Abstract
Rotaviruses are leading causes of gastroenteritis in the young of many species. Molecular epidemiological studies in children suggest that interspecies transmission contributes to rotavirus strain diversity in people. However, population-based studies of rotaviruses in animals are few. We investigated the prevalence, risk factors for infection, and genetic diversity of rotavirus A in a cross-sectional survey of cats housed within 25 rescue catteries across the United Kingdom. Morning litter tray fecal samples were collected during the winter and summer in 2012 from all pens containing kittens and a random sample of those housing adult cats. Group A rotavirus RNA was detected by real-time reverse transcription-PCR, and positive samples were G and P genotyped using nested VP4 and VP7 PCR assays. A total of 1,727 fecal samples were collected from 1,105 pens. Overall, the prevalence of rotavirus was 3.0% (95% confidence interval [CI], 1.2 to 4.9%). Thirteen out of 25 (52%; 95% CI, 31.3 to 72.2%) centers housed at least one rotavirus-positive cat. The prevalence of rotavirus was associated with season (odds ratio, 14.8 [95% CI, 1.1 to 200.4]; P = 0.04) but not age or diarrhea. It was higher during the summer (4.7%; 95% CI, 1.2 to 8.3%) than in winter (0.8%; 95% CI, 0.2 to 1.5%). Asymptomatic epidemics of infection were detected in two centers. G genotypes were characterized for 19 (33.3%) of the 57 rotavirus-positive samples and P genotypes for 36 (59.7%). Two rotavirus genotypes were identified, G3P[9] and G6P[9]. This is the first population-based study of rotavirus in cats and the first report of feline G6P[9], which questions the previous belief that G6P[9] in people is of bovine origin.
INTRODUCTION
Rotavirus A (RVA), a species of the Rotavirus genus and the family Reoviridae, is an important pathogen causing acute diarrhea in the young of many animal species, including people (1, 2). With the advent of modern sequencing techniques, the extent of the contribution of interspecies transmission and reassortment to rotavirus genetic diversity is increasingly being realized (3–5). Despite the many reports of potential zoonotic infections in people (6, 7), due to the scarcity of rotavirus surveillance programs in animals, little is known about the prevalence of potential anthroponotic and zoonotic strains in animals. Particularly, our knowledge and understanding of rotaviruses circulating in companion animal populations is minimal, which is remiss when we consider the extent of contact that occurs between pets (especially cats and dogs) and children in developed countries.
Although infections with feline rotaviruses (FRVs) rarely cause severe illness in cats (8–11), FRVs have captured attention as perpetuating, albeit infrequent, sources of human disease. Human RVAs with genetic homology to feline RVAs have been isolated from widespread geographical locations, including Japan (12, 13), Israel (14, 15), Tunisia (16), and the United States (17). Additionally, putative human/feline reassortant rotaviruses have been identified in children in Italy (18).
Despite the reports of FRV isolations from people, there have been no recent studies or surveillance of rotaviruses in cats. FRV infection was first identified by serology in cats in 1978 (19). Experimental infections have given inconsistent results, with some showing an association between rotavirus and reduced fecal quality (increased water content and suboptimal stool conformation) in kittens (8), while others failed to link infection with disease (9, 10, 20). Currently, rotavirus is considered to play a minor role in clinical disease and is not routinely screened for in diarrheic cases in small-animal veterinary practices (21).
The prevalence of rotavirus infection in cats has been investigated by serum antibody titers and fecal electron microscopy; seroprevalence studies indicated that exposure ranged from 3.5% to 100% (8–11, 22), while electron microscopy indicated 3 to 6% of cats were infected (9, 10, 20). These studies involved small numbers of cats and looked at convenience samples of subpopulations (e.g., veterinary hospital admissions, single premises, and research colonies) rather than a representative sample of the national feline population.
The aim of this study was to examine, using a systematic population-based approach irrespective of diarrhea status, the prevalence and genotypes of RVA circulating in domestic cats in the United Kingdom.
MATERIALS AND METHODS
Study population.
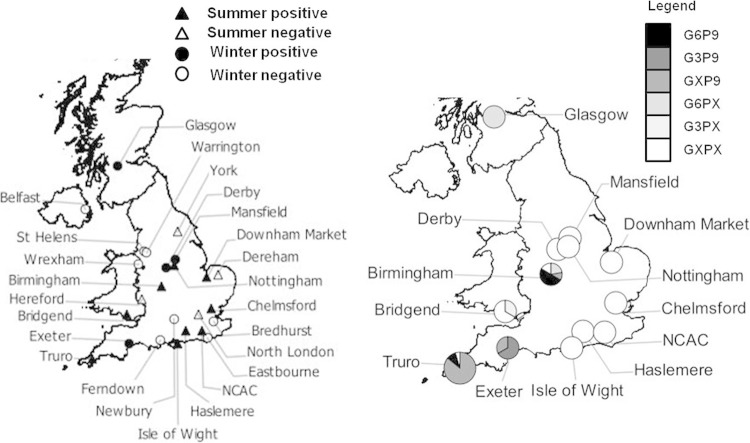
The study population comprised cats held in the 25 rehoming or adoption centers in the United Kingdom run by Cats Protection, the United Kingdom's largest feline welfare charity. Each year, it rehomes or reunites approximately 50,000 cats with owners. These centers are widely distributed geographically, with 21 in England, two in Wales, one in Northern Ireland, and one in Scotland (Fig. 1, left). The centers vary in size and their construction. The number of cat accommodation spaces (pens) are fixed and range from 16 in a specially adapted city house to 202 in the purpose-built National Cat Adoption Centre (NCAC).
FIG 1.
