Abstract
Chikungunya fever is a mosquito-borne disease of key public health importance in tropical and subtropical countries. Although severe joint pain is the most distinguishing feature of chikungunya fever, diagnosis remains difficult because the symptoms of chikungunya fever are shared by many pathogens, including dengue fever. The present study aimed to develop a new immunochromatographic diagnosis test for the detection of chikungunya virus antigen in serum. Mice were immunized with isolates from patients with Thai chikungunya fever, East/Central/South African genotype, to produce mouse monoclonal antibodies against chikungunya virus. Using these monoclonal antibodies, a new diagnostic test was developed and evaluated for the detection of chikungunya virus. The newly developed diagnostic test reacted with not only the East/Central/South African genotype but also with the Asian and West African genotypes of chikungunya virus. Testing of sera from patients suspected to have chikungunya fever in Thailand (n = 50), Laos (n = 54), Indonesia (n = 2), and Senegal (n = 6) revealed sensitivity, specificity, and real-time PCR (RT-PCR) agreement values of 89.4%, 94.4%, and 91.1%, respectively. In our study using serial samples, a new diagnostic test showed high agreement with the RT-PCR within the first 5 days after onset. A rapid diagnostic test was developed using mouse monoclonal antibodies that react with chikungunya virus envelope proteins. The diagnostic accuracy of our test is clinically acceptable for chikungunya fever in the acute phase.
INTRODUCTION
Chikungunya virus (CHIKV), the causative agent for chikungunya fever (CF), belongs to the genus Alphavirus of the family Togaviridae. It is an enveloped virus with a single-stranded positive-sense RNA genome (1). There are three genotypes of CHIKV: West African, Asian, and East/Central/South African (ECSA) (2). CF is characterized by the abrupt onset of fever, headache, vomiting, rash, myalgia, and severe arthralgia (3). Early diagnosis of CHIKV infection remains difficult because the clinical symptoms of CF are similar to those of dengue fever (DF). CF and DF are mosquito-borne diseases of public health importance in tropical and subtropical countries (4). These two diseases now cocirculate in many countries (5). Differentiating between CF and DF is paramount not only for its diagnostic and epidemiological relevance but also for the significantly different prognoses of these diseases. However, in resource-limited settings, sophisticated laboratory tests to distinguish between these infections may be unavailable or costly, necessitating epidemiological and symptom-based approaches for diagnosis.
Several methods have been used to diagnose CHIKV infection. Enzyme-linked immunosorbent assay (ELISA), real-time PCR (RT-PCR), and virus isolation can be performed to arrive at a definitive diagnosis or to clarify the immune response, but these methods are not widely performed in hospitals because they require specialist equipment and laboratory skills. An anti-CHIKV IgM detection kit is used to support clinical findings in the assessment of patients with suspected CHIKV infection (6). However, the sensitivity of IgM detection kits is limited for the majority of patients in the acute stage of illness (days 1 to 5) (7). For the serological diagnosis to justify the infection, paired sera are needed to confirm the rising of specific antibody titer in convalescence serum. Therefore, the development of new antigen-based diagnostic assays is critical for a rapid and reliable clinical diagnosis on admission.
The immunochromatographic (IC) assay with monoclonal antibodies (MAbs) is used as a tracer to detect antigens. This assay has been widely applied for the diagnosis of several human diseases, such as dengue virus infection (8), rotavirus infection (9), norovirus infection (10), and rabies (11). Considering the successful application of this system in other diseases, we developed a rapid antigen detection test using the IC method, with MAbs against the envelope protein of CHIKV. The performance of the IC test was evaluated using clinical isolates and human serum samples and was compared with the results of other diagnostic methods for CHIKV. Our data indicated that the diagnostic accuracy of the IC test targeting CHIKV antigen was sufficient to consider this assay a clinically acceptable method for the diagnosis of CHIKV infection in the acute phase.
MATERIALS AND METHODS
Cells and virus.
Vero, BHK-21, and B7 (BALB/c mouse cell line) cells (12) were maintained in Eagle's minimum essential medium (HyClone Laboratories, Inc., UT) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, Inc.). Mouse myeloma PAI cells were cultured in RPMI 1640 (HyClone) containing 10% FBS. All cell lines were cultured at 37°C with 5% CO2, according to the method detailed by Masrinoul et al. (13).
CHIKV was isolated from patients' plasma samples collected during the 2010 epidemic in Thailand and was used to infect Vero cells (14). Sequence analysis confirmed that the genotype of the isolate clustered within the ECSA lineage (26). SL11131 (ECSA genotype) and S27 (ECSA genotype) were kindly provided by Chang-Kweng Lim, National Institute of Infectious Diseases, Tokyo, Japan (15). CHIKV isolates SBY59/10 (Asian genotype) and B143-09 (West African genotype) were isolated from the sera of patients from Surabaya, Indonesia (16) and Kedougou, Senegal (JQ943719), respectively. Sindbis virus (SV; R68 strain), another alphavirus, was kindly provided by Kohji Moriishi, University of Yamanashi. These alphaviruses were maintained in BHK-21 cells. Dengue virus serotype 2 (DENV2; 16681 strain) and Japanese encephalitis virus (JEV; Nakayama strain) were maintained in C6/36 cells. Infectivity titers were estimated according to the number of PFU in Vero cells, as previously described (14).
