Abstract
The emergence of drug-resistant forms of tuberculosis (TB) represents a major public health concern. Understanding the transmission routes of the disease is a key factor for its control and for the implementation of efficient interventions. Mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) marker typing is a well-described method for lineage identification and transmission tracking. However, the conventional manual genotyping technique is cumbersome and time-consuming and entails many risks for errors, thus hindering its implementation and dissemination. We describe here a new approach using the QIAxcel system, an automated high-throughput capillary electrophoresis system that also carries out allele calling. This automated method was assessed on 1,824 amplicons from 82 TB isolates and tested with sets of markers of 15 or 24 loci. Overall allele-calling concordance between the methods from 140 to 1,317 bp was 98.9%. DNA concentrations and repeatability and reproducibility performances showed no biases in allele calling. Furthermore, turnaround time using this automated system was reduced by 81% compared to the conventional manual agarose gel method. In sum, this new automated method facilitates MIRU-VNTR genotyping and provides reliable results. Therefore, it is well suited for field genotyping. The implementation of this method will help to achieve accurate and cost-effective epidemiological studies, especially in countries with a high prevalence of TB, where the high number of strains complicates the surveillance of circulating lineages and requires efficient interventions to be carried out in an urgent manner.
INTRODUCTION
Tuberculosis (TB) remains a major public health concern worldwide. The widespread emergence of multidrug resistant (MDR-TB) strains and extensively and extremely drug-resistant (XDR-TB and XXDR-TB) strains has hampered the management of treatment of the disease and the control of TB outbreaks (1). Many epidemiological questions remain unresolved, particularly in regard to the risk factors associated with the transmission of the bacilli to the host and to the prevalence of strain reactivation versus exogenous reinfection. Various genotype lineages of Mycobacterium tuberculosis have been described that can be associated with drug resistance, a higher rate of transmission, a higher rate of progression to disease, virulence, and vaccine escape (2, 3). The emergence of whole-genome sequencing (WGS) technologies can provide a complete genomic picture of lineage characterization and drug-resistant mutations. However, these technologies are presently not affordable for most laboratories, considering the costs and specialized skills required for data analysis. Thus, methods on the bench still have their value, as they have a faster turnaround time and can handle a higher batch capacity at a lower cost. One of these in particular, mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) marker typing, is commonly used for effective genotyping. There are numerous MIRU-VNTR loci that have been described since 1997 (4, 5), and their combination allows us to accurately discriminate among lineages. Thus, MIRU-VNTR typing remains a relevant method for lineage identification and transmission tracking (6). Additionally, this method is one of the few that are capable of detecting mixtures of strains, which are not rare clinically (6). Strain mixtures hinder accurate genotyping when using methods such as spoligotyping or IS6110 restriction fragment length polymorphism (RFLP) (7).
Conventionally, MIRU-VNTR typing involves PCR amplifications of each targeted locus, followed by processing through high-resolution electrophoresis to estimate the size of each amplified locus and thus infer the number of repeats it contains. The combination of all loci allele callings results in a barcode profile that can be used to identify and compare strains. A standard approach consists of investigating sets of 15 or 24 loci for epidemiological and phylogenetic studies, respectively (8). This straightforward method, which has fewer biohazard constraints than IS6110 RFLP, is technically easier to implement than other highly discriminating methods. Nevertheless, it takes longer and is more cumbersome to perform. In addition, the numerous manual steps (e.g., pipetting, gel reading, and interpretation) are sources of substantial errors. A few studies have investigated the intra- and interlaboratory reliability of MIRU-VNTR typing. In particular, they confirmed that manual methods can present discrepancies and allele-calling failures (9). These factors hinder the wider implementation of this method.
The proposed automated methodology we assessed and describe here aims to obtain reliable MIRU-VNTR strain patterns and to facilitate technical steps. The QIAxcel technology platform can promote a wider use of genotyping, especially in countries with middle- and high-burden TB, in response to the need for an affordable and robust field genotyping methodology. We describe here (i) the comparison between this automated method versus the conventional manual method and (ii) the lineages identified in countries such as Georgia, Haiti, and Laos, where there are no or few genotypic data (10–13). The overall outcome is to facilitate comprehensive investigations of TB epidemiology for transmission tracking and better understanding of the prevalence of TB reactivation versus TB reinfection.
