Abstract
Human infections caused by toxigenic corynebacteria occur sporadically across Europe. In this report, we undertook the epidemiological and molecular characterization of all toxigenic corynebacterium strains isolated in England between January 2007 and December 2013. Epidemiological aspects include case demographics, risk factors, clinical presentation, treatment, and outcome. Molecular characterization was performed using multilocus sequence typing (MLST) alongside traditional phenotypic methods. In total, there were 20 cases of toxigenic corynebacteria; 12 (60.0%) were caused by Corynebacterium ulcerans, where animal contact was the predominant risk factor. The remaining eight (40.0%) were caused by Corynebacterium diphtheriae strains; six were biovar mitis, which were associated with recent travel abroad. Adults 45 years and older were particularly affected (55.0%; 11/20), and typical symptoms included sore throat and fever. Respiratory diphtheria with the absence of a pharyngeal membrane was the most common presentation (50.0%; 10/20). None of the eight C. diphtheriae cases were fully immunized. Diphtheria antitoxin was issued in two (9.5%) cases; both survived. Two (9.5%) cases died, one due to a C. diphtheriae infection and one due to C. ulcerans. MLST demonstrated that the majority (87.5%; 7/8) of C. diphtheriae strains represented new sequence types (STs). By adapting several primer sequences, the MLST genes in C. ulcerans were also amplified, thereby providing the basis for extension of the MLST scheme, which is currently restricted to C. diphtheriae. Despite high population immunity, occasional toxigenic corynebacterium strains are identified in England and continued surveillance is required.
INTRODUCTION
Toxigenic diphtheria can be caused by three Corynebacterium species, namely, C. diphtheriae, C. ulcerans, and C. pseudotuberculosis. Classic respiratory diphtheria is characterized by a gray throat membrane (“pseudomembrane”) and bull-neck appearance, while cutaneous diphtheria is characterized by chronic, nonhealing ulcers (1, 2). While C. diphtheriae is spread through direct contact, respiratory droplets, and aerosols from infected individuals, C. ulcerans and C. pseudotuberculosis are less common globally and are usually associated with farm animal contact and dairy products (3, 4). Diphtheria toxin can affect the myocardium and nervous and adrenal tissues, causing paralysis and cardiac failure (3, 4).
Although diphtheria is still endemic in some parts of the world, the global incidence of diphtheria has decreased substantially as a result of the introduction of a highly effective vaccine with increased vaccination coverage. High immunization coverage across the United Kingdom (since 1991, at least 93% of children completed the primary course by their second birthday) resulted in only a few cases of diphtheria being reported over the last decades (5, 6). In 2009, serological surveys indicated that 75% of the United Kingdom population had at least basic protection against diphtheria (≥0.01 IU/ml), compared to 60% in 1996 (6).
In the United Kingdom, diphtheria is a statutory notifiable disease, where reporting is based upon clinical and microbiological diagnosis. The majority of isolates are nontoxigenic strains which require no public health action (5, 6); however, toxigenic strains continue to be isolated occasionally (7), and an understanding of the characteristics of patients infected is important to inform risk assessments and a proportionate public health response to individual cases, as well as immunization policy.
Diphtheria is still endemic in many countries and is a potentially resurgent disease, so it is important to maintain the ability to genotype corynebacteria. Molecular typing has an important role in public health: The application of appropriate typing methods is essential not only in outbreak investigations to monitor the evolution and spread of epidemic clones of C. diphtheriae but also in understanding and predicting epidemics. The selection of an appropriate typing method depends on a number of factors, including the scale of the investigation and the financial and technical resources available. Several typing methods have been applied to C. diphtheriae genotyping, but many are laborious and time-consuming or require expensive equipment. Additionally, their use may be hindered by limited portability and, in some instances, poor reproducibility. Multilocus sequence typing (MLST) is able to circumvent these limitations by directly analyzing nucleotide information within selected housekeeping genes (8).
