Abstract
Strains of Escherichia coli O26:H11 that were positive for stx2 alone (n = 23), which were not epidemiologically related or part of an outbreak, were isolated from pediatric patients in France between 2010 and 2013. We were interested in comparing these strains with the new highly virulent stx2a-positive E. coli O26 clone sequence type 29 (ST29) that has emerged recently in Europe, and we tested them by multilocus sequence typing (MLST), stx2 subtyping, clustered regularly interspaced short palindromic repeat (CRISPR) sequencing, and plasmid (ehxA, katP, espP, and etpD) and chromosomal (Z2098, espK, and espV) virulence gene profiling. We showed that 16 of the 23 strains appeared to correspond to this new clone, but the characteristics of 12 strains differed significantly from the previously described characteristics, with negative results for both plasmid and chromosomal genetic markers. These 12 strains exhibited a ST29 genotype and related CRISPR arrays (CRISPR2a alleles 67 or 71), suggesting that they evolved in a common environment. This finding was corroborated by the presence of stx2d in 7 of the 12 ST29 strains. This is the first time that E. coli O26:H11 carrying stx2d has been isolated from humans. This is additional evidence of the continuing evolution of virulent Shiga toxin-producing E. coli (STEC) O26 strains. A new O26:H11 CRISPR PCR assay, SP_O26_E, has been developed for detection of these 12 particular ST29 strains of E. coli O26:H11. This test is useful to better characterize the stx2-positive O26:H11 clinical isolates, which are associated with severe clinical outcomes such as bloody diarrhea and hemolytic uremic syndrome.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) is responsible for gastrointestinal diseases such as diarrhea or bloody diarrhea and can lead to hemolytic uremic syndrome (HUS). The most common EHEC serotype associated with human disease is O157:H7. However, a growing number of human EHEC infections are caused by non-O157 EHEC strains (1–4). Among non-O157 EHEC strains, O26:H11 has emerged as the most common serotype associated with severe diarrhea and HUS worldwide (1–6).
EHEC O26:H11 strains are very dynamic; they can undergo frequent genetic rearrangements in their chromosome, virulence plasmids, and pathogenicity islands. They also have the ability to rapidly lose and acquire stx-carrying phages (7), which makes them highly adaptable and may account for their global spread. Until recently, EHEC O26:H11 strains isolated from humans mostly harbored Shiga toxin 1 (Stx1) (Stx1a subtype) only or, more rarely, Stx1a associated with the Stx2a subtype. In the middle 1990s, however, a new EHEC O26:H11 clone carrying the Shiga toxin Stx2a subtype alone emerged in Europe (8–18). This new clone has also been observed in South America (19) and in the United States (1). Shiga toxin-producing E. coli (STEC) strains carrying the stx2 gene are usually associated with more severe outcomes (20). Indeed, this new O26:H11 clone appears highly virulent and is significantly associated with HUS (8, 9).
Multilocus sequence typing (MLST) analysis shows that the stx2a-harboring E. coli O26:H11 strains are mostly divided into 2 related phylogenetic groups, i.e., sequence type 21 (ST21) (which also contains the EHEC O26:H11 strains harboring stx1a alone or in combination with stx2a) and ST29 (which contains the new highly pathogenic clone carrying stx2a only) (9). The large EHEC plasmids encoding enterohemolysin (ehxA), catalase peroxidase (katP), serine protease (espP), and type II effector (etpD) can be found in most EHEC O26:H11 strains (8, 9), and the presence of these specific plasmid virulence determinants can be used to distinguish the 2 clones. ST21 is characterized by the plasmid gene combination ehxA+/katP+/espP+/etpD−, while ST29 exhibits the distinctive combination ehxA+/katP−/espP−/etpD+ (9).
Chromosomally encoded virulence factors such as the locus of enterocyte effacement (LEE) effectors and some type III secretion system effectors were also found to be conserved in the phylogenetic group ST21 (21). In the context of a molecular risk assessment strategy, we previously described a combination of molecular markers for specific identification of EHEC and EHEC-like O26:H11 strains. Assays for these markers included wzxO26, fliCH11, eae-β, stx, espK, and arcA single-nucleotide polymorphism (SNP) genotyping (21), as well as a set of PCR tests (SP_O26_C and SP_O26_D) targeting the clustered regularly interspaced short palindromic repeat (CRISPR) locus of EHEC O26:H11 (22).
Although they have been observed all over Europe, the stx2-harboring O26:H11 strains circulating in France have not been thoroughly characterized. In the present study, we analyzed 23 stx2-harboring E. coli O26 strains isolated from different sporadic infections that occurred in France between 2010 and 2013. We characterized their chromosomal and plasmid virulence gene contents to compare them with STEC O26 strains circulating in other countries in Europe. We also performed multilocus sequence typing (MLST), to examine phylogenetic relatedness, and CRISPR typing, to look for genetic diversity among the strains.