Preparation and characterization of mouse MAbs.
CK47 (IgG2a subtype) and CK119 (IgG1) were generated as described previously (13). Briefly, spleen cells from mice immunized with B-7 cells infected with CHIKV Thai isolates (ECSA genotype) (12) were fused with PAI myeloma cells. The hybridomas producing antibodies were screened by an immunofluorescence assay (IFA) using CHIKV Thai isolate-infected Vero cells. Positive hybridomas were cloned twice by limiting dilution. Antibody isotypes were determined by immunochromatography using the IsoQuick kit (Sigma, St. Louis, MO, USA). To generate ascitic fluid, pristane-primed mice were intraperitoneally injected with the hybridomas. The ascitic fluid was then purified on protein G Sepharose 4 Fast Flow (GE Healthcare, Uppsala, Sweden) according to the manufacturer's protocol. The antibody concentrations were measured using the Bradford protein assay (Bio-Rad Laboratories, Hercules, CA). Characterization of these MAbs by an IFA and Western blotting was conducted as described previously (13).
Assembly for rapid diagnostic testing using the IC method.
The IgG fraction purified from the ascitic fluid of inoculated mice was used to develop the IC test, using a system from Alfresa Pharma Corporation, Osaka, Japan. The anti-CHIKV specific MAbs, CK47, prepared in this study were immobilized onto a nitrocellulose membrane for the test line to capture CHIKV protein. To prepare the control line, an anti-mouse IgG antibody (Nippon Biotest Laboratories, Tokyo, Japan) was immobilized onto a nitrocellulose membrane to capture mouse IgG. A conjugated pad containing the CK119 MAb used for the test line was labeled with colloidal gold, impregnated onto glass fibers, dried, and placed between the test line and the sample-dropping region.
ELISA.
Serum IgM levels were determined by ELISA using a CHIKV IgM μ-capture ELISA kit (Nova Tec, Dietzenbach, Germany), according to the manufacturer's protocol.
RNA (RT-PCR) assay.
Total RNA was extracted directly from the viral culture supernatant (140 μl) and sera (140 μl) of CHIKV-suspected patients. RNA was extracted using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. The QuantiTect SYBR green RT-PCR kit (Qiagen) was used for quantitative RT-PCR (qRT-PCR) with the primers CHIKV-E1-F (CTCATACCGCATCCGCATCAG) and CHIKV-E1-R (ACATTGGCCCCACAATGATATTG) for CHIKV samples from Thailand and Indonesia. The qRT-PCR for samples from Laos and Senegal followed the protocol of Pastorino et al. (17). The amplification was performed as described previously (13), under the following conditions: reverse transcription at 50°C for 30 min; then inactivation of the enzyme at 95°C for 15 min; followed by 44 cycles of denaturation at 95°C for 20 s, annealing at 55°C for 30 s, and extension at 72°C for 30 min; and a final extension step at 65°C for 5 s (CFX96 real-time system C1000 thermal cycler, Bio-Rad, Tokyo, Japan). In addition, RT-PCR was performed with primers targeting a 557-bp region of the E2 gene, namely, CHIK_F1 (GAAACTCTGACCGGTGGGATTCAC) and CHIK_R2 (GAGTGTTGGGTGGTCAGGATACAG) for CHIKV, as described previously (14). Samples that gave a positive result by qRT-PCR or RT-PCR were considered positive by RT-PCR assay for CHIKV infection. Quantification of viral RNA was performed using the cycle threshold (CT) values of the samples. The viral genome copy number, as determined by qRT-PCR of viral RNA compared with standard synthetic RNA, was compared with the viral copy number, as determined by PFU/ml.
Evaluation of the IC test using virus culture supernatants.
Supernatant samples for each virus (CHIKV, SV, DENV2, and JEV) were used to confirm the specificity and sensitivity of the IC test. Serial 10-fold dilutions of each virus supernatant by healthy donor serum, corresponding to 1 × 106 to 1 × 100 PFU/ml, were subjected to RT-PCR and tested using the IC test. Serum samples (20 μl) and sample diluent (120 μl) were applied to sample wells of the IC test. The test signal for each IC test was read by visualization after 15 min. The rapid tests were evaluated independently by two investigators, and the results were considered positive when at least one of the investigators read a test as positive.
Evaluation of the IC test using clinical samples.