In this report, we examined all toxigenic corynebacteria from respiratory and cutaneous diphtheria cases isolated in England between January 2007 and December 2013. The epidemiology of toxigenic diphtheria in the United Kingdom during 1986 to 2008 has previously been reported, showing the increasing role of C. ulcerans (5). Here, we provide an update on the current epidemiology of toxigenic diphtheria, including demography, risk factors, clinical presentation, diphtheria antitoxin administration, and outcome. Strains were characterized using traditional phenotypic methods as well as MLST. We further demonstrated that the C. diphtheriae housekeeping genes can also be amplified in C. ulcerans using slightly modified primers, thereby providing proof of concept that the diphtheria MLST scheme can be extended to incorporate both C. diphtheriae and C. ulcerans.
MATERIALS AND METHODS
Epidemiological and molecular data were collected on toxigenic diphtheria cases reported to Public Health England (PHE), Colindale, London. Routine surveillance for diphtheria is based on clinical and laboratory notifications. Under the Infectious Disease (Notification) Act of 1889 and the updated 2010 regulations, doctors in England have a statutory duty to notify of all forms of diphtheria diagnosed clinically, including cutaneous presentations (9). Also under these regulations, laboratories have a duty to notify of isolates of C. diphtheriae and C. ulcerans. Public Health England (PHE) also requests notification of isolates of C. pseudotuberculosis. Laboratories notify the local Health Protection Teams in PHE Centres, and all such isolates are referred to the Respiratory and Vaccine Preventable Bacteria Reference Unit (RVPBRU), which is the national reference laboratory for toxigenicity testing (9). In addition to notifications and mandatory laboratory reporting, PHE has conducted enhanced surveillance of all toxigenic cases since 1986.
About three-quarters (16/20) of toxigenic C. diphtheriae and C. ulcerans isolates investigated were isolated from throat/nasal swabs or from wounds/ulcers. Microbiological procedures were conducted according to the World Health Organization manual for the laboratory diagnosis of diphtheria (10). Briefly, Gram-positive bacilli consistent with corynebacteria were subcultured onto Tinsdale, tellurite, and blood agar medium and biochemically identified using the API Coryne strip according to the manufacturer's instructions (bioMérieux, Durham, NC) (11). All strains described in this report produced positive toxigenicity results using the modified Elek test, which was performed according to the work of Engler et al. (12). The genetic relationship between the C. diphtheriae isolates from Britain was further characterized using the MLST scheme as described by Bolt et al. (8). Briefly, extracted DNA was amplified by PCR using the corresponding primers (Table 1) for the seven C. diphtheriae housekeeping genes atpA, dnaE, dnaK, fusA, leuA, odhA, and rpoB. After amplification, the respective genes were sequenced, and allelic profiles and sequence type (ST) designations for each studied strain were obtained by submitting the generated DNA alleles to the PubMLST database curated by the Pasteur Institute Paris (http://pubmlst.org/cdiphtheriae/). A novel ST designation was given to all unique allelic profiles, while isolates with identical profiles belonged to the same ST.
TABLE 1.
PCR primer sequences for amplification of the seven MLST housekeeping genesa
| Gene | Gene function | Amplification primer |
Sequencing primer |