MATERIALS AND METHODS
Specimen collection.
After culturing clinical specimens and thermal lysis, 81 strain isolates of patients from Southeast Asia (Laos, n = 14), the Caribbean (Haiti, n = 8), Europe (France, n = 2), and Caucasia (Georgia, n = 57) and the laboratory reference strain H37Rv were processed to get a range of various lineages. Fifty of the clinical isolates bore a wide range of resistance patterns (including MDR and XDR-TB). TB culture isolates were processed to France after thermal lysis at 95°C for 20 min and transported according to current International Air Transport Association (IATA) regulations.
Amplification.
Simplex PCR amplifications were performed on 96-well microplates according to standard protocol (14) on 1:10 diluted thermal lysates. Two controls were included in each run: one laboratory strain (H37Rv) and one PCR blank (molecular grade water). PCR products were quantified using a Qubit double-stranded DNA high-sensitivity (dsDNA HS) assay kit (Life Technologies, USA).
Electrophoresis and allele calling using the conventional or manual MIRU-VNTR typing method.
A total of 2 μl of PCR products was loaded on NuSieve 3:1 3% agarose (Lonza, Switzerland) using the intercalating agent GelRed (Biotium Inc., USA) and low-range-mass DNA ladder (Euromedex, France) according to standard protocol (14).
Electrophoresis and allele calling using the automated MIRU-VNTR method.
The automated system used was the QIAxcel advanced system (Qiagen AG, Germany), a high-resolution capillary electrophoresis device for up to 96 samples per run. In this system, PCR products are automatically loaded from microplates to a high-resolution cartridge for DNA electrophoresis and detection, using its size marker of 100 to 2,500 bp and alignment marker of 15 to 3,000 bp, as well as the OM1700 instrument method specifically designed for our application. With OM1700, PCR products are automatically injected over 10 s at 5 kV and separation occurs over 1,700 s at 2 kV. They are labeled with a dye to be detected by a fluorescence detector and then converted to an electropherogram and gel image. Sample consumption is <0.1 μl per analysis, allowing further downstream testing. The ScreenGel software version 1.2.0 was customized by adding the reference tables (14) for inferring the number of repeats from the observed size, in order to automatically perform allele calling on selected amplicons. Next, the corresponding number of repeats was retained when the deviation from the expected size was <95% of a half repeat as a tolerance filter. A plug-in was developed to generate a final spreadsheet file to be exported for further genotype analysis in the MIRU-VNTRplus database, for example.
Sizing reference method.
Sanger sequencing was used as the reference method to determine the size of amplicons. Amplicons were cloned with the Topo TA cloning kit for sequencing (Life Technologies, USA) before being sequenced using universal primers and sent to GATC Biotech AG (Germany).
In-house size marker.
An in-house size marker was prepared to be used as the ladder for experiments. It included DNA fragments of 100, 206, 382, 438, 562, 639, 681, and 1,065 bp that were confirmed with Sanger sequencing.
Lineage identification.
MIRU-VNTR profiles were submitted to the MIRU-VNTRplus online database, which contains 186 distinct MIRU-VNTR patterns assigned to lineages (15), for genotype determination and analysis of the diversity of the strains.
Turnaround time and cost.
For 24 loci-processed isolates, the unit costs and turnaround times for the conventional and automated methods were compared in our setting, based on a processing batch of 94 samples and 2 controls (i.e., one 96-well microplate format). For cost computation, we considered reagents and consumables such as Tris-borate-EDTA (TBE) buffer, NuSieve 3:1 agarose, intercalating agents, size markers, tubes, microtips, and specific consumables for automated systems (e.g., nitrogen cartridge, alignment marker). On the other hand, we calculated equipment costs that consisted of required days of use multiplied by depreciation cost. Depreciation cost was based on the overall equipment cost to be used for 7 years, considering 215 working days a year, regardless of whether the device was used. Briefly, to get days of use, we considered the processing of 3 conventional gels (including 3 size markers and 17 PCR products each, including controls) and 3 runs of QIAxcel (95 PCR products per run, including controls) per working day that we combined with the daily price of the equipment. For conventional equipment, the cost included the power supply, gel imager, and complete electrophoresis device Sub-Cell GT (Bio-Rad); for the automated method, the cost included the QIAxcel system and ScreenGel software. Handling time, i.e., operator intervention, was also considered for post-PCR steps. For the manual method, this included gel and buffer preparations, pipetting PCR products to load them on the gel, taking photos with the camera, and gel interpretation. The automated method included only microplate loading on the system and a few software operations for data interpretation.