MATERIALS AND METHODS
Bacterial strains.
The surveillance of STEC infections in France is based on the surveillance of HUS in children <15 years of age. A nationwide surveillance system, relying on voluntary reporting of pediatric HUS cases by the pediatric nephrology units from a network of 31 public hospitals, was set up in 1996. Clinical strains used in this study are from the strain collection of the pediatric Hôpital Robert-Debré (Paris, France), which is the associated national reference laboratory for E. coli. Between 2010 and 2013, 46 STEC O26 strains were isolated from HUS in children <15 years of age. Except for 3 household transmissions, all O26-based infections were sporadic, and no food source was identified. During this time period, no O26 outbreak was reported. The 23 STEC O26:H11 strains studied were originally isolated from stool specimens or rectal swabs from patients with HUS in various part of France. They were not epidemiologically related or part of an outbreak. They were selected to include only strains positive for stx2 alone.
Genetic characterization by real-time PCR.
The 23 STEC O26:H11 strains were tested by real-time PCR for the presence of wzxO26, fliCH11, stx1, stx2, stx2a, eae, eae-β, ehxA, katP, espP, etpD, arcA allele 2, espK, espV, Z2098, O26:H11 CRISPR (assays SP_O26_C, SP_O26_D, and SP_O26_E), and wecA (as a genetic marker of E. coli), using a Biomark (Fluidigm, San Francisco, CA) or LightCycler Nano (Roche Diagnostics, Meylan, France) thermocycler. All primers and probes were described previously (21–24) except for the probe for arcA allele 2, which was modified from the report by Bugarel et al. (21) with locked nucleic acid (LNA)-substituted nucleotides to specifically bind arcA allele 2 (arcA2-Taq, 5′-CAAAATTCAGTCCC+G+TGC-3′ [+ indicates LNA-substituted nucleotides]), and SP_O26_E, for which the reverse primer (SP_O26-R, 5′-ATCAACATGCAGCGCGAACG-3′) was used with the previously described SP_O26_D forward primer and probe (22). The primers and probe for stx2a were as follows: stx2a-F, 5′-TTCTGTTAATGCAATGGCGGCG-3′; stx2a-R, 5′-CCAGTATTCTTTCCCGTCAACCTTC-3′; stx2a-Taq, 5′-AATGTGTCATCCTCATTATACTTGG-3′.
CRISPR typing.
Sequence polymorphisms of the CRISPR loci in the strains were examined using the nomenclature of CRISPR1 and CRISPR2a (25). The CRISPR loci were amplified by PCR as described previously (22). Amplicons were double-strand sequenced (Eurofins MWG Operon, Courtaboeuf, France) and the CRISPR sequences of the strains were assembled using BioEdit version 7.1.3.0. Analysis of the CRISPR loci was as described by Feng et al. (26).
Multilocus sequence typing.
Multilocus sequence typing (MLST) was performed for the 23 strains using seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA), as described by Wirth et al. (27) and modified by Bielaszewska et al. (7). The alleles and sequence types (STs) were assigned in accordance with the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli).
Partial sequencing of stx2 genes.
Partial sequencing of the stx2 genes of the 7 strains negative for stx2a was performed according to the method described by Scheutz et al. (28), with primers F4 and R1. Nucleotide sequences were assembled using BioEdit version 7.1.3.0 and were analyzed by comparison with the established reference sequences. The nucleotide sequences were translated to amino acids and compared using CLC Sequence Viewer version 7.0.2.
RESULTS
Strains and patients.
The 23 STEC O26:H11 strains studied were selected to include only strains positive for stx2 alone. These strains were isolated mainly from female patients (65%), rather than male patients (35%). The patients' median age at the time of isolation was 16 months (Table 1).
TABLE 1.