||||
|---|---|---|---|---|---|---|---|
| Sequence |
Size (bp) | Sequence |
Size (bp) | ||||
| Fwd | Rev | Fwd | Rev | ||||
| atpA | ATP synthase alpha chain | GCGATTGCGAACTACACC | CTCGAGGAATACCTRACC | 1,029 | AGAAGGCGACGAAGTMAAGC | CRGAATCAGAAGCTGGWGCA | 378 |
| dnaE | DNA polymerase III alpha subunit | TGCGTCATCTGATTGAAA; C. ulcerans, TCCGAAACCTCATCGAGA | CGGTCCAATAAGACACCA; C. ulcerans, CAGTCCAATAAGAAACTA | 858 | GTGCGACAAGCTGGTGTG; C. ulcerans, GTGCGCCAAGCAGGTGTC | GGCTTWCGGCCATTYTTG; C. ulcerans, GGTTTACGGCCATTCTTG | 354 |
| dnaK | Chaperone protein DnaK | ACTTGGGTGGCGGTACTT; C. ulcerans, ACCTCGGCGGCGGAACCT | TGGTGAACGTCTCGGAAC; C. ulcerans, TGGTAAAGGTCTCAGAAC | 696 | AGATGGCTATGCAGCGTCT; same for C. ulcerans | GATGAGCTTGGTCATCACG; C. ulcerans, GATCAGCTTGGTCATCACG | 345 |
| fusA | Elongation factor G | TACCGCGAGAAGCTCGTT | GAAGGTTGGGTCCTCTTC | 683 | CGTAAGCTGACCGTTAACTC | CCATGGACTCRAGGATGA | 360 |
| leuA | 2-Isopropylmalate synthase | CGTGCACTTCTACAACTC | ACCGTGATCGGTCTTCAT | 865 | CCYATCATCATCAAYCTGCC | CAGCTGGTTGCAGTAYTC | 384 |
| odhA | 2-Oxoglutarate dehydrogenase | CGGCAAGGAAASCATGAC | GTTGTCGCCRAACATCTG | 505 | TBCAAGATCGCATYGARRC | TWGGCTCGATGTGKCCTTC | 382 |
| rpoB | RNA polymerase beta chain | AAGCGCAAGATCCAGGAC | TCGAACTCGTCGTCATCC | 845 | CGWATGAACATYGGBCAGGT | TCCATYTCRCCRAARCGCTG | 342 |
The dnaE and dnaK primer sequences specific for C. ulcerans are underlined; all other primers were the same for both C. diphtheriae and C. ulcerans.
Eleven nontoxigenic C. diphtheriae strains (nine of biovar gravis and two of biovar mitis) taken from recent isolates were also investigated to assess whether they have similar allelic profiles compared to the toxigenic strains or compared with each other.
To extend the MLST scheme to include C. ulcerans, we initially attempted to amplify the seven C. ulcerans housekeeping genes with the C. diphtheriae PCR primers, using a panel of 10 C. ulcerans strains, which were representative of all of the isolates received from England. Five of the seven genes (atpA, fusA, leuA, odhA, and rpoB) had highly conserved sequences in C. ulcerans and C. diphtheriae, and these genes could also be amplified in C. ulcerans using the C. diphtheriae primers. However, two genes (dnaE and dnaK) had more divergent sequences and uniformly failed to amplify across all C. ulcerans strains. The C. diphtheriae-specific primer sequences (Table 1) were therefore modified according to the corresponding dnaE and dnaK sequences from C. ulcerans (Table 1). These primers targeted exactly the same genetic regions as in C. diphtheriae. All genes across the 10 tested C. ulcerans isolates could thereby be amplified with the primers listed in Table 1, including the modified dnaE and dnaK primers, and a lowered annealing temperature of 45°C for all seven genes (all other PCR parameters were as previously described [8]).
RESULTS
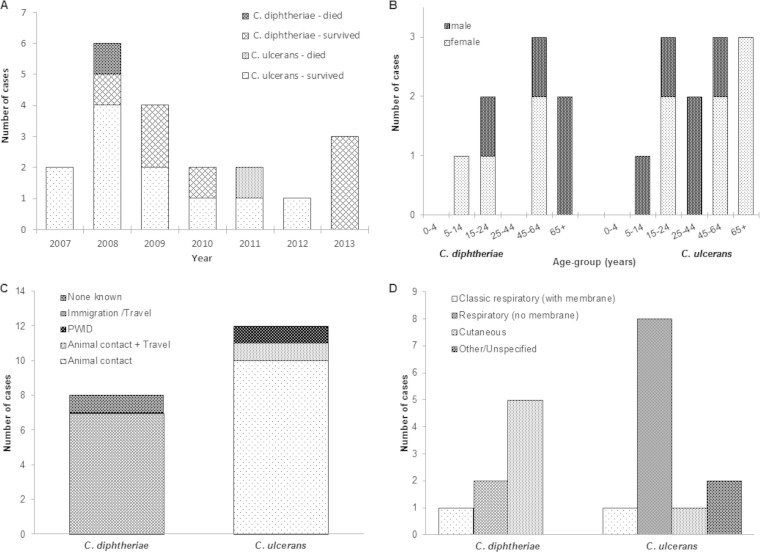
Clinical and epidemiological data for the 20 toxigenic diphtheria cases with onset dates between January 2007 and December 2013 are shown in Fig. 1. The number of cases each year ranged from one to six cases, with a peak in 2008 (Fig. 1A). Eleven of the 20 patients were female (Fig. 1B). Older adults were particularly affected: five patients were >65 years old, six were 45 to 64 years, two were 25 to 44 years, five were 14 to 24 years, and two were 5 to 14 years (Fig. 1B). Two-fifths (8/20) of all toxigenic diphtheria infections recorded in England were observed in London. All but two patients recovered; the case fatality ratio was 10.0% (Fig. 1A).