Cats Protection Adoption Centres and feline rotavirus distribution in the United Kingdom. (Left) Distribution of Cats Protection Adoption Centres across the United Kingdom and their feline rotavirus (FRV) status in 2012. (Right) Distribution of feline rotavirus (FRV) genotypes between the 12 FRV-positive Cats Protection Adoption Centres in the United Kingdom. The pie chart markers represent strain distribution within a center, but their size does not indicate prevalences between centers. X, unable to determine genotype. The maps were generated using QGIS 2.0.1 (Dufour).
The populations of cats in the centers are in constant flux. The relinquishment and adoption rates are such that centers operate at capacity throughout the year, although stocking density (cats per pen) will vary. The demographic pattern in the centers changes with the seasonal breeding pattern of cats, such that a higher proportion of the population are kittens during the summer months (June to August) (Fig. 2). Additionally, the centers will not put cats originating from different sources in the same pen, so relinquishment events (single cats or multicat households) will also influence the total population size and stocking density in each center.
FIG 2.
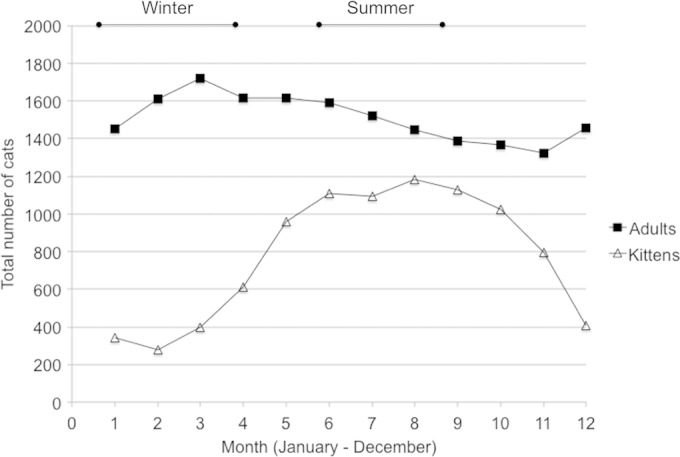
Seasonal demographics of the cat population of the Cats Protection Adoption Centres. Cats are seasonal breeders (spring to autumn), resulting in high numbers of kittens over the summer months. Demographic data were used from 2010 to plan the collection periods (indicated as winter and summer at the top of the graph); this pattern remains relatively constant between the years. The data shown are from 2012, indicating the true distribution of the cat population sampled.
Study design.
Two cross-sectional studies were undertaken to account for the seasonal changes in demography. The first was in the United Kingdom during the winter months (3 February to 30 March 2012), and the second was in the summer months (29 May to 17 August 2012). The centers were stratified by size (small, medium, and large) and randomly allocated to summer and winter collection periods (Fig. 1, left).
The unit of sampling was the pen. These samples were selected from pen occupancy data obtained the day prior to sampling. All pens containing at least one kitten were selected. In addition, a random sample of those housing one or more adult cats was also selected; the sample size was chosen to allow 95% confidence of detecting one positive pen if the prevalence of fecal shedding was 2%, assuming a test with 100% specificity and 95% sensitivity. This also allowed a prevalence of 2% to be estimated, with 95% confidence and 1.3 to 1.9% precision.
Recording sheets were used to transcribe demographic data from a number of sources, including center admission records, pen data recording sheets, veterinary paper records, the internal database of Cats Protection (PAWS), and from observations of pen content and construction, unit structure, hygiene precautions, and center management.
Sample collection and processing.
Fecal samples were collected from litter trays and, where necessary, the pen floor between 6.30 a.m. and 12.00 p.m. (noon) on the first day of the study visit; where feces were not present, the cats were monitored through the rest of the working day, and a collection was made if feces were passed. In large centers, where the number of cats necessitated a longer visit for data transcription, cats who defecated only once every 2 to 3 days were observed, and samples were collected from these individuals when feces were eventually passed.
Fecal samples were collected in sterile 30-ml universal tubes using either individual disposable gloves or wooden applicator sticks. The color, consistency, and number of complete deposits of feces in and outside the litter tray were recorded. The color was recorded as brown, green, yellow, black, or other (which was described). Consistency was graded as 1 (watery) to 6 (hard, dry), using a modified version of the Bristol Stool Scale (Meyers Scale) (23). Where the number of fecal deposits was equal to the number of cats, it was assumed that each was from a different cat (as an adult cat is highly unlikely to defecate twice in the morning unless it has large intestinal diarrhea or diffuse gastrointestinal disease resulting in a high volume of feces); the deposits were then randomly assigned to each cat. Occasionally, cats were observed defecating, or dual-occupancy pens contained cats that reliably produced differently graded feces; these deposits were specifically assigned to an individual cat. In the maternity pens, adult stools were easily differentiated from kitten stools by their size. In single-occupancy pens, where there was more than one deposit, a sample was taken from each and the sample recorded as “pooled from an individual.” In multiple-occupancy pens, where the number of samples exceeded the number of cats, the sample was recorded as “pooled sample from more than one cat.” The fecal samples were also examined for the presence or absence of worms, mucus, and blood. The samples were transferred at 4°C or −20°C when all pens in a particular building had been sampled. They were transported to the laboratory at this temperature and either aliquoted into two 1.5-ml cryovials the following day or kept at −80°C until aliquoted. The samples were kept at −80°C until testing.
Detection and typing of rotavirus.
Ten percent fecal suspensions were prepared, clarified by centrifugation at 12,000 rpm for 10 min, and a 250-μl aliquot was used for RNA extraction (QIAamp RNA kit; Qiagen, Manchester, United Kingdom), according to the manufacturer's instructions, and eluted into 50 μl of RNase-free water. A 20-μl aliquot was used for reverse transcription using random primers and SuperScript II reverse transcriptase (Invitrogen, Paisley, United Kingdom), according to the manufacturer's instructions, giving a final cDNA volume of 35 μl. Rotavirus NSP3-specific quantitative PCR (qPCR) (24) was performed, and samples with a threshold cycle (CT) value of <40 were considered positive. The qPCR mixture per reaction was 12.5 μl of Invitrogen Platinum master mix, 8 pmol NSP3-F/NSP3-R primers, and 3 fmol NSP-3 probe with 2 μl of cDNA, made up to 25 μl with RNase-free water. The assays were run on a Rotor-Gene Q 6000 thermal cycler at 50°C for 2 min, 95°C for 2 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min.