The IC test was evaluated with serum/plasma samples of CF-suspected patients clinically diagnosed during the acute phase. Cases were defined according to clinical criteria, including acute onset of fever >38.5°C and severe/incapacitating arthralgia not explained by other clinical conditions. The serum samples from these patients indicated negative results for DENV infection by RT-PCR and/or the SD Bioline dengue NS1 Ag + Ab combo kit (Standard Diagnostics, Inc., Kyonggi-do, South Korea). The samples were collected from Pattani province, Thailand, in 2008 (50 cases); Songkla province, Thailand, in 2011 (13 cases, 35 samples); Champasak province, Laos, in 2012 and 2013 (54 cases); Surabaya, Indonesia, in 2013 (2 cases); and Kedougou, Senegal, in 2009 (6 cases). The 13 suspected CF patients from Songkla province were serially collected every few days (2, 3, or 4 times) after onset. Each sample was tested using the IC test, RT-PCR assay, and an IgM μ-capture ELISA for CHIKV. The results of the RT-PCR assay were used as the gold standard. Ten of the serum samples collected in Thailand were positive for DENV infection and negative for CHIKV infection by virus isolation, and these samples were also tested using the IC test.
Ethics.
The research protocols for human samples were approved by the ethics committee of the Ministry of Public Health of Thailand, the National Ethics Committee for Health Research of the Ministry of Health of Lao PDR, and Airlangga University in Indonesia. The mouse experiments for the preparation of MAbs were approved by the ethics committee of Mahidol University, Thailand.
RESULTS
Characterization of MAbs against CHIKV.
Characterization of the anti-CHIKV MAbs was carried out as described previously (13). CK47 reacted with a 50-kDa protein, which had a molecular mass similar to that of the E1 or E2 envelope protein, whereas CK119 did not react. In transfected cells, both of these MAbs reacted with recombinant CHIKV E1 protein but not with E2 protein, as determined by an IFA. These data suggest that CK47 recognizes the linear epitope and CK119 recognizes the conformational epitope.
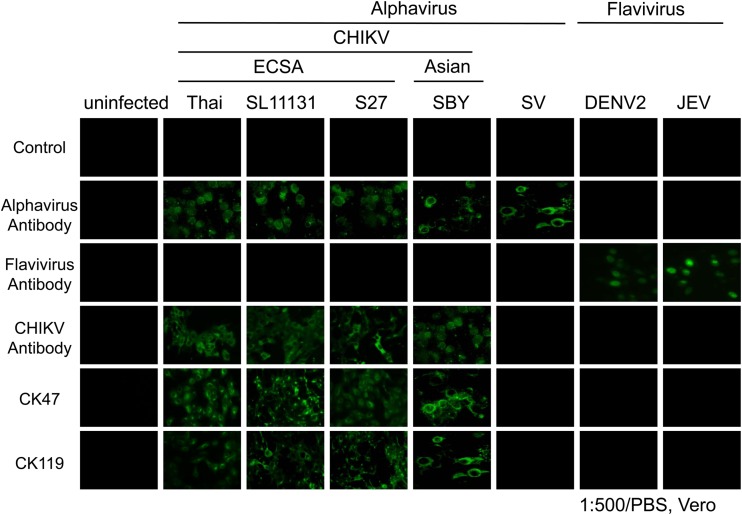
Next, the cross-reactivity of these MAbs was characterized by an IFA using several strains/genotypes of CHIKV and three other viruses: SV, DENV, and JEV. These MAbs only reacted with the CHIKV strains (Thai strain, SL11131, and S27) but not with SV, DENV, or JEV, according to the IFA (Fig. 1). The IC test was therefore developed using these two MAbs.
FIG 1.
Characterization of mouse monoclonal antibodies against chikungunya virus (CHIKV) by an immunofluorescence assay. Four CHIKV strains were included (Thai, SL11131, S27, and SBY). SV, Sindbis virus; DENV2, dengue virus serotype 2; JEV, Japanese encephalitis virus. Alphavirus antibody, Santa Cruz Sc-293153; flavivirus antibody, 4G2; anti-flavivirus monoclonal antibody; anti-CHIKV antibody, ATCC VR-1241AF (CHIKV immune ascetic fluid).
Evaluation of the IC test using virus produced in culture supernatant.
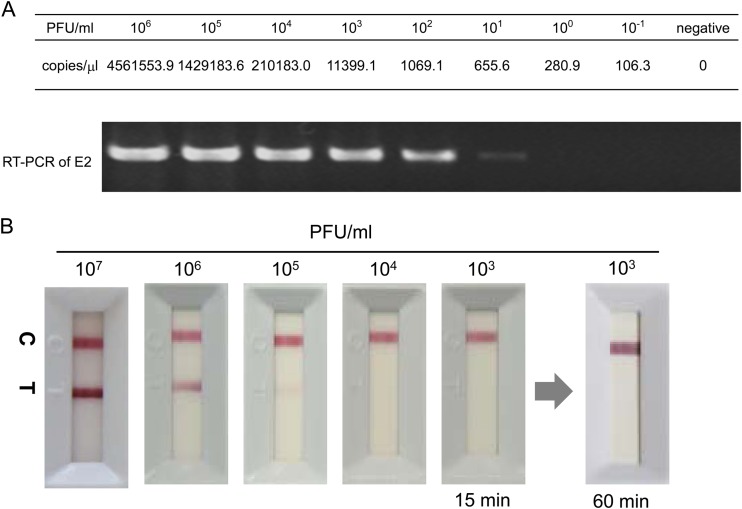
Serial 10-fold dilutions of the CHIKV Thai strain solution with healthy donor serum, corresponding to 1 × 106 to 1 × 100 PFU/ml, were subjected to RT-PCR. The detection limits of the RT-PCR assay were 280 copies/μl by qRT-PCR and 655.6 copies/μl by RT-PCR (Fig. 2A). To determine the detection limits of the IC test, the same samples were applied to the test. The CHIKV Thai strain resulted in the appearance of a band on the test line of the IC test within 15 min (Fig. 2B, band T). The detection limits of the IC test were ≥1.0 × 105 PFU/ml for the CHIKV Thai strain. No nonspecific bands appeared on the test line after 60 min (Fig. 2B).