Assessment criteria.
Automated-method performances for MIRU-VNTR typing were measured in terms of sizing and allele-calling accuracy, the effect of DNA concentration, repeatability and reproducibility, and turnaround time compared to the manual-method performance.
First, the accuracy of sizing and allele calling was measured according to the in-house ladder data and the isolate data; to allow for proper allele calling, parameters were set to call the closest allele from theoretical abacus. Second, as it is established that electrophoresis migration is affected by high DNA concentrations, sizing was assessed within a range of concentrations of PCR products. Amplicons at high DNA concentrations were diluted, and sizing by the two typing methods was compared. Third, to assess the repeatability and reproducibility of the system for this application, we prepared an in-house ladder to test the performance of the 12 capillaries of the cartridge that can be reused 100 times during its 9-month shelf life. Intrarun sizing repeatability was done at the beginning, middle, and end of the shelf life of the cartridges. Interrun sizing reproducibility was also assessed considering three time points within the cartridge shelf life and two cartridge batches. Fourth, the turnaround times of the two typing methods were calculated for a batch of 94 PCR products and 2 controls.
RESULTS
Description of loci and sizes processed.
DNA from 81 isolates and 1 reference strain H37Rv were processed using a 15-locus panel. Among the isolates, 66 were processed for 9 auxiliary loci, to complete the 24-locus panel. This represented a total of 1,824 assays. The large diversity of detected alleles represented sizes ranging from 140 to 1,317 bp (Table 1), of which 50% were between 400 and 650 bp, with a median of 496 bp. Fourteen allele mixtures were detected on 13 loci for 5 isolates using both methods. These results were all confirmed after reprocessing the lysate dilution, PCR, and electrophoresis steps.
TABLE 1.
Summary of MIRU-VNTR allele sizes and distribution observeda
| MIRU-VNTR panel | Alias/locus | Size (bp) |
Allele distribution |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Minimum | Maximum | 0 | 1 | 2 | 3–3s | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 15 | Mixtureb | ||
| 15 locic (n = 82) | MIRU04/580 | 250 | 757 | 2 | 63 | 3 | 7 | 2 | 3 | 2 | ||||||
| MIRU10/960 | 590 | 961 | 8 | 48 | 17 | 2 | 3 | 2 | 1 | 1 | ||||||
| MIRU16/1644 | 565 | 738 | 1 | 4 | 19 | 57 | 1 | |||||||||
| MIRU26/2996 | 335 | 649 | 4 | 8 | 10 | 6 | 40 | 2 | 11 | 1 | ||||||
| MIRU31/3192 | 492 | 816 | 1 | 10 | 24 | 3 | 40 | 3 | 1 | |||||||