Limit of detection.
The limit of detection of the NSP3 qPCR assay was investigated using a simian rotavirus positive control, strain SA11 (25). This was diluted in a 2-fold series from 90 infectious virus particles (IVP)/reaction to 3 IVP/reaction, and repeatability was assessed with five replicates of a 10-fold dilution series from 1 × 105 IVP/reaction to 1 × 101 IVP/reaction and with two replicates of a 2-fold dilution series from 1,000 to 16 IVP/reaction. The assay was validated further for the limit of detection of feline RVs FRV-1 (26) and FRV-64 (27).
Genotyping.
Rotavirus-positive samples were further characterized using heminested VP7 (G-type) and VP4 (P-type) PCR assays, which included primers specific for genotype 1 (G1), G2, G3, G4, G8, G9, G10, and G12, and P[4], P[6], P[8], P[9], P[10], and P[11] (28, 29). To increase the G typing sensitivity, a fragment of the VP7 gene segment from culture-adapted FRV strains from this study was sequenced, using the consensus VP7 primers from the G typing first amplification round. The sequence data generated were used to characterize the G types of the strains that could be propagated in cell culture, and also to design feline G6- and G3-specific primers. These primers were subsequently used in single typing assays to test all rotavirus-positive samples. A total of 20 pmol primers G6F (AACGAGGATGATGGACTACA) (nucleotides [nt] 126 to 145) and G3R (TARATAGATCCTGTTGGCC) (nt 347 to 329) were used in separate heminested reactions, with VP7R or VP7F first-round consensus primers, respectively, using Top Taq master mix (Qiagen, United Kingdom) and an annealing temperature of 50°C.
Statistical analysis.
Prevalence was estimated at the cat and center levels for the combined and separate collection periods. The overall prevalence at the cat level was estimated using the svy commands in Stata statistical software (release 11; 2009, Stata Corp LP, College Station, TX) to adjust for stratification by season and clustering by center and pen. The sampling weights were adjusted for the different sampling strategies (i.e., for pens with kittens and those with adult cats) and for those cats that did not defecate on the day of collection. The prevalence at the center level was the proportion of centers with at least one positive cat, with exact binomial confidence intervals. Fisher's exact tests were used to compare the center prevalences in the summer and winter. Hierarchical univariable and multivariable logistic regression analyses were used to examine the associations between infection and age, season, and diarrhea using the melogit commands in Stata. A three-level model was used, incorporating the center, pen, and individual. Age was modeled as a continuous variable and also as a binary variable. Three categories were used for kittens: cats age ≤5 months, ≤3 months, or ≤2 months. Fecal score was also reduced to a binary variable, with a cutoff at both scores 2 and 3. Cats with a score ≤2 or 3 were considered to have diarrhea.
Geographical distribution maps were constructed using QGIS 2.0.1 (Dufour).
Ethics approval.
This study was approved by the University of Liverpool Veterinary Research Ethics Committee (VREC20) and the Cats Protection ethics review committee.
RESULTS
Population structure.
A total of 1,727 fecal samples were collected from 1,105 pens across the 25 centers. The median number of occupied pens per center was 41 (interquartile range [IQR], 30 to 79). The number of samples from each center varied from 8 in the North London center to 224 in Bridgend. The ages of cats sampled ranged from 1 week to 21.5 years of age. Overall, approximately one-third (30.9%; confidence interval [CI], 29 to 32.8% [683/2,213]) of the Cats Protection Adoption Centre population was kittens (<6 months). The proportion of cats that were kittens was significantly greater during the summer collection period (Fig. 2), at 44.6% compared with 13.9% in the winter period (P < 0.0005). This was also reflected in the sample, where the proportion of kittens was 33.6% overall (CI, 31.4 to 35.9% [581/1,727]), at 48.2% in the summer and 11.6% in the winter. The median ages of the cats sampled in the summer were 9.0 months (IQR, 3.0 to 38.0 months) compared with 36 months (IQR, 10.5 to 84.0) for the winter-sampled population (P < 0.0005).
A total of 85.6% (95% CI, 83.6 to 87.8%) of the cats defecated during the collection period. These cats were of similar age to those that did not defecate. The median ages were 16.5 months (IQR, 3.3 to 62 months) and 26.5 months (IQR, 1.5 to 84.5 months), respectively. Of the fecal samples, 1,686/1,727 (97.6%) were scored for consistency. The median fecal score was 5 (ideal) (IQR, 4.0 to 5.0). Of the samples, 5.6% (95/1,686) had a fecal score of 6 (dry/hard), 11.9% (201/1,686) had a score of ≤3 (diarrhea), and 2.4% (41/1,686) were ≤2 (watery diarrhea). Of the samples, 60.2% (1,040/1,727) were collected during the summer.
Rotavirus prevalence.
The overall estimated prevalence of rotavirus qPCR-positive fecal samples in the population was 3.0% (95% CI, 1.2 to 4.9% [57/1,727]), and 52% of the centers (CI, 31.3 to 72.2% [13/25]) housed at least one positive cat (Fig. 1, left). There were differences in the prevalences between the two collection periods and individual centers (Table 1). The prevalence was 4.7% (CI, 1.2 to 8.3%) in the summer collection period compared with 0.8% (95% CI, 0.2 to 1.5%) in the winter period (P < 0.0001). The proportion of centers with rotavirus-positive cats was also higher during the summer (69.2%; CI, 44.3 to 89.4%) than that in winter (33.3%; CI, 11.6 to 62.3%), but this difference was not statistically significant (P = 0.16).
