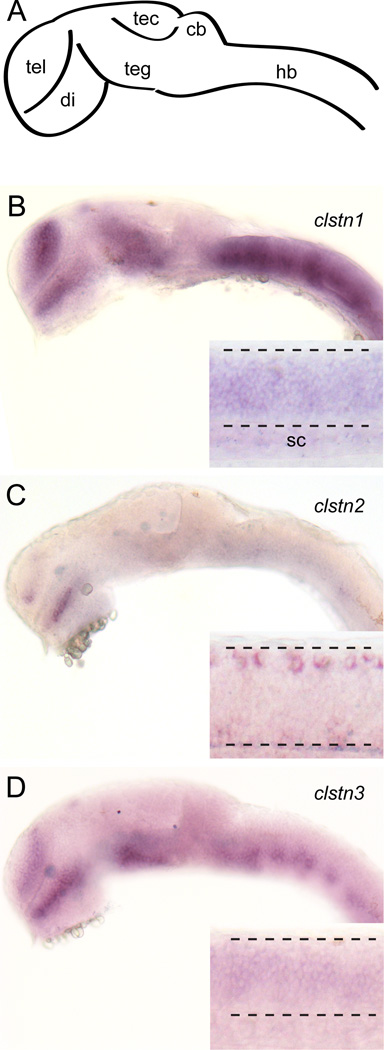
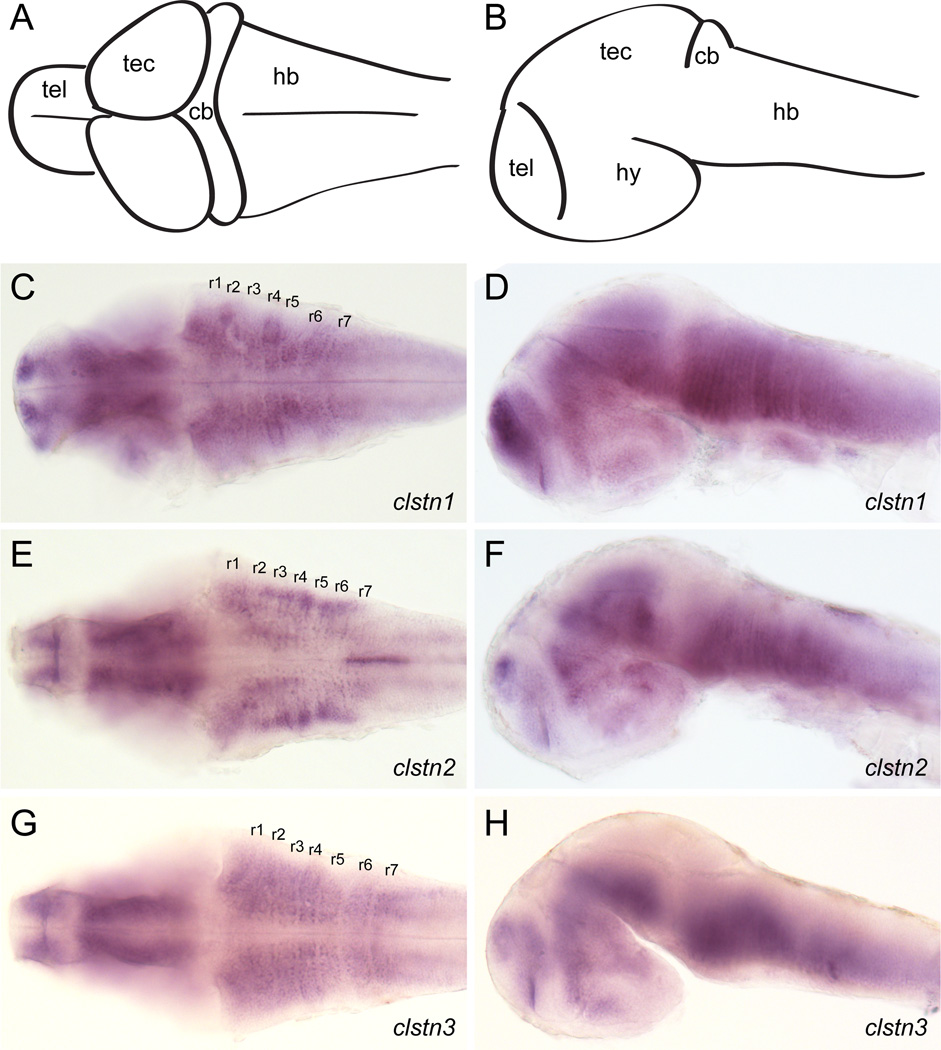
As a first step in investigating the roles of the calsyntenins in neural development, we determined their temporal and spatial expression using RT-PCR and in situ hybridization. To characterize the temporal expression of the calsyntenins, we performed RT-PCR using primers directed against the intracellular domains (Fig. 3). Expression of each calsyntenin is detected at 4.5 hour post-fertilization (hpf), the earliest timepoint assayed, and continues into the larval stage (Fig. 3). Though present at early stages, the expression of clstn3 is initially weak and steadily increases as development proceeds. At 24 hpf, the three clstn genes exhibit similar, yet distinct patterns. The expression pattern for clstn1 is similar to what was described previously (Ponomareva et al., 2014). Clstn1 and clstn3 are expressed strongly in the forebrain, midbrain and hindbrain (Fig. 4). Clstn1 expression appears broader that that of clstn3 in the telencephalon and extends more dorsally in the midbrain. By contrast, the expression of clstn2 is weak in the brain, with some expression detectable in the telencephalon and diencephalon (Fig. 4). In the spinal cord, both clstn1 and clstn3 are expressed broadly (Fig. 4B & D, insets), while clstn2 expression is restricted to an array of large dorsal neurons and small clusters of ventral neurons, likely Rohon-Beard cells and Primary motoneurons, respectively (Fig. 4C, inset). By 48 hpf, the expression of each clstn is expressed broadly in the brain (Fig. 5). Each is expressed strongly in the telencephalon and the midbrain, though clstn3 expression is absent in the optic tectum (Fig. 5H). All three clstn genes exhibit segmental expression in the hindbrain, with distinct patterns (Fig. 5C,E,G).