FIG 2.
Typical banding profile of the rapid diagnostic test for chikungunya virus (CHIKV). (A) Serial 10-fold dilutions of the CHIKV Thai strain (East/Central/South African [ECSA] genotype) culture supernatant were subjected to qRT-PCR/RT-PCR amplification of the E2 envelope gene. (B) Representative profile of the immunochromatography (IC) test using serial 10-fold dilutions of the CHIKV Thai strain (ECSA genotype) culture supernatant. RT-PCR, real-time PCR; C, control band; T, test band.
To evaluate the reactivity of the IC test, several strains/genotypes of CHIKV and other disease-causing viruses related to CHIKV (SV, DENV, and JEV) were simultaneously tested. Positive bands were observed for the ECSA, Asian, and West African genotypes of CHIKV (data not shown), and the detection limits of the IC test were ≥1.0 × 105 PFU/ml for these genotypes (Table 1). None of the non-CHIKVs showed positive bands (data not shown).
TABLE 1.
Cross-reactivities and detection limits of the immunochromatography test
| Virus strain/genotype | Detection limit (PFU/ml) |
|---|---|
| Thailand/Laos/ECSAa | ≥1 × 105 |
| SL11131/Sri Lanka/ECSA | ≥1 × 105 |
| S27/African prototype/ECSA | ≥1 × 105 |
| SBY/Indonesia/Asian | ≥1 × 105 |
| B143-09/Senegal/West African | ≥1 × 105 |
| Sindbis virus | <107 |
| Dengue virus | <106 |
| Japanese encephalitis virus | <106 |
| Negative serum |
ECSA, East/Central/South African.
Evaluation of the IC test using clinical specimens from patients with suspected CF.
To evaluate the applicability of the IC test for diagnosing CHIKV infection in humans, clinical serum samples from suspected CF cases in Thailand, Laos, Indonesia, and Senegal were examined using the IC test, RT-PCR, and IgM ELISA. Of the 112 samples tested, the IC test produced 68 true-positive (60.7%), 34 true-negative (30.3%), 2 false-positive (1.8%), and 8 false-negative (7.1%) results (Table 2). Of the 50 samples from Thailand, the IC test yielded 31 true-positive (62%), 15 true-negative (30%), 1 false-positive (2%), and 3 false-negative (6%) results (Table 2). Sequence analysis of the viral E1 gene showed that these patients were infected with the ECSA genotype of CHIKV (M. Grandadam, personal communications). One sample that tested positive with the IC test tested negative by RT-PCR assay but also tested positive by IgM ELISA for CHIKV. The viral copy number for this sample was 28.1 copies/μl. The sensitivity and specificity of the IC test and the overall agreement between the results from the IC test and those from the RT-PCR assay were 91.2%, 93.8%, and 92.0%, respectively. The 10 Thai serum samples that were positive for DENV infection did not react with the IC test (data not shown). Similarly, of the 54 samples from Laos, the IC test yielded 29 true-positive (53.7), 19 true-negative (35.2), 1 false-positive (1.9%), and 5 false-negative (9.3%) results (Table 2). Sequence analysis showed that these patients were infected with the ECSA genotype of CHIKV. One sample that tested negative for CHIKV by RT-PCR assay and IgM ELISA showed a positive reaction with the IC test. The sensitivity and specificity of the IC test and the overall agreement between the results from the IC test and those from the RT-PCR assay were 85.3%, 95.0%, and 88.9%, respectively. Two samples from Indonesia and 6 samples from Senegal diagnosed as CF on the basis of clinical symptoms were tested by RT-PCR assay, the IC test, and IgM ELISA. These samples were collected within 4 days of presentation of fever. Sequence analysis of the viral E1 gene showed that these patients were infected with the Asian (Indonesia) and West African (Senegal) genotypes of CHIKV, respectively. These samples tested positive for CHIKV by the RT-PCR assay and the IC test but negative by IgM ELISA.
TABLE 2.