| MIRU40/802 | 398 | 630 | 8 | 9 | 42 | 10 | 12 | 1 | ||||||||
| Mtub04/424 | 602 | 784 | 1 | 25 | 12 | 38 | 6 | |||||||||
| ETR C/577 | 270 | 445 | 3 | 14 | 63 | 2 | ||||||||||
| Mtub21/1955 | 160 | 1,053 | 1 | 16 | 14 | 8 | 42 | 1 | ||||||||
| QUB-11b/2163b | 140 | 626 | 6 | 12 | 6 | 11 | 2 | 38 | 5 | 1 | 1 | |||||
| ETR A/2165 | 274 | 732 | 1 | 6 | 17 | 50 | 2 | 5 | 1 | |||||||
| Mtub30/2401 | 248 | 481 | 1 | 7 | 22 | 52 | ||||||||||
| Mtub39/3690 | 387 | 783 | 7 | 53 | 8 | 9 | 2 | 1 | 1 | 1 | ||||||
| QUB-26/4052 | 385 | 1,317 | 1 | 6 | 4 | 7 | 17 | 11 | 29 | 4 | 1 | 2 | ||||
| QUB-4156/4156 | 560 | 809 | 4 | 8 | 56 | 8 | 6 | |||||||||
| 24 locid (n = 66) | MIRU 02/154 | 456 | 513 | 3 | 62 | 1 | ||||||||||
| MIRU 20/2059 | 508 | 605 | 2 | 64 | ||||||||||||
| MIRU 23/2531 | 306 | 472 | 1 | 59 | 5 | 1 | ||||||||||
| MIRU 24/2687 | 439 | 505 | 59 | 7 | ||||||||||||
| MIRU 27/3007 | 644 | 665 | 65 | 1 | ||||||||||||
| MIRU 39/4348 | 584 | 755 | 1 | 17 | 47 | 1 | ||||||||||
| Mtub29/2347 | 451 | 579 | 1 | 7 | 58 | |||||||||||
| ETR B/2461 | 403 | 647 | 1 | 57 | 1 | 6 | 1 | |||||||||
| Mtub 34/3171 | 386 | 492 | 1 | 64 | 1 | |||||||||||
n = 1,824. Data were obtained from automated and manual methods. In case of noncongruence, Sanger sequencing was used as a reference method for accurate sizing of amplicons in order to infer the correct number of repeats to be retained.
Number of PCR products containing allele mixtures.
Sampling is 81 isolates and 1 reference strain.
Sampling is 65 isolates and 1 reference strain.
Sizing and VNTR allele-calling accuracy.
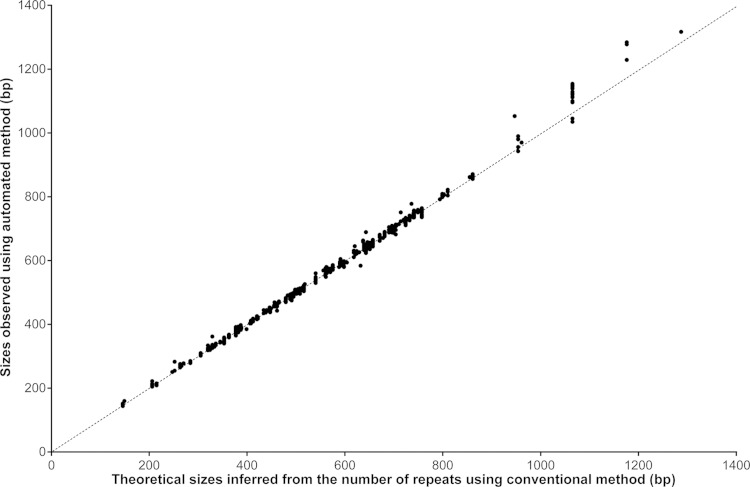
A total of 82 and 66 isolates were processed using 15-locus and 24-locus panels, respectively. The overall concordance on allele calling observed between the automated and manual methods was 98.9% (1,803/1,824) (Fig. 1), with a perfect match for 21 of the 24 tested loci. For the size range 140 to 900 bp (98% of processed amplicons), the concordance was 99.9% (1,780/1,781).
FIG 1.
Sizing correlation between automated and conventional method. The diagonal line indicates the virtual equal correlation (n = 1,824).
Discrepancies were observed for Mtub39/VNTR3690 (1 case presenting 8 alleles, i.e., 736 bp), Mtub21/VNTR1955 (1 case presenting 15 alleles, i.e., 947 bp), and QUB-26/VNTR4052 (19 cases presenting 7, 8, and 9 alleles, i.e., 1,065, 1,176, and 1,287 bp) (Table 2), showing that the automated method overestimated sizes that exceeded 900 bp. For of all these cases, Sanger sequencing unambiguously confirmed the allele calling given by the manual method as shown in Table 2.
TABLE 2.