FIG 1.
Epidemiological and clinical information for human diphtheria cases recorded in Britain. (A) Annual diphtheria cases October 2006 to December 2013. (B) Age group and gender of cases. (C) Risk factors for cases. PWID, person who injects drugs. (D) Diagnosis.
The 20 toxigenic isolates were taken from throat/nasal swabs, wounds/ulcers, sputum, blood culture, tissue, or other sources (Table 2). Twelve toxigenic isolates were identified as C. ulcerans, and eight were identified as C. diphtheriae (Table 2).
TABLE 2.
Clinical characteristics and MLST profiles for toxigenic and nontoxigenic C. diphtheriae and C. ulcerans isolates, 2007 to 2014a
| Yr | Organism | Age (yr) | Sex | Type of sample | Allelic profile | ST |
|---|---|---|---|---|---|---|
| 2007 | C. ulcerans | 54 | Male | Throat swab | 20-19-11-47-43-37-7 | 287 |
| 2007 | C. ulcerans | 54 | Female | Throat swab | 20-6-11-47-43-37-17 | 288 |
| 2008 | C. ulcerans | 42 | Male | Other | ||
| 2008 | C. ulcerans | 17 | Female | Throat swab | 20-6-11-47-43-37-17 | 288 |
| 2008 | C. ulcerans | 20 | Male | Throat swab | 20-25-11-11-43-37-17 | 289 |
| 2008 | C. diphtheriae biovar mitis | 7 | Female | Other (lavage) | 2-12-4-1-41-23-2 | 266 |
| 2008 | C. diphtheriae biovar mitis | 16 | Male | Throat swab | 3-2-3-6-41-8-2 | 267 |
| 2008 | C. ulcerans | 89 | Female | Throat swab | 20-6-11-47-43-37-17 | 288 |
| 2009 | C. ulcerans | 30 | Male | Throat swab | 20-19-11-42-41-35-17 | 286 |
| 2009 | C. ulcerans | 82 | Female | Other (blood) | 20-30-72-42-29-35-17 | 285 |
| 2009 | C. diphtheriae biovar mitis | 74 | Male | Skin swab | 13-4-8-44-3-23-13 | 261 |
| 2010 | C. diphtheriae biovar gravis | 15 | Female | Throat swab | 5-2-7-1-3-5-8 | 10 |
| 2010 | C. ulcerans | 19 | Female | Throat swab | ||
| 2010 | C. diphtheriae biovar mitis | 57 | Female | Skin swab | 3-8-28-16-3-23-2 | 262 |
| 2011 | C. ulcerans | 59 | Female | Other (sputum) | 20-6-11-47-43-37-17 | 288 |
| 2011 | C. ulcerans | 67 | Female | Other (tissue) | 20-19-11-47-43-37-7 | 287 |
| 2012 | C. ulcerans | 10 | Male | Throat swab | 20-19-11-47-43-37-7 | 287 |
| 2013 | C. diphtheriae biovar mitis | 65 | Male | Skin swab | 4-10-3-1-3-23-27 | 263 |
| 2013 | C. diphtheriae biovar mitis | 48 | Female | Skin swab | 30-23-63-25-3-23-28 | 265 |
| 2013 | C. diphtheriae biovar mitis | 54 | Male | Skin swab | 2-4-8-19-3-23-9 | 264 |
| 2006 | Nontoxigenic C. diphtheriae biovar gravis | 19 | Male | Skin swab | 3-1-72-4-43-23-5 | 276 |
| 2007 | Nontoxigenic C. diphtheriae biovar gravis | 2-4-72-1-43-23-5 | 277 | |||
| 2007 | Nontoxigenic C. diphtheriae biovar gravis | 11-1-53-4-43-23-24 | 278 | |||
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 19 | Female | Skin swab | 3-1-72-4-43-37-32 | 279 |
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 19 | Male | Skin swab | 3-1-72-4-43-23-5 | 276 |
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 19 | Female | Skin swab | 3-1-72-4-43-23-32 | 281 |
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 30 | Male | Skin swab | 3-1-72-4-43-40-32 | 280 |
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 24 | Female | 3-1-72-4-43-40-5 | 282 | |
| 2014 | Nontoxigenic C. diphtheriae biovar mitis | 56 | Female | Skin swab | 2-10-72-4-43-23-2 | 283 |
| 2014 | Nontoxigenic C. diphtheriae biovar mitis | 29 | Male | Skin swab | 2-4-72-1-19-40-5 | 284 |
| 2014 | Nontoxigenic C. diphtheriae biovar gravis | 14 | Female | Other (tissue) | 3-1-72-4-43-40-5 | 282 |
All STs were novel, with the exception of ST 10 (bold).