TABLE 1.
Prevalence of fecal rotavirus in different centersa
| Center | Prevalence (% [no. with rotavirus/total no.]) | 95% CI (%)b |
|---|---|---|
| Summer | ||
| Birmingham | 10.2 (18/176)c | 8.2–12.7 |
| Bridgend (Wales) | 1.3 (4/224) | 0.9–2.0 |
| Chelmsford | 3.5 (2/57) | 1.5–8.1 |
| Dereham | 0 (0/75) | 0–3.9 |
| Downham Market | 2.6 (2/85) | 1.6–4.1 |
| Haslemere | 3.9 (1/31) | 1.2–11.9 |
| Hereford | 0 (0/43) | 0–8.2 |
| IOWd | 1.5 (1/73) | 0.9–2.6 |
| NCAC | 0.6 (1/172) | 0–2.1.7 |
| North London | 0 (0/8) | 0–36.9 |
| Nottingham | 8.9 (2/24) | 4.9–15.6 |
| Truro | 61.3 (21/34)c | 57.6–64.8 |
| York | 0 (0/38) | 0–9.3 |
| Winter | ||
| Belfast (Ireland) | 0 (0/65) | 0–5.4 |
| Bredhurst | 0 (0/93) | 0–3.8 |
| Derby | 0.8 (1/134) | 0.4–1.4 |
| Eastbourne | 0 (0/46) | 0–7.7 |
| Exeter | 2.9 (3/97) | 1.7–5.1 |
| Ferndown | 0 (0/27) | 0–12.8 |
| Glasgow (Scotland) | 1.2 (1/69) | 0.4–3.4 |
| Mansfield | 2.9 (1/30) | 1.2–6.5 |
| Newbury | 0 (0/25) | 0–13.7 |
| St. Helens | 0 (0/31) | 0–11.2 |
| Warrington | 0 (0/47) | 0–7.6 |
| Wrexham (Wales) | 0 (0/23) | 0–14.8 |
Centers in Ireland, Scotland, and Wales are indicated; the rest of the centers are in England.
95% CI, 95% confidence interval.
Centers with prevalences significantly higher than those in other centers, P < 0.0001.
IOW, Isle of Wight.
The higher prevalence in the summer was a reflection of two centers (Table 1), Truro and Birmingham, in which the prevalences were 61.2% (CI, 57.5 to 65.1%) and 10.2% (7.9 to 12.5%), respectively. These prevalences were significantly higher than those in other centers (P < 0.0001) and were considered to be infection epidemics. The prevalences in the remaining centers ranged from 0 to 8.9%. There was no significant difference in the prevalences in the centers sampled in the winter period, which ranged from 0 to 2.9%.
Lack of association between rotavirus and diarrhea.
The presence of rotavirus RNA in feces was not associated with diarrhea. The median fecal score for rotavirus-positive feces was 5.5 (IQR, 4 to 5), compared with 5 (IQR, 4 to 5) for rotavirus-negative feces. This difference was not statistically significant (P = 0.17). When a binary variable was created, with a fecal score of either 3 or ≤2 considered to be diarrhea, there was still no association with the presence of rotavirus. The odds ratios (OR) were 1.83 (95% CI, 0.60 to 5.53; P = 0.28) and 4.48 (CI, 0.79 to 25.49; P = 0.09), respectively. Intraclass correlation coefficients indicated that diarrhea was highly correlated with the same pen within a center; 53 and 60% of the variation in the probability of having a diarrhea score of 3 or 2 was associated with pens in the same center. In contrast, it was only slightly correlated with the same center alone, accounting for 7 and 10% of the total residual variances, respectively.
Risk factors for fecal rotavirus.
In univariable analysis, the presence of rotavirus in feces was associated with season. The OR for rotavirus presence in the summer collection period was 12.3 (CI, 1.07 to 141.5; P = 0.04) compared with that in the winter period. There was no association with age, whether entered in the model either as a continuous variable in months (OR, 1.0; CI, 0.99 to 1.02; P = 0.8) or as a binary variable defining a kitten of 5 months (OR, 1.41; CI, 0.52 to 3.85; P = 0.5), 3 months (OR, 1.32; CI, 0.46 to 3.80; P = 0.6), or ≤2 months (OR, 1.17; CI, 0.30 to 4.64; P = 0.82) of age. When season, kittens, and an interaction term between season and kittens were entered into a multilevel model, only season remained significant (OR, 14.8; CI, 1.1 to 200.4; P = 0.04) (Table 2). Intraclass correlation coefficients, estimated by including season, center, and pen in four-level intercept-only models, indicated that 76% of the variance was associated with pens within the centers, 41% with centers within seasons, and 6.3% with season alone.
TABLE 2.
Multivariable analysis of the effects of age and season on rotavirus sheddinga
| Variable | Odds ratio | 95% CI | P value | Coefficient | 95% CIb |
|---|---|---|---|---|---|
| Season (summer) | 14.78 | 1.09–200.4 | 0.04 | ||
| Age (kittens) | 3.82 | 0.17–87.1 | 0.4 | ||
| Age × season | 0.29 | 0.11–7.67 | 0.46 | ||
| Center | 4.64 | 1.52–14.21 | |||
| Center > pen no. | 5.35 | 1.71–16.68 |
LR test versus logistic regression: chi-square = 111.5 (P = 0.0000, chi-square).
95% CI, 95% confidence interval.
Limit of detection.
The limit of detection for SA11 in the two-step NSP3 qPCR was between 16 and 32 IVP/reaction. Intra-assay repeatability was good, with a coefficient of variation of 0.015. The limits of detection for feline rotavirus genomes belonging to the FRV-64 genogroup (K9 genogroup) and the FRV-1 genogroup were similar.