Investigation of discrepant results between automated and manual methods and comparison with Sanger sequencing
| Alias/locus | No. of isolates | Automated method |
Inferred no. of repeats for manual methoda | Sanger sequencingb |
||
|---|---|---|---|---|---|---|
| Sizing (bp) | Inferred no. of repeats | Sizing (bp) | Inferred no. of repeats | |||
| Mtub21/VNTR1955 | 1 | 1,053 | 17 | 15 | 948 | 15 |
| Mtub39/VNTR3690 | 1 | 778 | 9 | 8 | 740 | 8 |
| QUB-26/VNTR4052 | 14 | 1,114–1,153 | 8 or 9 | 8 | 1,051 | 8 |
| QUB-26/VNTR4052 | 4 | 1,168–1,251 | 9 or 10 | 9 | 1,173 | 9 |
| QUB-26/VNTR4052 | 1 | 1,317 | 10 or 11 | 10 | 1,284 | 10 |
Sizing data are not indicated for manual method, as gel reading cannot provide digital data.
Sanger sequencing used as reference method to accurately determine the length of amplicons and infer the number of repeats.
Effect of DNA concentration on sizing.
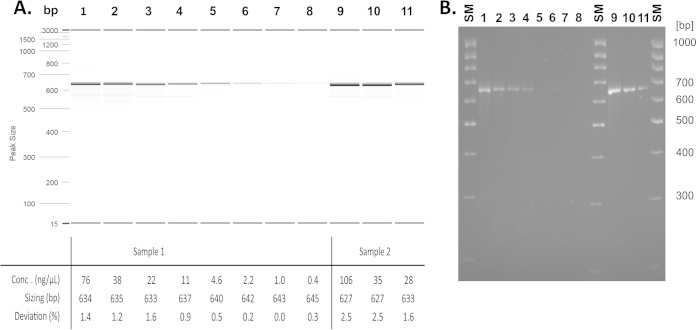
To assess if there is a sizing bias due to DNA concentration, 2 PCR products of locus MIRU10 both presenting 3 repeats were diluted from 106 to 0.4 ng/μl and processed using the two typing methods. As shown in Fig. 2, sizes varied from 627 to 645 bp using the automated method, for a 643-bp expected size. However, no change in the allele calling was observed for either method. At the same time, the automated method showed a lower detection limit of 0.4 ng/μl, while the manual method was unable to detect concentrations of <2.2 ng/μl, corresponding to 0.8 ng and 4.4 ng, respectively, deposited on the gel.
FIG 2.
Automated (A) and conventional (B) sizing comparison with various DNA concentrations from 2 PCR products presenting 3 repeats on MIRU10. Lanes 1 to 8, sample 1 dilutions with DNA concentrations from 76 to 0.4 ng/μl. Lanes 9 to 11, sample 2 dilutions with DNA concentrations from 106 to 1.6 ng/μl. The expected size for all PCR products is 643 bp, and the accepted deviation is ±3.3%, i.e., 21 bp. The automated method was performed with a DNA high-resolution cartridge (OM1700 instrument method and alignment marker 15 to 3,000 bp), with an estimated PCR product consumption of <0.1 μl; conventional electrophoresis gel was performed loading 2 μl of PCR products on a NuSieve 3:1 3% gel with low-range-mass DNA ladder.
Reproducibility and repeatability.
To estimate the robustness of the automated system, reproducibility and repeatability on the 12 capillaries of cartridge were assessed. The in-house amplicon ladder with fragments of 100, 206, 382, 438, 562, 639, 681, and 1,065 bp was processed on the cartridge as a sample at the beginning, middle, and end of the 9-month shelf life of two different cartridges. This therefore represented 6 different runs of 12 tests each. Intrarun sizing repeatability was calculated on each expected size and for each run separately, whereas reproducibility was calculated on all 6 runs. Considering repeatability, the maximum sizing deviation observed was between 1.9 bp (for the 100-bp smallest marker) and 6.5 bp (for the 1,065-bp largest marker), i.e., 1.9% to 0.6%. Reproducibility showed a variation from 1.2 to 5.7 bp, i.e., 1.2% to 0.5%. These deviation ranges are much lower than those with manual gel performance, as the readout cannot reach such accuracy. For a proper allele calling, the deviation in base pairs must not exceed half of the shortest repeat length, i.e., 25.5 bp, which is half of the 51-bp repeat length of both MIRU26 and Mtub04/VNTR424; the 6.5-bp deviation observed using the automated system fits these criteria. Indeed, the variability in reproducibility and repeatability had no impact on allele calling.