C. diphtheriae.
Among the eight cases with laboratory-confirmed toxigenic C. diphtheriae infection, the median age at time of illness was 51 years (range, 7 to 74 years); 4 patients (50%) were female (Fig. 1B). Vaccination histories were available for six cases: five were known to have received diphtheria toxoid-containing vaccines, and one was unimmunized. Seven cases had a recent history of travel/immigration (Fig. 1C); six had stayed in a country where diphtheria was endemic. Five were diagnosed with cutaneous diphtheria (Fig. 1D), and three respiratory cases presented with sore throats, one of whom had “classical” diphtheria with a pseudomembrane. All cases were treated with antibiotics; none were given diphtheria antitoxin. An unimmunized school-aged child presented with symptoms consistent with laryngeal diphtheria, although this was not recognized at the time of treatment and was diagnosed only postmortem.
C. ulcerans.
The median age of the 12 cases with laboratory-confirmed toxigenic C. ulcerans infections was 48 years (range, 10 to 89 years); seven (58.3%) were female (Fig. 1B). Seven cases had received diphtheria toxoid-containing vaccines; however, none were fully immunized, two were unimmunized, and immunization status was unknown in three cases. Animal contact was the predominant risk factor for C. ulcerans infection (Fig. 1C), and 11 cases had contact with companion animals (dogs, cats, or rabbits), three of whom also had contact with farm and/or wild animals. Three-quarters (9/12) of the cases presented with sore throat and were diagnosed with respiratory diphtheria, one of whom had a “classical” presentation with a pseudomembrane (Fig. 1D). Of the three remaining cases, one presented with a necrotic patch diagnosed as cutaneous diphtheria, another presented with a sore throat and endocarditis, and one presented with an oozing wound following surgery (Fig. 1D). All were known to have been treated with antibiotics. Two cases also received diphtheria antitoxin: one was a fatal case in an unimmunized adult who had “classical” respiratory diphtheria with systemic involvement, and the other had mild respiratory diphtheria without membrane or systemic involvement. The second fatal case had an unknown immunization status and presented with respiratory diphtheria and stridor.
MLST analyses.
While the clinical and public health management methods are identical for all toxigenic strains of C. diphtheriae, four biovars can generally be distinguished biochemically: gravis, intermedius, mitis, and belfanti. Biovar mitis (75.0%; 6/8) was the most prevalent C. diphtheriae strain; the remaining two strains belonged to biovar gravis. One C. pseudotuberculosis strain was recorded (5) but was later classified as an atypical C. ulcerans strain as confirmed by real-time PCR (RT-PCR), and rpoB gene sequencing (data not shown). This strain was further confirmed as C. ulcerans using matrix-assisted laser deionization–time of flight (MALDI-TOF) analysis (Bruker MALDI Biotyper Microflex, Biotyper software, sample pretreatment by extraction with formic acid [70%] and acetonitrile [ACN], both SR Taxonomy and Bruker Taxonomy library used for sample run). Interestingly, some of the biochemical results for this specimen, which was isolated from the patient's aortic root vegetation, were atypical for C. ulcerans (API Coryne results were trehalose and glycogen negative, which are characteristic of C. pseudotuberculosis).