Figure 3. Temporal expression of calsyntenins during zebrafish development.
RT-PCR was performed using cDNA prepared from 4.5 hpf to 72 hpf embryos. Expression was evident as early as 4.5 hpf and continued throughout all developmental stages analyzed. Actin was used as a control.
Figure 4. Expression of calsyntenins in 1 day old zebrafish embryos.
A. Diagram of a lateral view of a 1 day old zebrafish brain. Abbreviations: tel, telencephalon; di, diencephalon; teg, tegmentum; tec, optic tectum; cb, cerebellum; hb, hindbrain.
B–D. Expression of clstn1 (B), clstn2 (C), and clstn3 (D) in the brain and spinal cord of 1 day zebrafish embryos, as determined by in situ hybridization (inset; sc, spinal cord).
Figure 5. Expression of calsyntenins in 2 day old zebrafish embryos.
A, B. Diagrams of a lateral view (A) and dorsal view (B) of a 2 day old zebrafish brain. Abbreviations: tel, telencephalon; hy, hypothalamus; tec, optic tectum; cb, cerebellum; hb, hindbrain.
C, D. Expression of clstn1 in the brain of 2 day zebrafish embryos, as determined by in situ hybridization. Expression is evident in all the major subdivisions of the brain and is particularly strong in the telencephalon, as well as showing a ladder-like expression in the hindbrain. Rhombomeres are labeled r1–r7.
E, F. Expression of clstn2 in the brain of 2 day zebrafish embryos, as determined by in situ hybridization. Like clstn1, expression is evident in all the major subdivisions of the brain. Segmental labeling of lateral cell clusters is apparent in the hindbrain.
G, H. Expression of clstn3 in the brain 2 day zebrafish embryos, as determined by in situ hybridization. The overall pattern of expression is comparable to the other calsyntenins, though clstn3 appears to be absent from the optic tectum.