Summarized results from the IC test with specimens from suspected chikungunya fever cases
| Country | No. of patients | Genotype | RNA result (n) | IC test result |
IgM ELISA result |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. positive | No. negative | Sensitivity (%) | Specificity (%) | OAAa | No. positive | No. negative | Sensitivity (%) | Specificity (%) | OAAa | ||||
| Thailand | 50 | ECSAb | Positive (34) | 31 | 3 | 91.2 | 93.8 | 92.0 | 6 | 12 | 17.6 | 37.5 | 24.0 |
| Negative (16) | 1 | 15 | 26 | 6 | |||||||||
| Laos | 54 | ECSA | Positive (34) | 29 | 5 | 85.3 | 95.0 | 88.9 | 2 | 3 | 5.8 | 85.0 | 35.2 |
| Negative (20) | 1 | 19 | 32 | 17 | |||||||||
| Indonesia | 2 | Asian | Positive (2) | 2 | 0 | 0 | 0 | ||||||
| Negative (0) | 0 | 0 | 0 | 2 | |||||||||
| Senegal | 6 | West African | Positive (6) | 6 | 0 | 0 | 0 | ||||||
| Negative (0) | 0 | 0 | 0 | 6 | |||||||||
| Total | 112 | Positive (76) | 68 | 8 | 89.4 | 94.4 | 91.1 | 8 | 15 | 10.5 | 86.1 | 34.8 | |
| Negative (36) | 2 | 34 | 58 | 31 | |||||||||
OAA, overall agreement with RT-PCR assays.
ECSA; East/Central/South African.
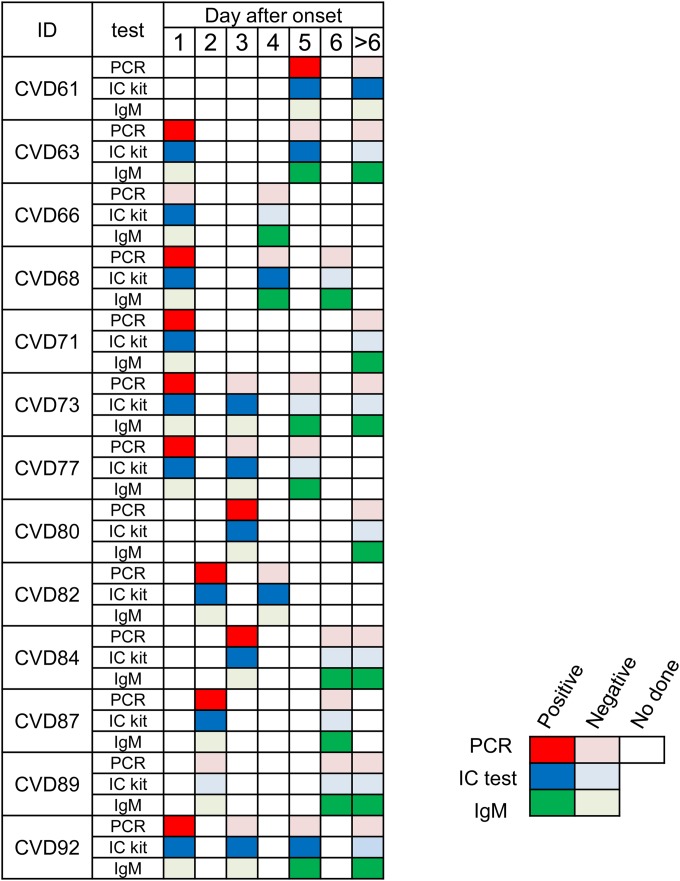
To determine the reactivity of the IC test at different time points of the infection after onset (day 1 to day 14), 35 serial samples from 13 patients with suspected CF were used (Fig. 3, Table 3, and Table 4). The IC test and the RT-PCR assay gave positive results for samples collected within 5 days but not for those collected after 6 days. Sensitivities of the IC test were higher than those of the RT-PCR assay within 5 days (Table 4). It is noteworthy that 11 samples were positive with the RT-PCR assay and IC test at the first collection; however, 7 samples among them were negative with the RT-PCR assay but still positive with the IC test (Table 3; Fig. 3, CVDs 61, 63, 68, 73, 77, 82, and 92). IgM-positive samples started to be detectable 4 days after symptom onset. Interestingly, among the 17 IgM-positive samples, 3 were positive with the IC test (Table 3; Fig. 3, CVDs 63, 68, and 92), but none were positive with the RT-PCR assay.
FIG 3.
The effect of fever days on the results of RT-PCR assay, immunochromatography (IC) test, and IgM ELISA with serial specimens from suspected CF cases in Songkla province, Thailand, 2011. Serial samples (35 samples) from 13 patients (CVD) with suspected CF, collected at different time points after onset of the infection (days 1 to >6), were tested with RT-PCR assay, IC test, and IgM ELISA.
TABLE 3.
Results from the IC test with serial specimens from suspected CF cases in Songkla province, Thailand, 2011
| No. of patients | RNA result (n) | IC test result (n) |
IgM ELISA result (n) |
||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| 35 | Positive (11) | 11 | 0 | 0 | 11 |
| Negative (24) | 9 | 15 | 17 | 7 | |
TABLE 4.