MLST demonstrated that the large majority of C. diphtheriae isolates represented new STs (Table 2). One strain of biovar gravis, isolated in 2010, was shown to represent ST 10, identical with a strain previously isolated in England in 1993 (8). No associations were observed between ST and epidemiological characteristics. Eleven nontoxigenic C. diphtheriae isolates were included as reference strains. The nontoxigenic strains also had allelic profiles and STs similar to those of each other. Unlike the findings for the toxigenic C. diphtheriae strains, sequence analysis revealed two pairs of identical STs, ST 276 and ST 282. However, all of the nontoxigenic reference strains differed from toxigenic strains (Table 2).
In addition to the C. diphtheriae strains described above, preliminary MLST analysis of 10 representative toxigenic C. ulcerans strains was performed. By modifying the dnaE and dnaK primers, the seven MLST genes could be amplified in C. ulcerans. A sequence comparison revealed two distinct clusters; four isolates were designated ST 288 and three isolates were ST 287 (Table 2). There was no direct epidemiological link apparent between the patients in either cluster; there was at least 6 months between each case and no known contact between the cases or their companion animals. Sequence comparison also showed that several strains differed by only a few alleles (Table 2). There were no significant associations between ST and epidemiological or clinical characteristics. It should be noted that the listed allele types and STs for C. ulcerans are currently provisional, i.e., the identified C. ulcerans STs are currently included in the C. diphtheriae database but might eventually be part of a separate database specific for C. ulcerans.
DISCUSSION
Diphtheria vaccination is extremely effective, and high vaccination coverage across the United Kingdom resulted in only a few cases of diphtheria being reported over the last 2 decades. The current vaccine coverage for the routine childhood vaccination program has been maintained at around 95% for the last 2 decades, so the majority of cases in the United Kingdom present as mild infections either in partially immunized individuals or occasionally in adults who have been fully immunized but have waning immunity. According to serological surveillance studies, the proportion of individuals susceptible to diphtheria infections remains high, even in vaccinated populations (6, 13). Our study showed that a total of 20 diphtheria cases occurred in England between January 2007 and December 2013. All but two patients recovered. Two cases received diphtheria antitoxin, which is a relatively scarce equine polyclonal antibody preparation that neutralizes the bacterial toxin before it binds to tissue (14).
The main risk factor for C. diphtheriae infection was travel or contact with travelers returning from an area of endemicity (5, 7). In contrast, the main risk factor for acquiring C. ulcerans infection was animal contact. Toxigenic C. ulcerans infections are frequently acquired from companion animals, including dogs and cats (15). The epidemiology of diphtheria in England is consistent with that in the earlier study by Wagner et al. (5), with more toxigenic isolates of C. ulcerans than C. diphtheriae infection. Sporadic importations of toxigenic C. diphtheriae cases and indigenous C. ulcerans infections emphasize the need to maintain routine vaccination coverage at or above 95%, as recommended by WHO.
We observed here that older generations in particular are at higher risk of contracting diphtheria. This confirms previous observations, e.g., it has recently been estimated that 70 to 75% of those aged 50 to 60 years old across the United Kingdom show inadequate protection levels (5). Another study in 2009 (6) further confirmed the risk for older generations, as the age group with the largest proportion of individuals susceptible to diphtheria and tetanus was >70 years (>32% susceptible). Clinicians could theoretically use routine consultations as opportunities to check the immunization status of elderly patients who may not have received diphtheria immunizations during childhood and of adult patients born before 1980 who would not have been offered a routine booster dose of diphtheria toxoid-containing vaccine at school-leaving age (introduced in 1995), although this of course has major cost implications and might not be feasible. However, the opportunity to ensure that patients are up to date with the recommended doses of diphtheria toxoid-containing vaccine should not be missed at pretravel consultations, particularly when travelers are going to countries where diphtheria is endemic.
All toxigenic strains were characterized using traditional phenotypic methods and MLST. All but one of the C. diphtheriae isolates represented new STs, but a single toxigenic isolate comprising biovar gravis was identified as ST 10, identical with a strain previously isolated in the United Kingdom in 1993 (8). While it is possible that this strain may have circulated in England over the last 2 decades, it is more likely that these cases were due to imported infections which originated from an area where C. diphtheriae, and ST 10, is endemic.