Calsyntenins mediate homophilic adhesion through their cadherin domains
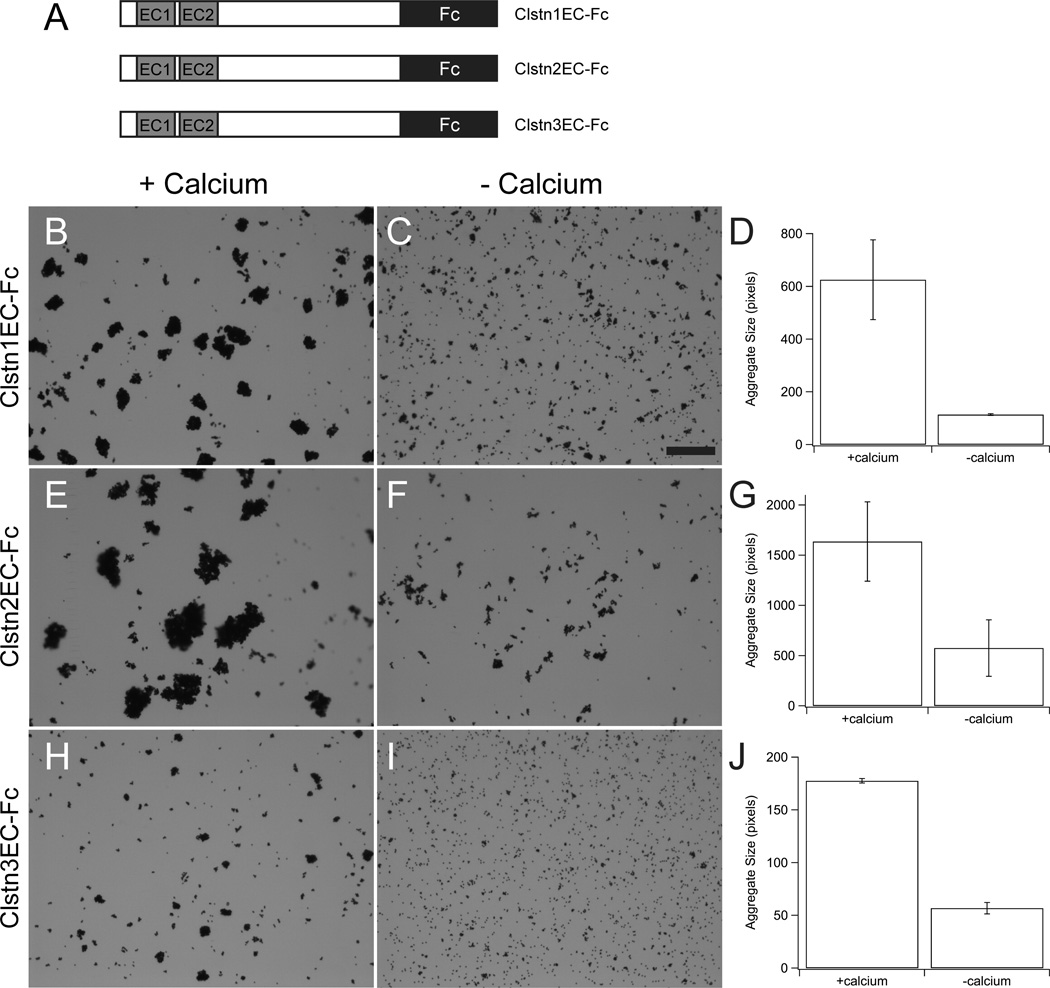
As members of the cadherin superfamily, it is possible that the calsyntenins mediate homophilic cell adhesion. To test this, we fused the ectodomain sequences of Clstn1, Clstn2 and Clstn3 to the Fc region of human IgG (Fig. 6A). Plasmids encoding each of these fusion proteins were transfected into HEK293 cells and the secreted ClstnEC-Fc fusions were captured on magnetic Protein-A beads. Using standard bead aggregation assays (Emond and Jontes, 2014), each of the three ClstnEC-Fc fusions mediated calcium-dependent bead aggregation (Fig. 6B,E,H). However, in each case there appeared to be some residual aggregation in the absence of calcium (Fig. 6C,D,F,G,I,J).
Figure 6. Calsyntenin ectodomains mediate bead aggregation.
A. The ectodomain of each calsyntenin was fused to the Fc portion of human IgG and the CstnEC-Fc fusions were used in bead aggregation assays.