Effect of day after onset of fever on diagnosis of suspected CF cases in Songkla province, Thailand, 2011
| Days after onset | No. of patients | Positivity of suspected CF cases (%) witha: |
||
|---|---|---|---|---|
| RT-PCR | IC test | IgM ELISA | ||
| 1 | 7 | 85.7 | 100 | 0 |
| 2 | 3 | 66.7 | 66.7 | 0 |
| 3 | 5 | 40 | 100 | 0 |
| 4 | 3 | 0 | 66.7 | 66.7 (kit + 1b) |
| 5 | 5 | 20 | 60 | 80 (kit + 2b) |
| 6 | 4 | 0 | 0 | 100 |
| >6 | 8 | 0 | 0 | 87.5 |
CF, chikungunya fever; IC, immunochromatography.
No. of IC kit-positive sample.
DISCUSSION
In this study, we developed a rapid diagnostic test using mouse MAbs that react with CHIKV E1 proteins. This IC test showed high sensitivity and specificity, reacting with the ECSA genotype of CHIKV but not with other alphaviruses or nonalphaviruses. The serum samples, diagnosed as ECSA genotype CF in Thailand and Laos, Asian genotype CF in Indonesia, and West African genotype CF in Senegal, also tested positive with the IC test. Further studies are needed to confirm the usability of the IC test for the detection of CHIKV antigens, Asian and West African genotypes. Comparison between the data from the IC test and RT-PCR assay using serial suspected CF cases revealed that the diagnostic accuracy of the IC test targeting the CHIKV antigen may be clinically acceptable for the diagnosis of CHIKV infection during the acute phase, especially within the first 5 days after the onset of symptoms. Based on these performances, the IC test can be included in a global algorithm for CHIKV diagnosis in conjunction with the RT-PCR assay and serology, as has already been established for DF (18).
Clinical manifestations of CF are not specific and are difficult to differentiate from other febrile illnesses, especially DF. Mulyatno et al. (16) also detected five isolates (31.3%) of DF in 16 clinically suspected CF cases. These results suggested that suspected CF cases include a relatively large number of other febrile illnesses, including DF. An early confirmatory diagnosis of CHIKV infection is essential for the management and control of unprecedented epidemics. In this study, 15 (13.4%) of 112 clinically suspected CF cases were not found to be positive for CHIKV infection with our newly developed IC test, nor were they positive for CHIKV with the RT-PCR assay or IgM ELISA. However, they had already been confirmed as negative for DENV infection with the RT-PCR assay and/or the dengue NS1 Ag + Ab kit before we tested the samples. Therefore, these samples did not appear to be a misdiagnosis between DF and CF.
The anti-CHIKV IgM antibody-based assay with only acute-phase samples is not sufficient for the diagnosis of CHIKV infection (7). The sensitivity of the anti-CHIKV IgM-based assay is extremely low during the acute phase (during the first week of CF) (7, 19). Several reports have described long-lasting anti-CHIKV IgM antibodies despite short-lasting but high-level viremia (20, 21). In the majority of CF cases, anti-CHIKV IgM antibodies only reach detectable levels between day 4 and day 7 of illness (5, 22), resulting in false-negative results when early acute-phase sera are tested. Furthermore, anti-CHIKV IgM antibodies remain in the host for many months (21–24). Therefore, anti-CHIKV IgM detection may not always correlate with an acute CHIKV infection. To make a reliable diagnosis of CHIKV infection in the acute clinical setting, the assay should provide pathogen-specific detection (e.g., RNA, viral antigen, and specific antibodies) across the entire period of patient presentation. Several reports have described long-lasting anti-CHIKV IgM antibodies despite the short-lasting and high-level viremia (20, 21). In one report, the median viral load ranged from 2.9 × 104 to 1.6 × 108 PFU/ml during days 1 through 4 of the viremia period, and 70% of the CF patients visited the hospital during the first 4 days after symptom onset (25). The detection limit of the IC test was ≥1.0 × 105 PFU/ml; therefore, it may be able to detect CHIKV antigen during the acute phase. In the reports, the viral loads in CF peaked rapidly in the first 3 days of illness, after which they rapidly declined (20, 21, 25). They also demonstrated that CHIKV RNA persists for a long period, up to 8 days after the onset of illness in most cases. However, in our study, in the serial samples from Thailand, RNA was undetectable in all serum samples drawn after 5 days of illness, which may be related to the low number of samples collected after 5 days of fever. No detailed comparative data of viral kinetics in CHIKV-infected individuals based on the gold standard of viral culture (PFU), viral RNA measurement (viral copy number), and viral antigen measurement in blood samples were previously available. Further studies are needed to establish the kinetics of CHIKV antigenemia. In our study using serial samples, when tested within the first 5 days after onset, our IC test showed higher sensitivity than found with RT-PCR assay within 5 days. Thus, leading us to the possible explanation related not only with the difference of sensitivity of these tests but also with the preserving property of RNA and antigen in the serum samples.