Characteristics of E. coli O26:H11 clinical isolates and patients
| Strain | Serotype | stx1 | stx2 | eae | eae-β | Yr of isolation | Origin | Specimen | Patient sexa | Patient age |
|---|---|---|---|---|---|---|---|---|---|---|
| 30993 | O26:H11 | − | 2a | + | + | 2010 | HUS | Stool | F | 2 yr (25 mo) |
| 31131 | O26:H11 | − | 2a | + | + | 2010 | HUS | Stool | F | 22 mo |
| 31132 | O26:H11 | − | 2a | + | + | 2010 | HUS | Stool | M | 14 mo |
| 31493 | O26:H11 | − | 2a | + | + | 2010 | HUS | Stool | F | 11 yr |
| 32802 | O26:H11 | − | 2d | + | + | 2011 | HUS | Stool | M | 9 mo |
| 32876 | O26:H11 | − | 2a | + | + | 2011 | HUS | Stool | F | 2 yr (26 mo) |
| 33116 | O26:H11 | − | 2a | + | + | 2011 | HUS | Rectal swab | M | 13 mo |
| 33344 | O26:H11 | − | 2a | + | + | 2011 | HUS | Stool | F | 3 mo |
| 33618 | O26:H11 | − | 2d | + | + | 2012 | HUS | Stool | F | 16 mo |
| 34130 | O26:H11 | − | 2a | + | + | 2012 | HUS | Rectal swab | F | 13 mo |
| 34195 | O26:H11 | − | 2a | + | + | 2012 | HUS | Rectal swab | M | 2 yr (32 mo) |
| 34586 | O26:H11 | − | 2d | + | + | 2012 | HUS | Rectal swab | M | 8 mo |
| 34620 | O26:H11 | − | 2a | + | + | 2012 | HUS | Stool | M | 2 yr (25 mo) |
| 34629 | O26:H11 | − | 2d | + | + | 2012 | HUS | Stool | M | 2 yr (28 mo) |
| 34760 | O26:H11 | − | 2a | + | + | 2012 | HUS | Rectal swab | F | 12 mo |
| 34827 | O26:H11 | − | 2a | + | + | 2012 | HUS | Stool | F | 14 mo |
| 34870 | O26:H11 | − | 2a | + | + | 2012 | HUS | Stool | M | 13 mo |
| 36079 | O26:H11 | − | 2a | + | + | 2013 | HUS | Stool | F | 2 yr |
| 36084 | O26:H11 | − | 2a | + | + | 2013 | HUS | Stool | F | 2 yr |
| 36293 | O26:H11 | − | 2d | + | + | 2013 | HUS | Stool | F | 7 mo |
| 36348 | O26:H11 | − | 2d | + | + | 2013 | HUS | Stool | F | 19 mo |
| 36493 | O26:H11 | − | 2d | + | + | 2013 | HUS | Stool | F | 4 mo |
| 36708 | O26:H11 | − | 2a | + | + | 2013 | HUS | Stool | F | 4 yr |
F, female; M, male.
Multilocus sequence typing.
In order to better characterize the phylogenetic relationship of these strains and to compare them with the highly virulent new stx2a-harboring clone ST29, we performed MLST analysis (Fig. 1). Two STs were obtained. Seven isolates were grouped in ST21 and 16 isolates in ST29, both from clonal complex 29 (CC29).
FIG 1.
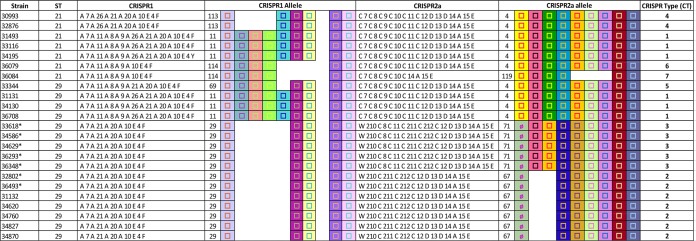
MLST and CRISPR typing of the O26:H11 clinical isolates. In the graphic representation of spacer arrangements in CRISPR1 and CRISPR2a, each unique spacer is represented by a unique combination of the center shape and background color. The shape in the center indicates the spacer length (□, 32 bp; Ø, 1,293 bp). Gaps were introduced to improve the alignment of similar CRISPR arrays. CRISPR1 and CRISPR2a allele numbers are shown in the respective columns to the left of the CRISPR arrays. Each unique combination of CRISPR1 and CRISPR2a alleles was assigned a CRISPR type (CT) number. *, stx2d-positive strains.
Plasmid gene profiles.
We characterized the plasmid gene contents of the 23 strains to compare them with STEC O26 strains circulating in other countries in Europe and described by others (8–10). The ehxA, katP, espP, and etpD genes were distributed in three different patterns (Table 2). One dominant profile was found in 12 strains (52% of isolated strains), in which all plasmid genes were absent. All strains in this profile belonged to ST29. One combination (lacking only etpD) was present in 7 strains, all of which belonged to ST21. The plasmid gene combination previously identified as characteristic of the new clone (ehxA+/katP-/espP-/etpD+) was found in 4 strains from ST29.
TABLE 2.