B–H. Each of the three Calsyntenin ectodomains, Clstn1EC-Fc (B, C, D), Clstn2EC-Fc (E, F, G) and Clstn3EC-Fc (H, I, J), are able to mediate calcium-dependent bead aggregation (n=3 for each condition). The aggregates formed by Clstn3EC-Fc (F) were smaller than those formed by either Clstn1EC-Fc (B) or Clstn2EC-Fc (D). Each fusion-protein also exhibits some calcium-independent aggregation (C, E, G). In each case, the calcium-dependent increase was statistically significant (p<0.05, Student's T-test). Scale bar = 50 µm.
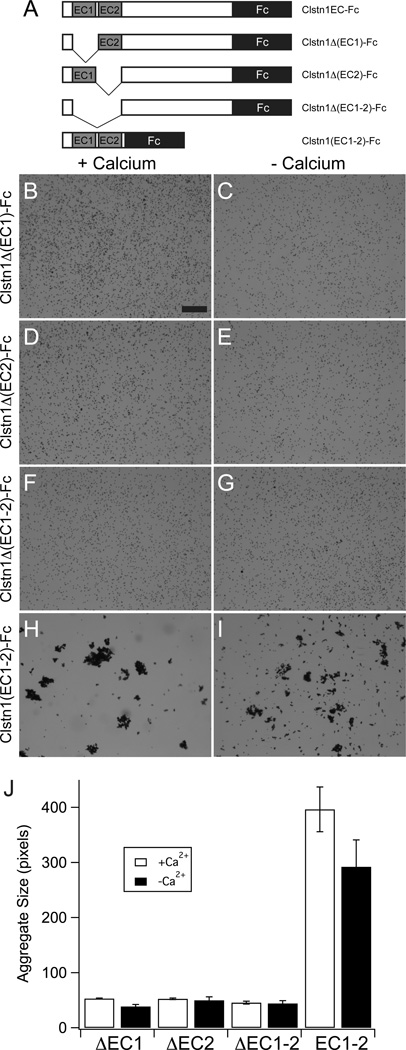
To determine whether the two amino-terminal cadherin domains are responsible for bead aggregation, we generated a domain deletion series for Clstn1EC (Fig. 7A). Calcium-dependent bead aggregation was completely abolished by deletion of either cadherin domain alone, or both together (Fig. 7B,D,F,J). Moreover, the low levels of calcium-independent adhesion were also eliminated (Fig.7C,E,G,J). To determine whether the cadherin repeats were sufficient for adhesion, we made Fc fusions using the coding sequence for the Clstn1 amino-terminal region, including the cadherin domains (Fig. 7A, Clstn1(EC1–2)-Fc). This truncated ectodomain was sufficient to mediate robust bead aggregation (Fig. 7H,J). We also observed substantial calcium-independent aggregation (Fig. 7I,J).
Figure 7. Cadherin repeats are required for bead aggregation by Calsyntenin-1.
A. A series of domain deletions of the Clstn1EC-Fc were generated to test the role of the cadherin repeats, EC1 and EC2.
B–G. Deletion of EC1 (B,C), EC2 (D,E) or EC1–EC2 (F,G) abolish calcium-dependent and calcium-independent bead aggregation. Scale bar = 50 µm.
H, I. The cadherin repeats EC1–EC2 are sufficient to mediate calcium-dependent bead aggregation. There is also an increase in calcium-independent bead aggregation.
J. Deletion of either cadherin repeat is sufficient to abolish bead aggregation, while the two cadherin repeats are sufficient for bead aggregation (n=3 for each condition). The two cadherin repeats exhibit only a modest, but significant, calcium-dependence (p=0.04, Student's T-test).