Molecular characterization.
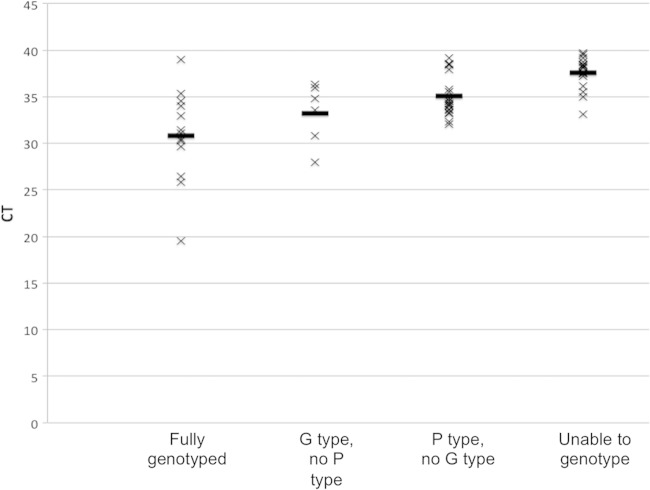
G genotypes were characterized for 19 (34%) of the positive samples and P genotypes for 34 (61%) of the positive samples. Two G genotypes were identified, G6 (16 [84%]) and G3 (3 [16%]). A single P genotype was recognized, P[9]. Combined G and P genotypes were found for 13 of the positive samples (23%); two of these were G3P[9], and 11 were G6P[9]. Higher NSP3 CT values correlated with a reduced likelihood of being able to determine a genotype (Fig. 3). Culture and sequencing of representative feline rotavirus strains, followed by primer and probe redesign, were essential to genotyping both G6P[9] and G3P[9] genotypes.
FIG 3.
Relationship between CT value and ability to genotype FRV strains. The horizontal bar indicates the average CT for each genotyping category. The ability to genotype improved as the CT value decreased. The majority of the FRV isolates had a CT value of ≥30.
Genotype G6 was isolated in both the winter and summer collection periods from three centers, Glasgow, Birmingham, and Truro, which span the length of the United Kingdom. Truro and Birmingham were centers at which epidemics of infection were observed. G3 was isolated from Exeter (winter) and Bridgend (summer) (Fig. 1, right, and Table 3).
TABLE 3.
Distribution and genotypes of feline rotaviruses isolated from all FRV-positive Cats Protection Adoption Centres in the United Kingdom during 2012
| Center | No. with genotypea: |
Total no. of positives | |||||
|---|---|---|---|---|---|---|---|
| G6P[9] | G6P[X] | G3P[9] | G3P[X] | GXP[9] | GXP[X] | ||
| Birmingham | 9 | 4 | 2 | 3 | 18 | ||
| Bridgend | 1 | 2 | 3 | ||||
| Chelmsford | 2 | 2 | |||||
| Derby | 1 | 1 | |||||
| Downham Market | 2 | 2 | |||||
| Exeter Axhayes | 2 | 1 | 3 | ||||
| Glasgow | 1 | 1 | |||||
| Haslemere | 1 | 1 | |||||
| Isle of Wight | 1 | 1 | |||||
| Mansfield | 1 | 1 | |||||
| NCAC | 1 | 1 | |||||
| Nottingham | 2 | 2 | |||||
| Truro | 2 | 18 | 1 | 21 | |||
| Total FRVs | 11 | 5 | 2 | 1 | 21 | 17 | 57 |
X indicates G or P rotavirus genotypes that were unable to be elucidated.
DISCUSSION
This study represents one of the first population-based studies of rotavirus prevalence in any animal species. Feline rotaviruses (G6P[9] and G3P[9]) were detected at a low prevalence (3.0%) in the cat population in the United Kingdom. This is similar to estimates from other countries (9, 10, 20) but showed interesting regional and seasonal variations. We report the first detection of epidemics of asymptomatic rotavirus infection (prevalences, 61.2% and 10.2%) in cats in spatially and temporally distinct locations (Table 1). The full details of these will be reported elsewhere. The low frequency of rotavirus epidemics in the cat population might be due to a combination of the low national prevalence of infection (3.0%) and the “snapshot” view that is afforded by cross-sectional sampling. Longitudinal sampling strategies based on this prevalence could be used to investigate transmission dynamics in more detail. Biosecurity measures for the Cats Protection Adoption Centres are of a high standard across the country, as they all adhere to strict hygiene guidelines. However, it is known in human hospitals that viral diseases, particularly gastrointestinal pathogens, are not easily contained, with rotavirus being one of the most frequent pathogens to transmit nosocomially and cause outbreaks, particularly in pediatric and neonatal wards (30). A return to Truro 6 weeks later found a complete turnover of cats and a reduction of the center prevalence from 61.2% (21/34) to 2.5% (1/40), despite the continued influx of new individuals, which might be thought to maintain epidemic infection. Therefore, it is unknown whether the observed epidemics arose from a spread within a center or reflected an intake of cats already infected with rotavirus due to an outbreak in the local area. The diversity and prevalence of feline calicivirus strains in United Kingdom cat shelters have been suggested to arise from cats sampling local strain diversity prior to entering a shelter, rather than infection occurring within a center (31).
The absence of an association between rotavirus infection and diarrhea in cats is in stark contrast to people, in whom diarrhea is a major cause of morbidity and mortality. Infection has also been associated with diarrhea and decreased productivity in cows (32, 33), suckling pigs (34–36), and horses (37, 38). Asymptomatic infections are reported, although their importance in transmission is not well understood due to the lack of population-based studies. The lack of an association between feline rotavirus infection and diarrhea may reflect cat behavior. Queens clean the perineal region of their kittens to stimulate defecation and consume the feces that are produced. Maternal coprophagy may reduce the risk of kitten diarrhea by preventing transmission within and between litters and by “orally vaccinating” the queen, increasing the titers of milk anti-rotavirus IgA antibodies.