In agreement with previous reports (20, 21, 25), IgM appeared after 4 days. Our IC test was able to detect 3 positive samples among the 17 IgM-positive samples that were negative by the RT-PCR assay. Taken together, the results of this serial-sample study suggest that the diagnostic accuracy of the IC test targeting CHIKV antigen is certainly clinically acceptable for the early diagnosis of CF within 5 days of disease onset.
In conclusion, we developed a rapid IC test for CHIKV antigen detection. The IC test has the advantages of being user friendly, rapid, easy, and deliverable, without requiring specialist equipment. The IC test approached 89.4% sensitivity, 94.4% specificity, and 91.1% agreement with an RT-PCR-based assay. This test can be used as a rapid tool for the confirmation of CHIVK infection during the acute phase of CF and the differentiation between CF and DF, facilitating more effective public health interventions.
ACKNOWLEDGMENTS
We take full responsibility for the content of the manuscript and thank BioEdit Ltd. for editorial assistance and useful comments.
We are grateful to Yaowalark Sukthana (Faculty of Tropical Medicine, Mahidol University), Naokazu Takeda (RCC-ERI, Research Institute for Microbial Diseases, Osaka University), and Yoshiharu Matsuura (Research Institute for Microbial Diseases, Osaka University) for their valuable support and help during this study.
This work was supported by a program of the Japan Initiative for Global Research Network on Infectious Diseases, directed by the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by Alfresa Pharma Corporation.
REFERENCES
- 1.Strauss JH, Strauss EG, Kuhn RJ. 1995. Budding of alphaviruses. Trends Microbiol 3:346–350. doi: 10.1016/S0966-842X(00)88973-8. [DOI] [PubMed] [Google Scholar]
- 2.Powers AM, Brault AC, Tesh RB, Weaver SC. 2000. Re-emergence of chikungunya and O'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81:471–479. [DOI] [PubMed] [Google Scholar]
- 3.Lumsden WH. 1955. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. II. General description and epidemiology. Trans R Soc Trop Med Hyg 49:33–57. [DOI] [PubMed] [Google Scholar]
- 4.Chen LH, Wilson ME. 2010. Dengue and chikungunya infections in travelers. Curr Opin Infect Dis 23:438–444. doi: 10.1097/QCO.0b013e32833c1d16. [DOI] [PubMed] [Google Scholar]
- 5.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. 2007. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 7:319–327. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 6.Mourya DT, Mishra AC. 2006. Chikungunya fever. Lancet 368:186–187. doi: 10.1016/S0140-6736(06)69017-X. [DOI] [PubMed] [Google Scholar]
- 7.Blacksell SD, Tanganuchitcharnchai A, Jarman RG, Gibbons RV, Paris DH, Bailey MS, Day NP, Premaratna R, Lalloo DG, de Silva HJ. 2011. Poor diagnostic accuracy of commercial antibody-based assays for the diagnosis of acute chikungunya infection. Clin Vaccine Immunol 18:1773–1775. doi: 10.1128/CVI.05288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watthanaworawit W, Turner P, Turner CL, Tanganuchitcharnchai A, Jarman RG, Blacksell SD, Nosten FH. 2011. A prospective evaluation of diagnostic methodologies for the acute diagnosis of dengue virus infection on the Thailand-Myanmar border. Trans R Soc Trop Med Hyg 105:32–37. doi: 10.1016/j.trstmh.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang BK, Song DS, Jung KI, Lee CS, Park SJ, Oh JS, An DJ, Yang JS, Moon HJ, Lee SS, Yoon YD, Park BK. 2007. Genetic characterization of canine rotavirus isolated from a puppy in Korea and experimental reproduction of disease. J Vet Diagn Invest 19:78–83. doi: 10.1177/104063870701900112. [DOI] [PubMed] [Google Scholar]
- 10.Takanashi S, Okame M, Shiota T, Takagi M, Yagyu F, Tung PG, Nishimura S, Katsumata N, Igarashi T, Okitsu S, Ushijima H. 2008. Development of a rapid immunochromatographic test for noroviruses genogroups I and II. J Virol Methods 148:1–8. doi: 10.1016/j.jviromet.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Nishizono A, Khawplod P, Ahmed K, Goto K, Shiota S, Mifune K, Yasui T, Takayama K, Kobayashi Y, Mannen K, Tepsumethanon V, Mitmoonpitak C, Inoue S, Morimoto K. 2008. A simple and rapid immunochromatographic test kit for rabies diagnosis. Microbiol Immunol 52:243–249. doi: 10.1111/j.1348-0421.2008.00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanai Y, Chittaganpitch M, Nakamura I, Li GM, Bai GR, Li YG, Ikuta K, Sawanpanyalert P. 2010. Distinct propagation efficiencies of H5N1 influenza virus Thai isolates in newly established murine respiratory region-derived cell clones. Virus Res 153:218–225. doi: 10.1016/j.virusres.