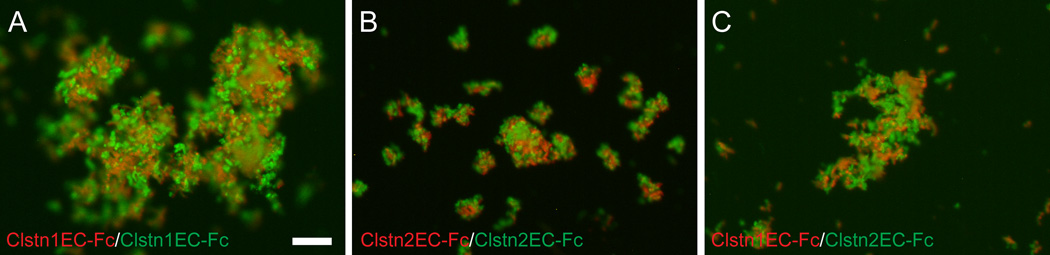
Classical cadherins are well known to exhibit homophilic preferences in their adhesive interactions. While cadherins do exhibit heterophilic interactions, different cadherins are able to mediate cell sorting. To determine whether the calsyntenins are preferentially homophilic, we used the Fc-fusions of Clstn1 and Clstn2 to perform bead sorting assays. The ClstnEC-Fc fusions for Clstn1 and Clstn2 were used to coat fluorescent Protein-A beads (Fig. 8). First, Clstn1EC-Fc was separately bound to red fluorescent beads and green fluorescent beads. These were dissociated in the absence of calcium, mixed, and then allowed to aggregate in the presence of calcium. As expected, the red and green beads, both of which were coated with Clstn1EC-Fc, formed intermixed aggregates (Fig. 8A). Similar results were obtained with Clstn2EC-Fc (Fig. 8B). To assess whether Clstn1 and Clstn2 would interact, we bound Clstn1EC-Fc to red fluorescent beads and Clstn2EC-Fc to green fluorescent beads. When allowed to aggregate in the presence of calcium, they formed intermixed aggregates (Fig. 8C), similar to what was observed for Clstn1EC-Fc (Fig. 8A) and Clstn2EC-Fc (Fig. 8B). Thus, Clstn1 and Clstn2 are able to interact heterophilically.
Figure 8. Heterophilic interactions by calsyntenin ectodomains.
A. The ectodomain of Clstn1 (Clstn1EC-Fc) was used to coat green or red fluorescent beads. Coated beads formed mixed aggregates. Scale bar = 20 µm.
B. As for Clstn1EC-Fc (A), green and red beads coated with Clstn2EC-Fc intermixed.
C. Red fluorescent beads were coated with Clstn1EC-Fc and green fluorescent beads were coated with Clstn2EC-Fc. When allowed to aggregate in the presence of calcium, the green and red beads intermixed, as in A and B.
Discussion
The classical cadherins were discovered on the basis of their ability to confer calciumdependent, homophilic cell-cell adhesion (Takeichi, 1977, Hatta et al., 1988, Nose et al., 1988). As revealed by extensive biophysical and structural analysis, classical cadherins mediate adhesion through interactions of their most membrane-distal cadherin motif (Boggon et al., 2002, Vendome et al., 2011). The primary mode of adhesion involves the formation of a "strand-swap" dimer, with a reciprocal exchange of the Trp2 residue of the Aβ strand of one cadherin embedded into a hydrophobic pocket on the EC1 of the partner cadherin (Boggon et al., 2002, Vendome et al., 2011). In contrast, the related protocadherins do not have the conserved tryptophan on their EC1 domains and evidence suggests that extracellular interactions employ divergent mechanisms. More recently, cadherin family members have been identified that mediate heterophilic interactions. The atypical cadherns Fat and Dachsous appear to interact heterophilically (Ishiuchi et al., 2009), as do Protocadherin-15 and Cadherin-23 (Kazmierczak et al., 2007, Sotomayor et al., 2012), which comprise the tip links of stereocilia. Thus, members of the cadherin superfamily may mediate extracellular interactions through a diverse array of mechanisms.
The Calsyntenins comprise a small family of atypical cadherins that are strongly expressed in the developing nervous system (Hintsch et al., 2002). They have been shown to localize to synaptic terminals and to play a role in memory, in both C. elegans and in human. In mammals, there are three clstn genes: clstn1, clstn2 and clstn3. In this study, we cloned clstn1, clstn2 and clstn3 from zebrafish and characterized both their patterns of expression in the developing nervous system, and the adhesive properties of their ectodomains. Although they are members of the cadherin superfamily, their adhesive interactions have not previously been investigated. Here, we show that: 1) the Clstn ectodomains mediate homophilic adhesive interactions, 2) these interactions are mediated by the two cadherin motifs present in the aminoterminal portion of the ectodomains, 3) the ectodomains exhibit both homophilic and heterophilic interactions, and 4) adhesion by the cadherin motifs is less calcium-dependent than has been found for other cadherin subfamilies. Thus, the adhesive properties of these molecules differ significantly from those of previously studied cadherins.