Turnaround time and cost.
We assessed the unit cost and turnaround time per isolate for the manual and automated methods based on a batch of 94 samples processed with the MIRU-VNTR 24-locus panel, with 2 additional controls. The costs greatly depend on the mode of computation and local settings. Under our conditions, the overall costs for the first step, PCR, for one sample (1 reaction for each of the 24 loci, per isolate) represent $31, to be added to the electrophoresis cost described below.
As shown in Table 3, the overall cost for the electrophoresis step is 16% cheaper using the automated method than using the manual method. Reagent and consumable costs are almost equivalent for both methods. We noticed that NuSieve agarose represents 60% of the reagent cost for the manual method, which still requires buffers, ladders, intercalating agent, loading buffer, and lots of tips, whereas the electrophoresis cartridge kit represents 90% of the reagent cost for the automated method. Considering equipment cost, QIAxcel daily use is more expensive than the manual method platform ($16 versus $8.8), but as the process is 70% shorter (1.1 h versus 3.7 h), it remains more profitable.
TABLE 3.
Comparison of unit cost and turnaround time for manual method for post-PCR steps
| Variablea | Conventional method data | Automated method data |
|---|---|---|
| Unit costsb ($) | ||
| All reagents and consumables | 22.6 | 21.6 |
| Equipment | 4.2 | 1.0 |
| Total | 26.8 | 22.6 |
| Unit turnaround time (h) | ||
| Handling time | 3.0 | 0.2 |
| Operating time | 3.7 | 1.1 |
| Total | 6.7 | 1.3 |
Computation was based on a processing batch of a full 96-well microplate: 94 samples to be genotyped and 2 controls.
As PCR cost is the same for both sizing methods, its cost was not included in this table.
On the other hand, the automated method showed substantial added value by reducing handling (i.e., steps requiring manual operation) and operating times (by 93% and 70%, respectively), providing an 81% shorter process. As handling time has to be associated to operator wages, which varies depending on the setting, this reduction in turnaround time will further contribute to significantly reduce the overall genotyping cost compared to that of the manual method.
Lineage identification.
Each MIRU-VNTR locus was successfully processed, resulting in complete MIRU-VNTR patterns for each strain. The corresponding profiles were then analyzed on the MIRU-VNTRplus website (www.miru-vntrplus.org) (15). As indicated in Table S1 in the supplemental material, 46 different profiles were detected. The identified lineages were Beijing (31 isolates, including 8 different patterns), LAM (3), EAI (2), and Ural (1). The H37Rv control strain was correctly assigned to its characterized pattern. Thirty-three other lineages from the 44 other isolates were detected with a phylogenetic distance of >0.09, i.e., >2 loci not matching to reference strains; in this case, we suggest that they cannot be assigned to a lineage. Finally, 46.4% (38/82) of the strains were assignable to a lineage.
DISCUSSION
The objective of this study was to assess a new automated approach for MIRU-VNTR typing in terms of reliability and practicability. Criteria such as sizing and allele-calling accuracy, effect of DNA concentration, repeatability and reproducibility of the automated method, and turnaround time were compared.
Overall size accuracy was 98.9%. The concordance reached 99.9% for the size range 100 to 900 bp, which corresponds to 98% of the processed data. These results are in line with previous observations (16) and demonstrate the capability of the automated method for MIRU-VNTR typing. However, the optimization of the automated sizing algorithm remains to be carried out to obtain full results for longer amplicons. Nevertheless, stutter peaks (due to stuttering of polymerase during amplification, which is not rare) or an allelic ladder can be efficiently used to circumvent this oversizing issue.