Plasmid and virulence gene profiles of E. coli O26:H11 clinical isolates
| Strain | stx2a | ehxA | katP | espP | etpD | arcA allele 2 | Z2098 | espK | espV | SP_O26_C | SP_O26_D | SP_O26_E |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 30993 | + | + | + | + | − | + | + | + | + | + | + | − |
| 32876 | + | + | + | + | − | + | + | + | + | + | + | − |
| 31493 | + | + | + | + | − | + | + | + | + | + | + | − |
| 33116 | + | + | + | + | − | + | + | + | + | + | + | − |
| 34195 | + | + | + | + | − | + | + | + | + | + | + | − |
| 36079 | + | + | + | + | − | + | + | + | + | + | + | − |
| 36084 | + | + | + | + | − | + | + | + | + | − | + | − |
| 33344 | + | + | − | − | + | + | + | + | + | + | + | − |
| 31131 | + | + | − | − | + | + | + | + | + | + | + | − |
| 34130 | + | + | − | − | + | + | + | + | + | + | + | − |
| 36708 | + | + | − | − | + | + | + | + | + | + | + | − |
| 33618a | − | − | − | − | − | − | − | − | − | − | + | + |
| 34586a | − | − | − | − | − | − | − | − | − | − | + | + |
| 34629a | − | − | − | − | − | − | − | − | − | − | + | + |
| 36293a | − | − | − | − | − | − | − | − | − | − | + | + |
| 36348a | − | − | − | − | − | − | − | − | − | − | + | + |
| 32802a | − | − | − | − | − | − | − | − | − | − | − | + |
| 36493a | − | − | − | − | − | − | − | − | − | − | − | + |
| 31132 | + | − | − | − | − | − | − | − | − | − | − | + |
| 34620 | + | − | − | − | − | − | − | − | − | − | − | + |
| 34760 | + | − | − | − | − | − | − | − | − | − | − | + |
| 34827 | + | − | − | − | − | − | − | − | − | − | − | + |
| 34870 | + | − | − | − | − | − | − | − | − | − | − | + |
stx2d-positive strain.
Virulence gene profiles.
We also tested all strains for the presence of a set of genetic markers previously postulated by Bugarel et al. (21) to be specific for EHEC and EHEC-like O26 strains (Table 2). All isolates were confirmed to be Escherichia coli through testing for the housekeeping gene wecA. All strains also tested positive for wzxO26, fliCH11, stx2, eae, and eae-β, while they all tested negative for stx1. Eleven strains (belonging to ST21 and ST29) tested positive for arcA allele 2, espK, espV, Z2098, and SP_O26_C and/or SP_O26_D.
Although positive for wzxO26, fliCH11, stx2, and eae-β, 12 strains exhibited peculiar genetic characteristics, as they appeared negative for arcA allele 2, espK, espV, Z2098, and SP_O26_C. Five of these strains were nonetheless positive for SP_O26_D. These 12 strains corresponded to those that were negative for all plasmid genes, and they belonged to ST29.
CRISPR typing.
All isolates were subtyped according to the sequences of the CRISPR1 and CRISPR2a loci. The spacer arrangements of CRISPR1 and CRISPR2a loci are shown in Fig. 1.
Five alleles were found for the CRISPR1 locus (21% allele diversity). These alleles are constituted by different modular arrangements of a set of 9 spacers. One CRISPR1 allele (allele 29) was found in 12/23 strains (52% of the strains), all of which belonged to ST29. This CRISPR1 allele was characterized by a reduced number of spacers (5 spacers). The strains carrying this CRISPR1 allele were also the ones in which all plasmid and virulence gene markers were absent. The next most frequent CRISPR1 allele (allele 11, found in reference strain 11368) was found in 6 strains (26%), which belonged to two different STs and plasmid profiles. Among the other less frequent CRISPR1 alleles, two alleles were found twice and one allele was found once.
We found four alleles for the CRISPR2a locus (17% allele diversity). In contrast to CRISPR1, the alleles of CRISPR2a are not variations of the same set of spacers but rather constitute 2 groups with a common core set of spacers and some unique spacers. The first group contained CRISPR2 alleles 67 and 71, which were found in the 12 strains that contained CRISPR1 allele 29 and in which all plasmid and virulence gene markers were absent. This group was characterized by the presence of a 1,260-bp transposon within the first spacer. These 2 CRISPR2 alleles otherwise differed by the presence of 2 additional spacers in allele 71. The second group, found in 11 strains, contained CRISPR2 allele 4 (found in reference strain 11368) and allele 119, which differed by the additional presence of 3 spacers in allele 4. Together, the CRISPR1 and CRISPR2a allele combinations form 7 CRISPR types (CTs).
Stx genotypes (subtypes).