The presence of cadherin repeats near the amino-terminus suggested that calsyntenins could play a role in cell-cell adhesion. Our data support the conclusion that calysntenins can mediate adhesive interactions through their cadherin repeats. Deletion of either cadherin motif abolishes bead aggregation by the Clstn1 ectodomain, and these motifs alone are sufficient for robust aggregation. Though each of the calsyntenins mediates homophilic adhesion, fluorescent bead sorting assays do not indicate a preference for homophilic over heterophilic interactions. In contrast, classical cadherins are also promiscuous in their interactions, but exhibit homophilic preferences. As bead sorting assays are not quantitative and do not measure relative affinities, they may not be suitable for revealing slight homophilic preferences among these proteins.
Cadherins were discovered through their role in mediating calcium-dependent cell adhesion. This is due to the binding of three calcium ions at the interfaces between successive cadherin repeats, which rigidifies the ectodomain. Although the calsyntenins share some of the conserved calcium-binding motifs, their adhesion is only modestly calcium-dependent. Moreover, this calcium-dependence disappears entirely if only the amino-terminal cadherin motifs are used: bead aggregates are of comparable size in the presence or absence of calcium. It appears that calsyntenin adhesive interactions are less sensitive to any changes in the conformational state of the protein brought about by loss of calcium.
The biological functions of calsyntenins are not well understood. Early in development, calsyntenins appear to play important roles in intracellular transport (Konecna et al., 2006, Araki et al., 2007, Ludwig et al., 2009, Vagnoni et al., 2012, Ponomareva et al., 2014, Ster et al., 2014). In more mature neurons, calsyntenins are enriched in postsynaptic compartments (Vogt et al., 2001) and may play roles in synapse formation (Pettem et al., 2013, Um et al., 2014). As occurs for many type-I transmembrane proteins, calsyntenins are subject to proteolytic processing and ectodomain shedding (Vogt et al., 2001, Araki et al., 2004, Maruta et al., 2012), and some evidence suggests that calsyntenins are cleaved in transit to the cell surface (Maruta et al., 2012). In C. elegans mutations in the casy-1 gene, encoding the sole calsyntenin in worms, affects several forms of learning (Ikeda et al., 2008, Hoerndli et al., 2009, Ohno et al., 2014). Casy-1 function is required in the mature neural circuit and can be rescued by the ectodomain alone (Ikeda et al., 2008). While some evidence suggests a potential role for calsyntenins in synaptogenesis (Pettem et al., 2013, Um et al., 2014), the extensive ectodomain shedding does not seem consistent with a primary role in stable cell-cell adhesion. It is possible that, rather than mediating extracellular interactions, the cadherin repeats could assemble cis-oligomers that would be important for intracellular trafficking. Thus, it isn't clear whether calsyntenins function as cell adhesion molecules, secreted ligands or transport chaperones, or what distinct biological functions that the intact and cleaved forms mediate. Our data are consistent with the ideas that membrane associated calsyntenins function as adhesion molecules and/or that the shed ectodomains act as homophilic or heterophilic signaling molecules.
Highlights.