The results also showed that DNA concentration impacts sizing but not allele-calling results using the automated method. Thus, DNA measurement is not required before electrophoresis under these experimental conditions. We determined that the automated method is more sensitive than the manual method (0.4 versus 2.2 ng/μl, respectively). This is an advantage for isolates with low bacterial load, i.e., lower PCR yield. Repeatability and reproducibility performances with the automated method for electrophoresis steps showed no significant deviation. Finally, the automated method, with its standardized high-throughput process, shortens the turnaround time, as both handling and operating times are reduced by 81%, yielding a cost competitive with the manual method. Reagent, consumable, and equipment costs are 14% reduced. In addition, by eliminating most manual steps, the risk of errors, including sample inversion and mixing during the numerous pipetting steps, gel misreading, and allele-calling misinterpretations are significantly reduced in the automated method compared to the manual method.
As widely described, another technology for TB genotyping is the ABI sequencer (8). This methodology is an excellent alternative to the manual method. The isolates can be processed on site if this equipment and commercial kits are available, or they can be sent to be processed by the Genoscreen company; both options provide reliable genotyping. The other alternative method described here, based on the QIAxcel system for electrophoresis and reading steps, aims to be more robust and reliable than the manual method. It also aims to be more easily implemented in limited-resource settings than the ABI-based method, which will contribute to increase the local capacity and to facilitate field epidemiological studies. This system can be enlarged to accommodate any standard DNA/RNA electrophoresis purpose.
The typing patterns and the lineages that we identified represented the genotypic diversity of our sampling cohort and proved the capability of the automated method to process the main TB lineages. Additionally, the substantial 53.6% of unassigned strains (44/82) by MIRU-VNTRplus computation suggests that this online database could be upgraded (the last update was released in 2011).
To ensure the optimal strain discrimination power, the MIRU-VNTR panel to be processed for a cohort has to be chosen carefully in addition to these robust improvements. Recent studies have proposed an optimized 24-locus panel (17), a new set of markers for Beijing sublineage discrimination (18), and a new set of markers for M. bovis genotyping (19). These new panels can be processed using the automated method we describe here; the sole modification is the implementation of new parameters for allele-calling instructions.
The MIRU-VNTR typing method was widely described as an appropriate approach for epidemiological use, especially for transmission tracking. This method is sufficiently discriminant compared to the widespread spoligotyping method (20). Undoubtedly, WGS may provide more comprehensive results from sequence data, allowing the most accurate strain discrimination. However, processes for WGS and its data analysis are not yet well standardized and still require skills and tools in bioinformatics that are out of reach for most laboratories today. Thus, the MIRU-VNTR typing method remains a good approach, especially if using an automated version, as described in this paper, which will provide more rapidly reliable results that can be easily interpreted.
In conclusion, TB and MDR-TB transmission routes are key components in the control of TB and its expansion. Our work shows that the automated method we assessed fulfills the overall requirements for MIRU-VNTR typing applications. This new approach should be considered for implementation in any TB laboratory that regularly performs TB genotyping, especially reference laboratories in countries with middle and high burdens of TB, where reliable techniques are indeed required in the field. This epidemiological information is crucial in building effective interventions to block the transmission of TB and thus eventually decrease morbidity and mortality due to this pandemic.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alain Rajoharison for PCR product cloning and sequencing and Alain Johnson for his support on the manuscript revision.
This work was funded by Fondation Mérieux. Qiagen AG kindly loaned the QIAxcel system and provided reagents.
We declare no conflict of interest, as no financial relationship was established between parties.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01611-14.