Low viral titers and virus strain type may also be a reason for the lack of association of feline rotavirus with diarrhea. Many of the CT values obtained for rotavirus-positive feline fecal samples, even in the epidemics of infection observed in this study, fell out of the clinically relevant range consistent with human symptomatic rotavirus infection (39). In cows, asymptomatic individuals shed similar viral titers to those of clinically infected individuals, and the roles of virulent and avirulent strains of rotavirus have been postulated (40, 41). In dogs and pigs, species in which asymptomatic infections are more frequently recognized (35, 42–45), no population-based studies of sufficient power exist to truly characterize the role of asymptomatic infection. Asymptomatic infection may be a reflection of the nature of the relationship of G6P[9] and G3P[9] with the feline host. It is possible that zoonotic infection with these strains is similarly associated with asymptomatic carriage or mild clinical signs in people, and if population studies of adults and children, particularly the pet-owning population and those working in contact with cats, were performed, we may see a greater prevalence than what we currently accept.
Rotavirus most commonly infects neonatal and young people, cattle, and pigs (6, 7, 46). However, this was not the case for the cat population in this study. Kittens may play a role as a multiplier of infection without this being detected because of maternal coprophagy. Kitten feces are not found in litter trays until they reach approximately 4 weeks of age. Ethically, per-rectum fecal sampling requires a home office license in the United Kingdom and was not possible. This age group, the equivalent of very young children, is not represented and therefore is part of a systematic bias in our study. However, rotavirus was not detected in the feces of any of the nursing queens.
FRV infection was strongly associated with season (OR, 14.8 [CI, 1.1 to 200.4]; P = 0.04). It was more common in the summer months, contrasting with the pattern observed in people and cattle in the United Kingdom, where rotavirus is considered a cold weather disease (32, 47, 48). In worldwide surveillance studies, country income level is a more reliable predictor of infection than is latitude or geographical location. Wealthier countries have a seasonal peak (49), the timing and spread of which are influenced by birth rate (50); poorer countries experience year-round disease associated with high transmission and birth rates (51). An increase in the susceptible population is therefore important, but it is possible that seasonality is also influenced by an increased density of individuals within housing and airspace. Cattle are brought into close contact by indoor housing over the winter, and people spend more time indoors over the winter in close proximity with others (52). In our cat population, although infection was not associated with age, it is possible that kittens play a role in seasonality but it is masked by population dynamics and cat behavior. Cats are seasonal and prolific breeders; an average adult queen can produce two litters of four kittens between spring and autumn (53, 54). During the breeding season, the increased number of animals per pen and the greater volume of stray kittens and pregnant queens might increase the risk for the transmission of infection.
We have identified two G and P genotype combinations carried by rotaviruses circulating in the cat population in the United Kingdom in 2012, G3P[9] and G6P[9]. While G3P[9] is a recognized feline genotype (AU-1-like and BA222-like genotype constellations [13, 14, 55]), this is the first report of G6P[9] detection in cats. G6 was the more prevalent genotype (84%) and was detected in Scotland, the Midlands, and Cornwall, which are geographically distinct regions encompassing the length of the United Kingdom. Further, it was identified in both the winter and summer collection periods. Although both rotavirus genotypes did not coexist within a single center, there is potential for spatial coexistence, as G and P genotyping was incomplete. (This is a common finding in the molecular epidemiology of rotavirus and most likely reflects low virus titers, although the possibility that a novel feline sequence reduced the efficiency of primer binding should also be considered). Both G3P[9] and G6P[9] genotypes have been isolated from people in other parts of the globe. Human G3P[9] is considered to be a direct result of transmission events from canine or feline rotaviruses and has been reported in human clinical surveillance samples worldwide, including Japan (13), Israel (56), Brazil (57), Thailand (58), Russia (59), and Hungary (60). G6P[9] was originally isolated from an Italian child with diarrhea (61) and has subsequently been reported in the United States (62), Hungary (60, 63), Japan (64), Australia (65, 66), and Tunisia (16). Yamamoto and colleagues (64) considered that their isolates represented reassortment events between bovine-like human rotaviruses and human/feline AU-1-like rotaviruses. G6 is a common genotype in cattle/buffalo (67–70), sheep (71–73), and goats (74, 75) and has been identified sporadically or at a low prevalence in rabbits (76) and pigs (77, 78). It is uncommon in people, and although a zoonotic origin is postulated (64, 79–83), it has not yet been convincingly proven whether such zoonotic strains spread among people. More recently, with the advent of advanced genome sequencing techniques and a more robust classification system (84), the possibility of G6 feline origin at some historical point has been proposed (16). With clustering of published human G6 genotypes with our feline G6 genotypes, rather than with published bovine G6 genotypes (85), our work strongly suggests that G6P[9] genotypes are examples of zoonotic or anthropozoonotic transmission between cats and people. Whole-genome sequencing to further explore the relationship between the G3P[9] and G6P[9] genotypes identified in this study and other human and animal rotaviruses is under way.
Interestingly, G3P[3] was not found in our study population. This genotype has been reported in cats (20, 86), people (14, 15, 17, 87–91), dogs (92–97), and other animals (98–102). This genotype may not be circulating in the cat population in the United Kingdom or may be at a prevalence too low for detection in our study (<2%). It is also possible that similar to human RVA, feline RVA strain diversity and the prevalences of different strains may fluctuate between years, but this can be confirmed only through sustained surveillance in consecutive years. Reassuringly, none of the common human rotaviruses were detected in cats, and no reassortment was observed between human and feline rotaviruses.