-
-
zebrafish calsyntenins exhibit distinct expression patterns in the developing zebrafish
-
-
zebrafsih calsyntenins mediate homophilic adhesion
-
-
the two, amino-terminal cadherin repeats are necessary and sufficient for adhesion
-
-
adhesion requires both cadherin repeats
-
-
adhesive interactions are promiscuous among the calsyntenins
ACKNOWLEDGMENTS
This work was supported by an NIH grant (R21MH098463) to J.D.J and a Neurosciences Core grant (P30 NS045758). H.O.-M. was supported by an NIH Postbacculaureate Research Education Program (PREP) grant R25 GM089571 to OSU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, Suzuki T. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Miyagi N, Kato N, Yoshida T, Wada S, Nishimura M, Komano H, Yamamoto T, De Strooper B, Yamamoto K, Suzuki T. Coordinated metabolism of Alcadein and amyloid beta-protein precursor regulates FE65-dependent gene transactivation. J Biol Chem. 2004;279:24343–24354. doi: 10.1074/jbc.M401925200. [DOI] [PubMed] [Google Scholar]
- Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. J Biol Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- Biswas S, Emond MR, Jontes JD. Protocadherin-19 and N-cadherin interact to control cell movements during anterior neurulation. J Cell Biol. 2010;191:1029–1041. doi: 10.1083/jcb.201007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins CJ, Emond MR, Biswas S, Jontes JD. Differential expression, alternative splicing, and adhesive properties of the zebrafish delta1-protocadherins. Neuroscience. 2011;199:523–534. doi: 10.1016/j.neuroscience.2011.09.061. [DOI] [PubMed] [Google Scholar]
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 2002;296:1308–1313. doi: 10.1126/science.1071559. [DOI] [PubMed] [Google Scholar]
- Chen X, Gumbiner BM. Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J Cell Biol. 2006;174:301–313. doi: 10.1083/jcb.200602062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Shifrin DA, Jr, Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, Kachar B, Tyska MJ. Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell. 2014;157:433–446. doi: 10.1016/j.cell.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Biswas S, Blevins CJ, Jontes JD. A complex of Protocadherin-19 and N-cadherin mediates a novel mechanism of cell adhesion. J Cell Biol. 2011;195:1115–1121. doi: 10.1083/jcb.201108115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emond MR, Jontes JD. Bead aggregation assays for the characterization of putative cell adhesion molecules. J Vis Exp. 2014 doi: 10.3791/51762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Nose A, Nagafuchi A, Takeichi M. Cloning and expression of cDNA encoding a neural calcium-dependent cell adhesion molecule: its identity in the cadherin gene family. J Cell Biol. 1988;106:873–881. doi: 10.1083/jcb.106.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintsch G, Zurlinden A, Meskenaite V, Steuble M, Fink-Widmer K, Kinter J, Sonderegger P. The calsyntenins--a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci. 2002;21:393–409. doi: 10.1006/mcne.2002.1181. [DOI] [PubMed] [Google Scholar]
- Hoerndli FJ, Walser M, Frohli Hoier E, de Quervain D, Papassotiropoulos A, Hajnal A. A conserved function of C. elegans CASY-1 calsyntenin in associative learning. PloS one. 2009;4:e4880. doi: 10.1371/journal.pone.0004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda DD, Duan Y, Matsuki M, Kunitomo H, Hutter H, Hedgecock EM, Iino Y. CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. PNAS. 2008;105:5260–5265. doi: 10.1073/pnas.0711894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Misaki K, Yonemura S, Takeichi M, Tanoue T. Mammalian Fat and Dachsous cadherins regulate apical membrane organization in the embryonic cerebral cortex. J Cell Biol. 2009;185:959–967. doi: 10.1083/jcb.200811030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen LK, Picciotto MR, Heath CJ, Mencl WE, Gelernter J. Allelic variation of calsyntenin 2 (CLSTN2) modulates the impact of developmental tobacco smoke exposure on mnemonic processing in adolescents. Biol Psychiatry. 2009;65:671–679. doi: 10.1016/j.biopsych.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Walker MA, Felsovalyi K, Vendome J, Bahna F, Mannepalli S, Cosmanescu F, Ahlsen G, Honig B, Shapiro L. Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca(2)+free interdomain linkers. PNAS. 2012;109:E127–E134. doi: 10.1073/pnas.1117538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, Kachar B. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- Konecna A, Frischknecht R, Kinter J, Ludwig A, Steuble M, Meskenaite V, Indermuhle M, Engel M, Cen C, Mateos JM, Streit P, Sonderegger P. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell. 2006;17:3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A, Blume J, Diep TM, Yuan J, Mateos JM, Leuthauser K, Steuble M, Streit P, Sonderegger P. Calsyntenins mediate TGN exit of APP in a kinesin-1-dependent manner. Traffic. 