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Masrinoul P, Puiprom O, Tanaka A, Kuwahara M, Chaichana P, Ikuta K, Ramasoota P, Okabayashi T. 2014. Monoclonal antibody targeting chikungunya virus envelope 1 protein inhibits virus release. Virology 464–465:111–117. doi: 10.1016/j.virol.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 14.Puiprom O, Morales Vargas RE, Potiwat R, Chaichana P, Ikuta K, Ramasoota P, Okabayashi T. 2013. Characterization of chikungunya virus infection of a human keratinocyte cell line: role of mosquito salivary gland protein in suppressing the host immune response. Infect Genet Evol 17:210–215. doi: 10.1016/j.meegid.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Lim CK, Nishibori T, Watanabe K, Ito M, Kotaki A, Tanaka K, Kurane I, Takasaki T. 2009. Chikungunya virus isolated from a returnee to Japan from Sri Lanka: isolation of two sub-strains with different characteristics. Am J Trop Med Hyg 81:865–868. doi: 10.4269/ajtmh.2009.09-0009. [DOI] [PubMed] [Google Scholar]
- 16.Mulyatno KC, Susilowati H, Yamanaka A, Soegijanto S, Konishi E. 2012. Primary isolation and phylogenetic studies of chikungunya virus from Surabaya, Indonesia. Jpn J Infect Dis 65:92–94. [PubMed] [Google Scholar]
- 17.Pastorino B, Bessaud M, Grandadam M, Murri S, Tolou HJ, Peyrefitte CN. 2005. Development of a TaqMan RT-PCR assay without RNA extraction step for the detection and quantification of African chikungunya viruses. J Virol Methods 124:65–71. doi: 10.1016/j.jviromet.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Blacksell SD, Jarman RG, Bailey MS, Tanganuchitcharnchai A, Jenjaroen K, Gibbons RV, Paris DH, Premaratna R, de Silva HJ, Lalloo DG, Day NP. 2011. Evaluation of six commercial point-of-care tests for diagnosis of acute dengue infections: the need for combining NS1 antigen and IgM/IgG antibody detection to achieve acceptable levels of accuracy. Clin Vaccine Immunol 18:2095–2101. doi: 10.1128/CVI.05285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rianthavorn P, Wuttirattanakowit N, Prianantathavorn K, Limpaphayom N, Theamboonlers A, Poovorawan Y. 2010. Evaluation of a rapid assay for detection of IgM antibodies to chikungunya. Southeast Asian J Trop Med Public Health 41:92–96. [PubMed] [Google Scholar]
- 20.Chopra A, Anuradha V, Lagoo-Joshi V, Kunjir V, Salvi S, Saluja M. 2008. Chikungunya virus aches and pains: an emerging challenge. Arthritis Rheum 58:2921–2922. doi: 10.1002/art.23753. [DOI] [PubMed] [Google Scholar]
- 21.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, Tallet F, Moiton MP, Gauzere BA, Bruniquet S, Jaffar Bandjee Z, Morbidelli P, Martigny G, Jolivet M, Gay F, Grandadam M, Tolou H, Vieillard V, Debre P, Autran B, Gasque P. 2010. Persistent chronic inflammation and infection by chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol 184:5914–5927. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- 22.Taubitz W, Cramer JP, Kapaun A, Pfeffer M, Drosten C, Dobler G, Burchard GD, Loscher T. 2007. Chikungunya fever in travelers: clinical presentation and course. Clin Infect Dis 45:e1–e4. doi: 10.1086/518701. [DOI] [PubMed] [Google Scholar]
- 23.Grivard P, Le Roux K, Laurent P, Fianu A, Perrau J, Gigan J, Hoarau G, Grondin N, Staikowsky F, Favier F, Michault A. 2007. Molecular and serological diagnosis of chikungunya virus infection. Pathol Biol (Paris) 55:490–494. doi: 10.1016/j.patbio.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, Guigand L, Dubreil L, Lebon P, Verrier B, de Lamballerie X, Suhrbier A, Cherel Y, Le Grand R, Roques P. 2010. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest 120:894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, Silpapojakul K. 2013. Viremic profiles in asymptomatic and symptomatic chikungunya fever: a blood transfusion threat? Transfusion 53:2567–2574. doi: 10.1111/j.1537-2995.2012.03960.x. [DOI] [PubMed] [Google Scholar]
- 26.Sasayama M, Benjathummarak S, Kawashita N, Rukmanee P, Sangmukdanun S, Masrinoul P, Pitaksajjakul P, Puiprom O, Wuthisen P, Kurosu T, Chaichana P, Maneekan P, Ikuta K, Ramasootan P, Okabayashi T, Singhasivanon P, Luplertlop N. 2014. Chikungunya virus was isolated in Thailand, 2010 Virus Genes 49(3):485–489. doi: 10.1007/s11262-014-1105-5. [DOI] [PMC free article] [PubMed] [Google Scholar]