An additional limitation of this study was using a shelter cat population as a sentinel for the cat population in the United Kingdom. Sampling a shelter cat population rather than an owned pet population may carry with it the additional stressors and opportunities for disease spread associated with mass housing of cats from different backgrounds and a high-throughput environment. However, the majority of cats housed in Cats Protection centers are healthy cats that have been relinquished from homes due to socioeconomic factors. True strays represent a minority of the population, and feral cats are only occasionally and briefly housed for trap-neuter-release programs. Although the nosocomial spread of viral disease is difficult to prevent, hygiene standards were very good, and the diversity of feline calicivirus within these centers has been suggested to reflect local strain distribution rather than within-center spread (31). Cats were also relinquished from geographically widespread postcode locations throughout the United Kingdom. Therefore, we considered it a valid comparison to use these centers as a reflection of the pet cat population in the United Kingdom.
G6P[9] is a relatively common feline rotavirus and, along with G3P[9], exists at low prevalence in this cat population in the United Kingdom. Diarrhea and age are not risk factors for infection, although infection increases in prevalence over the summer. Transmission events between cats and people in the United Kingdom likely exist, although they are infrequent and do not cause outbreaks of disease. The surveillance of rotavirus in our domestic pet population is important for investigating rotavirus genetic diversity, elucidating the role of asymptomatic carriage, exploring zoonotic risk, and monitoring the potential role of nonhuman rotaviruses in the evolution of rotavirus.
ACKNOWLEDGMENTS
This work was supported by an RCVS Charitable Trust Blue Sky award in virology (BSR11 1428) and internal funding from the Institute of Infection and Global Health, University of Liverpool.
We thank the Cats Protection for financial support of A.C.G.'s lectureship and the cooperation of their staff and cats with fecal sample and data collections in 2012. We also thank everyone involved with data entry, particularly Anna Edwards, Harriet Campbell, Anneka Summan, and Sabrina Knight.
N.C. has received grant support from GlaxoSmithKline (GSK) for rotavirus research and honoraria for participation in GSK rotavirus vaccine advisory board meetings. M.I.-G. has received funding from GSK and Sanofi Pasteur MSD in the form of unrestricted educational grants for research.
REFERENCES
- 1.WHO. 2009. Meeting of the immunization Strategy Advisory Group of Experts, April 2009–conclusions and recommendations. Wkly Epidemiol Rec. 84:220–236. http://www.who.int/wer/2009/wer8423.pdf. [PubMed] [Google Scholar]
- 2.World Health Organization. 2013. Rotavirus vaccines: WHO position paper–January 2013. Wkly Epidemiol Rec 88:49–64. http://www.who.int/wer/2013/wer8805.pdf. [PubMed] [Google Scholar]
- 3.Abe M, Ito N, Masatani T, Nakagawa K, Yamaoka S, Kanamaru Y, Suzuki H, Shibano K, Arashi Y, Sugiyama M. 2011. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J Gen Virol 92:952–960. doi: 10.1099/vir.0.028175-0. [DOI] [PubMed] [Google Scholar]
- 4.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gómara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Rahman M, Van Ranst M. 2008. Two out of the 11 genes of an unusual human G6P[6] rotavirus isolate are of bovine origin. J Gen Virol 89:2630–2635. doi: 10.1099/vir.0.2008/003780-0. [DOI] [PubMed] [Google Scholar]
- 6.Palombo EA. 2002. Genetic analysis of group A rotaviruses: evidence for interspecies transmission of rotavirus genes. Virus Genes 24:11–20. doi: 10.1023/A:1014073618253. [DOI] [PubMed] [Google Scholar]
- 7.Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet C. 2010. Zoonotic aspects of rotaviruses. Vet Microbiol 140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Snodgrass DR, Angus KW, Gray EW. 1979. A rotavirus from kittens. Vet Rec 104:222–223. doi: 10.1136/vr.104.10.222-a. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino Y, Baldwin CA, Scott FW. 1981. Isolation and characterization of feline rotavirus. J Gen Virol 54:313–323. doi: 10.1099/0022-1317-54-2-313. [DOI] [PubMed] [Google Scholar]
- 10.Marshall JA, Kennett ML, Rodger SM, Studdert MJ, Thompson WL, Gust ID. 1987. Virus and virus-like particles in the faeces of cats with and without diarrhoea. Aust Vet J 64:100–105. doi: 10.1111/j.1751-0813.1987.tb09638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mochizuki M, Nakagomi T, Nakagomi O. 1997. Isolation from diarrheal and asymptomatic kittens of three rotavirus strains that belong to the AU-1 genogroup of human rotaviruses. J Clin Microbiol 35:1272–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagomi O, Nakagomi T, Oyamada H, Suto T. 1985. Relative frequency of human rotavirus subgroups 1 and 2 in Japanese children with acute gastroenteritis. J Med Virol 17:29–34. doi: 10.1002/jmv.1890170105. [DOI] [PubMed] [Google Scholar]