REFERENCES
- 1.WHO. 2013. WHO global tuberculosis report 2013. WHO, Geneva, Switzerland. [Google Scholar]
- 2.Mathema B, Kurepina NE, Bifani PJ, Kreiswirth BN. 2006. Molecular epidemiology of tuberculosis: current insights. Clin Microbiol Rev 19:658–685. doi: 10.1128/CMR.00061-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parwati I, van Crevel R, van Soolingen D. 2010. Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis 10:103–111. doi: 10.1016/S1473-3099(09)70330-5. [DOI] [PubMed] [Google Scholar]
- 4.Supply P, Magdalena J, Himpens S, Locht C. 1997. Identification of novel intergenic repetitive units in a mycobacterial two component system operon. Mol Microbiol 26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- 5.Savine E, Warren RM, van der Spuy GD, Beyers N, van Helden PD, Locht C, Supply P. 2002. Stability of variable-number tandem repeats of mycobacterial interspersed repetitive units from 12 loci in serial isolated of Mycobacterium tuberculosis. J Clin Microbiol 40:4561–4566. doi: 10.1128/JCM.40.12.4561-4566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sougakoff W. 2011. Molecular epidemiology of multidrug-resistant strains of Mycobacterium tuberculosis. Clin Microbiol Infect 17:800–805. doi: 10.1111/j.1469-0691.2011.03577.x. [DOI] [PubMed] [Google Scholar]
- 7.Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey NC, van Helden PD. 2004. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med 169:610–614. doi: 10.1164/rccm.200305-714OC. [DOI] [PubMed] [Google Scholar]
- 8.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Hass P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Beer JL, Kremer K, Ködmön C, Supply P, van Soolingen D, Global Network for the Molecular Surveillance of Tuberculosis 2009 . 2012. First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains. J Clin Microbiol 50:662–669. doi: 10.1128/JCM.00607-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemann S, Diel R, Khechinashvili G, Gegia M, Mdivani N, Tang YW. 2010. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J Clin Microbiol 48:3544–3550. doi: 10.1128/JCM.00715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shamputa IC, Jugheli L, Sadradze N, Willery E, Portaels F, Supply P, Rigouts L. 2006. Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir Res 7:99. doi: 10.1186/1465-9921-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferdinand S, Sola C, Verdol B, Legrand E, Goh KS, Berchel M, Aubéry A, Timothée M, Joseph P, Pape JW, Rastogi N. 2003. Molecular characterization and drug resistance patterns of strains of Mycobacterium tuberculosis isolated from patients in an AIDS counseling center in Port-au-Prince, Haiti: a 1-year study. J Clin Microbiol 41:694–702. doi: 10.1128/JCM.41.2.694-702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ocheretina O, Merveille YM, Mabou MM, Escuyer VE, Dunbar SA, Johnson WD, Pape JW, Fitzgerald DW. 2013. Use of Luminex MagPlex magnetic microspheres for high-throughput spoligotyping of Mycobacterium tuberculosis isolates in Port-au-Prince, Haiti. J Clin Microbiol 51:2232–2237. doi: 10.1128/JCM.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Supply P. 2005. Multilocus variable number tandem repeat genotyping of Mycobacterium tuberculosis: technical guide. http://www.miru-vntrplus.org/MIRU/files/MIRU-VNTRtypingmanualv6.pdf.
- 15.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and user-strategy of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto T, Koshii Y, Sakane K, Murakawa T, Hirayama Y, Yoshida H, Kurokawa M, Tamura Y, Nagai T, Kawase I. 2013. A novel approach to automated genotyping of Mycobacterium tuberculosis using a panel of 15 MIRU VNTRs. J Microbiol Methods 93:239–241. doi: 10.1016/j.mimet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 17.de Beer JL, Akkerman OW, Schurch AC, Mulder A, van der Werf TS, van der Zanden AG, van Ingen J, van Soolingen D. 2014. Optimization of standard in-house 24-locus variable-number tandem-repeat typing for Mycobacterium tuberculosis and its direct application to clinical material. J Clin Microbiol 52:1338–1342. doi: 10.1128/JCM.03436-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allix-Beguec C, Wahl C, Hanekom M, Nikolayevskyy V, Drobniewski F, Maeda S, Campos-Herrero I, Mokrousov I, Niemann S, Kontsevaya I, Rastogi N, Samper S, Sng LH, Warren RM, Supply P. 2014. Proposal of a consensus set of hypervariable mycobacterial interspersed repetitive-unit variable-number of tandem-repeat loci for subtyping of Mycobacterium tuberculosis Beijing isolates. J Clin Microbiol 52:164–172. doi: 10.1128/JCM.02519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allix C, Walravens K, Saegerman C, Godfroid J, Supply P, Fauville-Dufaux M. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J Clin Microbiol 44:1951–1962. doi: 10.1128/JCM.01775-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidovec-Stojkovic U, Zolnir-Dovc M, Supply P. 2011. One year nationwide evaluation of 24-locus MIRU-VNTR genotyping on Slovenian Mycobacterium tuberculosis isolates. Respir Med 105(Suppl 1):S67–S73. doi: 10.1016/S0954-6111(11)70014-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.