The 11 nontoxigenic C. diphtheriae reference strains were shown to differ from the circulating toxigenic strains by several alleles. However, sequence analysis identified similarities between the nontoxigenic isolates, and two pairs of identical strains were observed, ST 276 and ST 282.
Furthermore, our preliminary MLST analysis for 10 C. ulcerans strains from England identified two clusters of patients with the same ST, ST 287 and ST 288, although there was no epidemiological link identified between these patients. Sequence comparison also demonstrated that most of the C. ulcerans strains had similar allelic profiles. While there were differences in the STs provisionally assigned to the C. ulcerans strains, several strains showed differences only in a few alleles. This might indicate that these strains are related, which would suggest that the C. ulcerans strains circulating in England are highly conserved. However, it should be emphasized that the allele types and STs assigned to C. ulcerans are only provisional. Further investigation of toxigenic C. ulcerans strains from England and elsewhere will shed light on how closely these strains are related.
The lack of statistical association between ST and clinical or demographic characteristics of the case is unsurprising given the comparatively small number of cases and the number of distinct STs identified. However, with the continued analysis of toxigenic Corynebacterium isolates, associations may become apparent.
Due to the occurrence of diphtheria in countries of endemicity and in countries with broad immunization coverage, typing tools for diphtheria surveillance are of great importance. While several typing techniques for C. diphtheriae have previously been developed, their use may frequently be hindered by limited reproducibility and subjective analysis, e.g., by visual inspection of gel band patterns. Recently, a comparison of the different typing techniques was performed (16), and ribotyping was shown to be the most discriminative method, allowing identification of 86 distinct ribotype patterns and cluster isolates associated with outbreaks in the former Soviet Union. However, ribotyping depends very much upon the use of a rigid standardized method, and without this, there might be difficulties in reproducibility. MLST overcomes some problems observed with ribotyping, previously considered the gold standard, by directly indexing nucleotide variation within several core metabolic genes, thereby providing high-resolution data appropriate for evolutionary and epidemiological investigations (17, 18). The extended MLST scheme incorporating both C. diphtheriae and C. ulcerans provides a valuable tool for monitoring and characterizing circulating strains in the United Kingdom and abroad.
ACKNOWLEDGMENTS
We thank the clinicians, microbiologists, and PHE Health Protection Teams who completed enhanced surveillance forms.
We declare no conflicts of interest.
REFERENCES
- 1.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. 2000. The pathology of diphtheria. J Infect Dis 181(Suppl 1):S116–S120. doi: 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 2.Popovic T, Mazurova IK, Efstratiou A, Vuopio-Varkila J, Reeves MW, De Zoysa A, Glushkevich T, Grimont P. 2000. Molecular epidemiology of diphtheria. J Infect Dis 181:S168–S177. doi: 10.1086/315556. [DOI] [PubMed] [Google Scholar]
- 3.Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, Azevedo V. 2006. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res 37:201–218. doi: 10.1051/vetres:2005056. [DOI] [PubMed] [Google Scholar]
- 4.Wagner KS, White JM, Neal S, Crowcroft NS, Kuprevičiene N, Paberza R, Lucenko I, Jöks U, Akbas E, Alexandrou-Athanassoulis H, Detcheva A, Vuopio J, von Hunolstein C, Murphy PG, Andrews N, Members of the Diphtheria Surveillance Network, Efstratiou A. 2011. Screening for Corynebacterium diphtheriae and Corynebacterium ulcerans in patients with upper respiratory tract infections 2007–2008: a multicentre European study. Clin Microbiol Infect 17:519–525. doi: 10.1111/j.1469-0691.2010.03269.x. [DOI] [PubMed] [Google Scholar]