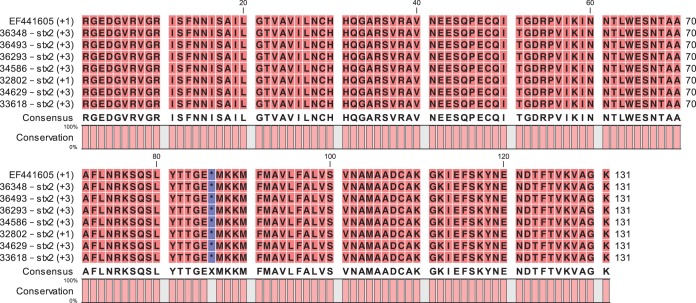
The stx2a subtype was tested in all strains by real-time PCR testing, and 16 strains were confirmed to be positive for stx2a. Surprisingly, 7 strains were found to be negative for stx2a although they were positive for stx2. To identify the stx2 subtype(s) of these strains, we determined the partial sequences of the stxAB2 operons of these 7 strains. The sequences were analyzed and compared with the published stx2 reference sequences (28). Through analysis of the nucleotide sequences, the stx2 gene for the 7 samples was identified as stx2d. The nucleotide sequences were 100% identical among the 7 samples and were most similar (one nucleotide difference) to that of strain 5905 (O55:H7; GenBank accession no. EF441605) (29). The sequences were translated to amino acids (Fig. 2), confirming the activatable type of the Stx2d toxin with the combined presence of the “activatable tail” in the Stx2A subunit (KSQSLYTTGE) and the END motif at positions 14 to 16 in the Stx2B subunit. As STEC can carry multiple stx2-subtype genes, we tested the 23 strains for all known stx2 subtypes using PCR assays described by Scheutz et al. (28). No strain was carrying multiple stx2-subtype genes. Strains were positive either for stx2a only or for stx2d only.
FIG 2.
Alignment with reference strain 5905 of the amino acid sequences of the C-terminal ends of the A subunits and the N-terminal ends of the B subunits of the 7 stx2a-negative and stx2d-positive strains. Numbers in parentheses indicate the reading frames used for amino acid translation. Asterisks indicate the separation of the amino acid sequences of subunits A and B of Stx2.
Development of CRISPR PCR test for detection of CRISPR2a alleles 67 and 71.
In order to detect the STEC O26:H11 strains that were negative for the previously published genetic markers arcA allele 2, espK, espV, Z2098, and O26 CRISPR (SP_O26_C), we developed a new real-time PCR assay targeting the CRISPR2a locus of the strains exhibiting CRISPR2a alleles 67 and 71, SP_O26_E. This assay was tested on all strains and, as expected, only strains with CRISPR2a alleles 67 and 71 were found to be positive (Table 2).
DISCUSSION
In France, 122 (in 2010) to 162 (in 2013) pediatric HUS cases were reported each year, 63% to 75% of which were confirmed by bacteriology or serology, between 2010 and 2013. Among these pediatric HUS patients, the majority (51%) were female (mean age, 2.6 years). During this time period, O26 was consistently the second most frequently isolated serogroup in France, representing 15% (in 2010) to 18% (in 2013) of isolated EHEC strains in HUS patients <15 years of age. Prior to the 1990s, the clinical EHEC O26:H11 strains isolated in Europe mainly possessed the stx1 gene alone or, more rarely, in association with stx2a. Over the past decade, a shift in the prevalence of EHEC O26:H11 strains recovered from humans has been observed. Since the middle 1990s, most of the clinical O26:H11 strains possess the stx2a gene alone or in association with stx1, and a new highly pathogenic clone carrying stx2a only has started to emerge in Europe (8–10).
In this study, we characterized the genetic properties of stx2-harboring O26:H11 clinical strains circulating in France between 2010 and 2013. We confirmed that the new highly pathogenic EHEC O26:H11 clone ST29 is circulating in France and constituted most of the stx2-harboring EHEC O26:H11 strains studied here, as 16 of the 23 strains appeared to correspond to this clone (ST29). This finding confirms observations made by others in different countries (3, 8–18). However, the characteristics of many of the strains analyzed here differed somewhat from the characteristics described previously.
Indeed, 12 strains were negative for all plasmid genes. This dominant profile (52% of the strains) was reported previously with a much lower prevalence (8–10). The possibility that the plasmid was lost during culture of the strains could not be excluded, however. Virulence genes present on the bacterial chromosome are more stable. However, the 12 strains that were negative for all plasmid genes were strikingly negative for the chromosomal genetic markers previously found to be specific for EHEC O26:H11 (21, 24, 30).