2009;10:572–589. doi: 10.1111/j.1600-0854.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- Maruta C, Saito Y, Hata S, Gotoh N, Suzuki T, Yamamoto T. Constitutive cleavage of the single-pass transmembrane protein alcadeinalpha prevents aberrant peripheral retention of Kinesin-1. PloS one. 2012;7:e43058. doi: 10.1371/journal.pone.0043058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumaru C, Saito Y, Ishikawa T, Yoshida T, Yamamoto T, Nakaya T, Suzuki T. Suppression of APP-containing vesicle trafficking and production of beta-amyloid by AID/DHHC-12 protein. J Neurochem. 2009;111:1213–1224. doi: 10.1111/j.1471-4159.2009.06399.x. [DOI] [PubMed] [Google Scholar]
- Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, Akutsu H, Yamaguchi T, Yagi T, Ikegami T. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem. 2006;281:33650–33663. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J Mol Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Nose A, Nagafuchi A, Takeichi M. Expressed recombinant cadherins mediate cell sorting in model systems. Cell. 1988;54:993–1001. doi: 10.1016/0092-8674(88)90114-6. [DOI] [PubMed] [Google Scholar]
- Ohno H, Kato S, Naito Y, Kunitomo H, Tomioka M, Iino Y. Role of synaptic phosphatidylinositol 3-kinase in a behavioral learning response in C. elegans. Science. 2014;345:313–317. doi: 10.1126/science.1250709. [DOI] [PubMed] [Google Scholar]
- Pettem KL, Yokomaku D, Luo L, Linhoff MW, Prasad T, Connor SA, Siddiqui TJ, Kawabe H, Chen F, Zhang L, Rudenko G, Wang YT, Brose N, Craig AM. The specific alpha-neurexin interactor calsyntenin-3 promotes excitatory and inhibitory synapse development. Neuron. 2013;80:113–128. doi: 10.1016/j.neuron.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva OY, Holmen IC, Sperry AJ, Eliceiri KW, Halloran MC. Calsyntenin-1 Regulates Axon Branching and Endosomal Trafficking during Sensory Neuron Development In Vivo. J Neurosci. 2014;34:9235–9248. doi: 10.1523/JNEUROSCI.0561-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuschhof C, Heekeren HR, Li SC, Sander T, Lindenberger U, Backman L. KIBRA and CLSTN2 polymorphisms exert interactive effects on human episodic memory. Neuropsychologia. 2010;48:402–408. doi: 10.1016/j.neuropsychologia.2009.09.031. [DOI] [PubMed] [Google Scholar]
- Redies C. Cadherins in the central nervous system. Prog Neurobiol. 2000;61:611–648. doi: 10.1016/s0301-0082(99)00070-2. [DOI] [PubMed] [Google Scholar]
- Sivasankar S, Zhang Y, Nelson WJ, Chu S. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–1081. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature. 2012;492:128–132. doi: 10.1038/nature11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ster J, Steuble M, Orlando C, Diep TM, Akhmedov A, Raineteau O, Pernet V, Sonderegger P, Gerber U. Calsyntenin-1 regulates targeting of dendritic NMDA receptors and dendritic spine maturation in CA1 hippocampal pyramidal cells during postnatal development. J Neurosci. 2014;34:8716–8727. doi: 10.1523/JNEUROSCI.0144-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Araki Y, Yamamoto T, Nakaya T. Trafficking of Alzheimer's disease-related membrane proteins and its participation in disease pathogenesis. J Biochem. 2006;139:949–955. doi: 10.1093/jb/mvj121. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol. 1977;75:464–474. doi: 10.1083/jcb.75.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherins: a molecular family important in selective cell-cell adhesion. Annu Rev Biochem. 1990;59:237–252. doi: 10.1146/annurev.bi.59.070190.001321. [DOI] [PubMed] [Google Scholar]
- Um JW, Pramanik G, Ko JS, Song MY, Lee D, Kim H, Park KS, Sudhof TC, Tabuchi K, Ko J. Calsyntenins function as synaptogenic adhesion molecules in concert with neurexins. Cell reports. 2014;6:1096–1109. doi: 10.1016/j.celrep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoni A, Perkinton MS, Gray EH, Francis PT, Noble W, Miller CC. Calsyntenin-1 mediates axonal transport of the amyloid precursor protein and regulates Abeta production. Hum Mol Genet. 2012;21:2845–2854. doi: 10.1093/hmg/dds109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, Honig B. Molecular design principles underlying beta-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18:693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt L, Schrimpf SP, Meskenaite V, Frischknecht R, Kinter J, Leone DP, Ziegler U, Sonderegger P. Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol Cell Neurosci. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Zhang H, Kranzler HR, Poling J, Gruen JR, Gelernter J. Cognitive flexibility is associated with KIBRA variant and modulated by recent tobacco use. Neuropsychopharmacology. 2009;34:2508–2516. doi: 10.1038/npp.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]