- 5.Wagner KS, White JM, Crowcroft NS, De Martin S, Mann G, Efstratiou A. 2010. Diphtheria in the United Kingdom, 1986–2008: the increasing role of Corynebacterium ulcerans. Epidemiol Infect 138:1519–1530. doi: 10.1017/S0950268810001895. [DOI] [PubMed] [Google Scholar]
- 6.Wagner KS, White JM, Andrews NJ, Borrow R, Stanford E, Newton E, Pebody RG. 2012. Immunity to tetanus and diphtheria in the UK in 2009. Vaccine 30:7111–7117. doi: 10.1016/j.vaccine.2012.09.029. [DOI] [PubMed] [Google Scholar]
- 7.Wagner KS, White JM, Lucenko I, Mercer D, Crowcroft NS, Neal S, Efstratiou A, Diphtheria Surveillance Network . 2012. Diphtheria in the postepidemic period, Europe, 2000–2009. Emerg Infect Dis 18:217–225. doi: 10.3201/eid1802.110987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolt F, Cassiday P, Tondella ML, Dezoysa A, Efstratiou A, Sing A, Zasada A, Bernard K, Guiso N, Badell E, Rosso ML, Baldwin A, Dowson C. 2010. Multilocus sequence typing identifies evidence for recombination and two distinct lineages within Corynebacterium diphtheriae. J Clin Microbiol 48:4177–4185. doi: 10.1128/JCM.00274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Public Health England (PHE) Diphtheria Guidelines Working Group. 2014. Public health control and management of diphtheria (in England and Wales). Interim guidelines 2014. Public Health England (PHE) Diphtheria Guidelines Working Group, London, United Kingdom: https://www.gov.uk/government/publications/diphtheria-public-health-control-and-management-in-england-and-wales. [Google Scholar]
- 10.Begg N. 1994. Manual for the management and control of diphtheria in the European region. Expanded Programme on Immunization in the European Region of WHO. WHO Regional Office for Europe, Copenhagen, Denmark. [Google Scholar]
- 11.Romney MG, Roscoe DL, Bernard K, Lai S, Efstratiou A, Clarke AM. 2006. Emergence of an invasive clone of nontoxigenic Corynebacterium diphtheriae in the urban poor population of Vancouver, Canada. J Clin Microbiol 44:1625–1629. doi: 10.1128/JCM.44.5.1625-1629.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engler KH, Glushkevich T, Mazurova IK, George RC, Efstratiou A. 1997. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J Clin Microbiol 35:495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmunds WJ, Pebody RG, Aggerback H, Baron S, Berbers G, Conyn-van Spaendonck MA, Hallander HO, Olander R, Maple PA, Melker HE, Olin P, Fievret-Groyne F, Rota C, Salmaso S, Tischer A, von Hunolstein C, Miller E. 2000. The sero-epidemiology of diphtheria in Western Europe. ESEN Project. European Sero-Epidemiology Network. Epidemiol Infect 125:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Both L, White J, Mandal S, Efstratiou A. 2014. Access to diphtheria antitoxin for therapy and diagnostics. Euro Surveill 19:pii:20830 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20830. [DOI] [PubMed] [Google Scholar]
- 15.Corti MA, Bloemberg GV, Borelli S, Kutzner H, Eich G, Hoelzle L, Lautenschlager S. 2012. Rare human skin infection with Corynebacterium ulcerans: transmission by a domestic cat. Infection 40:575–578. doi: 10.1007/s15010-012-0254-5. [DOI] [PubMed] [Google Scholar]
- 16.De Zoysa A, Hawkey P, Charlett A, Efstratiou A. 2008. Comparison of four molecular typing methods for characterization of Corynebacterium diphtheriae and determination of transcontinental spread of C. diphtheriae based on BstEII rRNA gene profiles. J Clin Microbiol 46:3626–3635. doi: 10.1128/JCM.00300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowe CF, Bernard KA, Romney MG. 2011. Cutaneous diphtheria in the urban poor population of Vancouver, British Columbia, Canada: a 10-year review. J Clin Microbiol 49:2664–2666. doi: 10.1128/JCM.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viguetti SZ, Pacheco LG, Santos LS, Soares SC, Bolt F, Baldwin A, Dowson CG, Rosso ML, Guiso N, Miyoshi A, Hirata R Jr, Mattos-Guaraldi AL, Azevedo V. 2012. Multilocus sequence types of invasive Corynebacterium diphtheriae isolated in the Rio de Janeiro urban area, Brazil. Epidemiol Infect 140:617–620. doi: 10.1017/S0950268811000963. [DOI] [PubMed] [Google Scholar]