Analysis of the CRISPR loci shows that the CRISPR1 locus of the analyzed O26 strains is formed from a single repertoire of spacers. Globally, the CRISPR1 locus is short, with a limited number of spacers. It is typically shorter than the prototypical O26:H11 stx1 CRISPR1 locus (from reference strain 11368) and those described by Delannoy et al. (22) and Yin et al. (25). It has already been suggested (31) that CRISPR array length may be inversely proportional to the virulence potential of a strain. Our data seem to corroborate this hypothesis, as the strains belonging to the highly virulent new ST29 clone appear to exhibit a shorter CRISPR1 allele length. However, such an observation is not true for the CRISPR2a allele length, as the CRISPR2a loci from the two groups did not differ significantly in length. The CRISPR2a locus of the 12 ST29 strains that are negative for all plasmid genes has an interesting feature, however, as it contains a large transposon within the first spacer. This transposon is found in CRISPR2 alleles 67 and 71. Allele 67 was previously described in an O26:H11 isolate from cattle carrying a stx2 gene (25). It was not found previously in human isolates. Allele 71 was observed previously in human stx-negative diarrheagenic E. coli O26 strains (32). It was not previously found to be related to stx-positive strains associated with bloody diarrhea and hemolytic uremic syndrome (HUS). Overall, CRISPR typing appears to be more discriminative than MLST, as a total of 7 CRISPR types (CTs) were identified for only 2 STs. Also, CRISPR typing but not MLST allowed identification of the strains that were negative for all plasmid and virulence genes. Thus, CRISPR typing provided the best strain profiling resolution, corroborating recent findings for other STEC serotypes such as E. coli O113:H21 (26). Moreover, our previous studies on E. coli O26 showed that O26 CRISPR loci can be used to differentiate clearly between EHEC and non-EHEC strains (22). Interestingly, we also showed in those previous studies that O26 strains with H-types other than H11 (O26:HND, O26:H31, O26:H34, and O26:H32) harbor significantly different CRISPR loci (22).
Although all strains seem to have closely related genetic backgrounds phylogenetically, as ST21 and ST29 differ by one nucleotide in the adk locus, their different CRISPR types suggest different evolutionary niches or pathways. CRISPR arrays are thought to act as an adaptive immune system, in which the spacer sequences of the arrays would be derived from foreign DNA following exposure to plasmids or phages (33, 34). Thus, different spacer compositions of CRISPR arrays may indicate exposure to different phage and plasmid environments. Similarly, STEC strains with different niches (i.e., exposed to different spectra of aggressors) will likely have different CRISPR arrays. Accordingly, all ST21 strains and the four ST29 strains (with the plasmid gene profile of ehxA+/katP−/espP−/etpD+ and the presence of all virulence genes) (CRISPR types CT1, CT4, CT5, CT6, and CT7) most probably share a background, as they exhibit similar CRISPR arrays. Similarly, the 12 ST29 strains (negative for all plasmid genes) have related CRISPR arrays and probably evolved in a common environment. This hypothesis is corroborated by the presence of the stx2d gene subtype in 7 of the 12 ST29 strains. To our knowledge, this is the first time that E. coli O26:H11 strains carrying stx2d have been described in humans. However, one stx2d-positive STEC O26 strain (D618/98) was isolated from cheese and cattle in Germany in 1998 (21, 30). The Stx2d-activatable toxin is usually produced by eae-negative STEC strains of serotypes O113:H21 and O91:H21 that carry stx2d alone and that were found to be associated with severe diseases such as bloody diarrhea and HUS (35).
The stx2d gene sequences in all 7 strains are identical and are most similar to reference strain 5905 (E. coli O55:H7), which was isolated from food in 1994. E. coli O26 is a highly dynamic group capable of acquiring stx-encoding phages (7). Although stx genes are generally highly conserved and show little sequence variation (29), it is likely that these stx2d-positive strains resulted from a single acquisition. Our findings highlight the continuing evolution of virulent STEC O26 strains and remind us that previously observed stable virulence gene combinations are subject to change (e.g., stx2d is now found in combination with eae). It was postulated by Bletz et al. that a diverse population of EHEC O26 strains circulated over a long period of time in an evolutionarily stable niche (source) and only a few strains were able to adapt during the transfer into a new niche (sink), with positive purifying selection (36). Highly pathogenic EHEC O26 strains of the new clones carrying solely stx2a or stx2d have only rarely been isolated outside humans. Further studies are necessary to elucidate potential reservoirs of these new clones and to understand the evolutionary dynamics between source and sink.
In an attempt to improve the current detection scheme for EHEC in food samples, we previously identified genetic markers associated with EHEC O26:H11 (21, 22, 24). Analysis of the plasmid and chromosomal virulence gene contents of the strains showed that 12 strains of ST29 were unexpectedly negative for all of the tested genetic markers. In order to detect these particular strains, including the E. coli O26:H11 strain carrying the stx2d gene, we developed a new CRISPR-based O26:H11 PCR assay, SP_O26_E. This new CRISPR-based O26:H11 PCR assay may be a useful addition for fully investigating STEC O26:H11 isolates that are associated with severe clinical outcomes, i.e., bloody diarrhea and HUS. A complete evaluation of this PCR assay with a large collection of strains should be performed in order to validate its use in STEC O26:H11 characterization. Further studies in France or in other countries will be necessary to better determine whether O26 strains with these profiles of stx types and genotypes are statistically significant in humans.
ACKNOWLEDGMENT
The project was partially financed by the French Joint Ministerial Program of R&D against CBRNE Risks.
REFERENCES
- 1.Brooks JT, Sowers EG, Wells JG, Greene KD, Griffin PM, Hoekstra RM, Strockbine NA. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J Infect Dis 192:1422–1429. doi: 10.1086/466536. [DOI] [PubMed] [Google Scholar]
- 2.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis 43:1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 3.European Food Safety Authority, European Centre for Disease Prevention and Control. 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J 10:2597. doi: 10.2903/j.efsa.2012.2597. [DOI] [Google Scholar]
- 4.European Food Safety Authority, European Centre for Disease Prevention and Control. 2014. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J 12:3547. doi: 10.2903/j.efsa.2014.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin L, Krause G, Zimmermann S, Kaulfuss S, Gleier K. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J Clin Microbiol 42:1099–1108. doi: 10.1128/JCM.42.3.1099-1108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vally H, Hall G, Dyda A, Raupach J, Knope K, Combs B, Desmarchelier P. 2012. Epidemiology of Shiga toxin producing Escherichia coli in Australia, 2000-2010. BMC Public Health 12:63. doi: 10.1186/1471-2458-12-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielaszewska M, Prager R, Kock R, Mellmann A, Zhang W, Tschape H, Tarr PI, Karch H. 2007. Shiga toxin gene loss and transfer in vitro and in vivo during enterohemorrhagic Escherichia coli O26 infection in humans. Appl Environ Microbiol 73:3144–3150. doi: 10.1128/AEM.02937-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang WL, Bielaszewska M, Liesegang A, Tschape H, Schmidt H, Bitzan M, Karch H. 2000. Molecular characteristics and epidemiological significance of Shiga toxin-producing Escherichia coli O26 strains. J Clin Microbiol 38:2134–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bielaszewska M, Mellmann A, Bletz S, Zhang W, Kock R, Kossow A, Prager R, Fruth A, Orth-Holler D, Marejkova M, Morabito S, Caprioli A, Pierard D, Smith G, Jenkins C, Curova K, Karch H. 2013. Enterohemorrhagic Escherichia coli O26:H11/H−: a new virulent clone emerges in Europe. Clin Infect Dis 56:1373–1381. doi: 10.1093/cid/cit055. [DOI] [PubMed] [Google Scholar]
- 10.Zweifel C, Cernela N, Stephan R. 2013. Detection of the emerging Shiga toxin-producing Escherichia coli O26:H11/H− sequence type 29 (ST29) clone in human patients and healthy cattle in Switzerland. Appl Environ Microbiol 79:5411–5413. doi: 10.1128/AEM.01728-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobieszczanska BM, Gryko R, Malek CW. 2000. Isolation of verotoxigenic strains of Escherichia coli O26 in Poland. Clin Microbiol Infect 6:227–229. doi: 10.1046/j.1469-0691.2000.00060-1.x. [DOI] [PubMed] [Google Scholar]
- 12.Paciorek J. 2002. Virulence properties of Escherichia coli faecal strains isolated in Poland from healthy children and strains belonging to serogroups O18, O26, O44, O86, O126 and O127 isolated from children with diarrhoea. J Med Microbiol 51:548–556. [DOI] [PubMed] [Google Scholar]
- 13.Allerberger F, Friedrich AW, Grif K, Dierich MP, Dornbusch HJ, Mache CJ, Nachbaur E, Freilinger M, Rieck P, Wagner M, Caprioli A, Karch H, Zimmerhackl LB. 2003. Hemolytic-uremic syndrome associated with enterohemorrhagic Escherichia coli O26:H infection and consumption of unpasteurized cow's milk. Int J Infect Dis 7:42–45. doi: 10.1016/S1201-9712(03)90041-5. [DOI] [PubMed] [Google Scholar]
- 14.Ethelberg S, Olsen KE, Scheutz F, Jensen C, Schiellerup P, Enberg J, Petersen AM, Olesen B, Gerner-Smidt P, Molbak K. 2004. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg Infect Dis 10:842–847. doi: 10.3201/eid1005.030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liptakova A, Siegfried L, Kmetova M, Birosova E, Kotulova D, Bencatova A, Kosecka M, Banovcin P. 2005. Hemolytic uremic syndrome caused by verotoxin-producing Escherichia coli O26: case report. Folia Microbiol (Praha) 50:95–98. doi: 10.1007/BF02931454. [DOI] [PubMed] [Google Scholar]
- 16.Kappeli U, Hachler H, Giezendanner N, Beutin L, Stephan R. 2011. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerg Infect Dis 17:180–185. doi: 10.3201/eid1702.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chase-Topping ME, Rosser T, Allison LJ, Courcier E, Evans J, McKendrick IJ, Pearce MC, Handel I, Caprioli A, Karch H, Hanson MF, Pollock KG, Locking ME, Woolhouse ME, Matthews L, Low JC, Gally DL. 2012. Pathogenic potential to humans of bovine Escherichia coli O26, Scotland. Emerg Infect Dis 18:439–448. doi: 10.3201/eid1803.111236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstraete K, de Reu K, van Weyenberg S, Piérard D, de Zutter L, Herman L, Robyn J, Heyndrickx M. 2013. Genetic characteristics of Shiga toxin-producing E. coli O157, O26, O103, O111 and O145 isolates from humans, food, and cattle in Belgium. Epidemiol Infect 141:2503–2515. doi: 10.1017/S0950268813000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivas M, Miliwebsky E, Chinen I, Roldan CD, Balbi L, Garcia B, Fiorilli G, Sosa-Estani S, Kincaid J, Rangel J, Griffin PM. 2006. Characterization and epidemiologic subtyping of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathog Dis 3:88–96. doi: 10.1089/fpd.2006.3.88. [DOI] [PubMed] [Google Scholar]
- 20.Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol 37:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl Environ Microbiol 77:2275–2281. doi: 10.1128/AEM.02832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delannoy S, Beutin L, Fach P. 2012. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J Clin Microbiol 50:4035–4040. doi: 10.1128/JCM.02097-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugarel M, Beutin L, Martin A, Gill A, Fach P. 2010. Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int J Food Microbiol 142:318–329. doi: 10.1016/j.ijfoodmicro.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Delannoy S, Beutin L, Fach P. 2013. Discrimination of enterohemorrhagic Escherichia coli (EHEC) from non-EHEC strains based on detection of various combinations of type III effector genes. J Clin Microbiol 51:3257–3262. doi: 10.1128/JCM.01471-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin S, Jensen MA, Bai J, Debroy C, Barrangou R, Dudley EG. 2013. The evolutionary divergence of Shiga toxin-producing Escherichia coli is reflected in clustered regularly interspaced short palindromic repeat (CRISPR) spacer composition. Appl Environ Microbiol 79:5710–5720. doi: 10.1128/AEM.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng P, Delannoy S, Lacher D, dos Santos LF, Beutin L, Fach P, Rivas M, Hartland E, Paton A, Guth BC. 2014. Genetic diversity and virulence potential of Shiga toxin-producing Escherichia coli O113:H21 strains isolated from clinical, environmental, and food sources. Appl Environ Microbiol 80:4757–4763. doi: 10.1128/AEM.01182-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JE, Reed J, Shields MS, Spiegel KM, Farrell LD, Sheridan PP. 2007. Phylogenetic analysis of Shiga toxin 1 and Shiga toxin 2 genes associated with disease outbreaks. BMC Microbiol 7:109. doi: 10.1186/1471-2180-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miko A, Lindstedt BA, Brandal LT, Lobersli I, Beutin L. 2010. Evaluation of multiple-locus variable number of tandem-repeats analysis (MLVA) as a method for identification of clonal groups among enteropathogenic, enterohaemorrhagic and avirulent Escherichia coli O26 strains. FEMS Microbiol Lett 303:137–146. doi: 10.1111/j.1574-6968.2009.01874.x. [DOI] [PubMed] [Google Scholar]
- 31.Toro M, Cao G, Ju W, Allard M, Barrangou R, Zhao S, Brown E, Meng J. 2014. Association of clustered regularly interspaced short palindromic repeat (CRISPR) elements with specific serotypes and virulence potential of Shiga toxin-producing Escherichia coli. Appl Environ Microbiol 80:1411–1420. doi: 10.1128/AEM.03018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazen TH, Sahl JW, Redman JC, Morris CR, Daugherty SC, Chibucos MC, Sengamalay NA, Fraser-Liggett CM, Steinsland H, Whittam TS, Whittam B, Manning SD, Rasko DA. 2012. Draft genome sequences of the diarrheagenic Escherichia coli collection. J Bacteriol 194:3026–3027. doi: 10.1128/JB.00426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR: a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 34.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 35.Bielaszewska M, Friedrich AW, Aldick T, Schurk-Bulgrin R, Karch H. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin Infect Dis 43:1160–1167. doi: 10.1086/508195. [DOI] [PubMed] [Google Scholar]
- 36.Bletz S, Bielaszewska M, Leopold SR, Köck R, Witten A, Schuldes J, Zhang W, Karch H, Mellmann A. 2013. Evolution of enterohemorrhagic Escherichia coli O26 based on single-nucleotide polymorphisms. Genome Biol Evol 5:1807–1816. doi: 10.1093/gbe/evt136. [DOI] [PMC free article] [PubMed] [Google Scholar]