- 13.Nakagomi T, Nakagomi O. 1989. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J Virol 63:1431–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagomi O, Ohshima A, Aboudy Y, Shif I, Mochizuki M, Nakagomi T, Gotlieb-Stematsky T. 1990. Molecular identification by RNA-RNA hybridization of a human rotavirus that is closely related to rotaviruses of feline and canine origin. J Clin Microbiol 28:1198–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aboudy Y, Shif I, Zilberstein I, Gotlieb-Stematsky T. 1988. Use of polyclonal and monoclonal antibodies and analysis of viral RNA in the detection of unusual group A human rotaviruses. J Med Virol 25:351–359. doi: 10.1002/jmv.1890250312. [DOI] [PubMed] [Google Scholar]
- 16.Fredj MBH, Heylen E, Zeller M, Fodha I, Benhamida-Rebai M, Van Ranst M, Matthijnssens J, Trabelsi A. 2013. Feline origin of rotavirus strain, Tunisia, 2008. Emerg Infect Dis 19:630–634. doi: 10.3201/eid1904.121383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagomi T, Nakagomi O. 2000. Human rotavirus HCR3 possesses a genomic RNA constellation indistinguishable from that of feline and canine rotaviruses. Arch Virol 145:2403–2409. doi: 10.1007/s007050070029. [DOI] [PubMed] [Google Scholar]
- 18.De Grazia S, Giammanco GM, Martella V, Ramirez S, Colomba C, Cascio A, Arista S. 2008. Rare AU-1-like G3P[9] human rotaviruses with a Kun-like NSP4 gene detected in children with diarrhea in Italy. J Clin Microbiol 46:357–360. doi: 10.1128/JCM.01593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNulty MS, Allan GM, Thompson DJ, O'Boyle JD. 1978. Antibody to rotavirus in dogs and cats. Vet Rec 102:534–535. doi: 10.1136/vr.102.24.534. [DOI] [PubMed] [Google Scholar]
- 20.Birch CJ, Heath RL, Marshall JA, Liu S, Gust ID. 1985. Isolation of feline rotaviruses and their relationship to human and simian isolates by electropherotype and serotype. J Gen Virol 66:2731–2735. doi: 10.1099/0022-1317-66-12-2731. [DOI] [PubMed] [Google Scholar]
- 21.Gaskell RM, Dawson S, Radford A. 2010. Other feline viral diseases, p 946–951. In Ettinger SJ, Feldman EC (ed), Textbook of veterinary internal medicine: diseases of the dog and the cat, 7th ed, vol 1 Saunders Elsevier, St. Louis, MO. [Google Scholar]
- 22.Yamaguchi N, Macdonald DW, Passanisi WC, Harbour DA, Hopper CD. 1996. Parasite prevalence in free-ranging farm cats, Felis silvestris catus. Epidemiol Infect 116:217–223. doi: 10.1017/S0950268800052468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis SJ, Heaton KW. 1997. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 32:920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 24.Zeng SQ, Halkosalo A, Salminen M, Szakal ED, Puustinen L, Vesikari T. 2008. One-step quantitative RT-PCR for the detection of rotavirus in acute gastroenteritis. J Virol Methods 153:238–240. doi: 10.1016/j.jviromet.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Malherbe HH, Strickland-Cholmley M. 1967. Simian virus SA-11 and the related O agent. Arch Gesamte Virusforsch 22:235–245. doi: 10.1007/BF01240518. [DOI] [PubMed] [Google Scholar]
- 26.Mochizuki M, Yamakawa M. 1987. Detection of rotaviruses in cat feces. Nihon Juigaku Zasshi 49:159–160. doi: 10.1292/jvms1939.49.159. [DOI] [PubMed] [Google Scholar]
- 27.Mochizuki M, Nakagomi O, Shibata S. 1992. Hemagglutinin activity of two distinct genogroups of feline and canine rotavirus strains. Arch Virol 122:373–381. doi: 10.1007/BF01317199. [DOI] [PubMed] [Google Scholar]
- 28.Eurorota: Rotavirus Surveillance Network. 2009. Appendix ii. Rotavirus detection and typing methods: nucleic acid extraction and reverse transcription. Virus Detection by PCR. Rotavirus VP7, VP4, VP6 and NSP4 genotyping. Version 4. Eurorota: Rotavirus Surveillance Network. See http://www.eurorota.net/docs.php. [Google Scholar]
- 29.Gouvea V, Ramirez C, Li B, Santos N, Saif L, Clark HF, Hoshino Y. 1993. Restriction endonuclease analysis of the vp7 genes of human and animal rotaviruses. J Clin Microbiol 31:917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Civardi E, Tzialla C, Baldanti F, Strocchio L, Manzoni P, Stronati M. 2013. Viral outbreaks in neonatal intensive care units: what we do not know. Am J Infect Control 41:854–856. doi: 10.1016/j.ajic.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne KP, Edwards D, Radford AD, Cripps P, Jones D, Wood JL, Gaskell RM, Dawson S. 2007. Longitudinal molecular epidemiological analysis of feline calicivirus infection in an animal shelter: a model for investigating calicivirus transmission within high-density, high-turnover populations. J Clin Microbiol 45:3239–3244. doi: 10.1128/JCM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhama K, Chauhan RS, Mahendran M, Malik SV. 2009. Rotavirus diarrhea in bovines and other domestic animals. Vet Res Commun 33:1–23. doi: 10.1007/s11259-008-9070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho YI, Han JI, Wang C, Cooper V, Schwartz K, Engelken T, Yoon KJ. 2013. Case-control study of microbiological etiology associated with calf diarrhea. Vet Microbiol 166:375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazaki A, Kuga K, Suzuki T, Kohmoto M, Katsuda K, Tsunemitsu H. 2013. Annual changes in predominant genotypes of rotavirus A detected in the feces of pigs in various developmental stages raised on a conventional farm. Vet Microbiol 163:162–166. doi: 10.1016/j.vetmic.2012.11.044. [DOI] [PubMed] [Google Scholar]
- 35.Fu ZF, Hampson DJ. 1987. Group A rotavirus excretion patterns in naturally infected pigs. Res Vet Sci 43:297–300. [PubMed] [Google Scholar]
- 36.Bohl EH. 1979. Rotaviral diarrhoea in pigs: brief review. JAVMA 174:613–615. [PubMed] [Google Scholar]
- 37.Monini M, Biasin A, Valentini S, Cattoli G, Ruggeri FM. 2011. Recurrent rotavirus diarrhoea outbreaks in a stud farm, in Italy. Vet Microbiol 149:248–253. doi: 10.1016/j.vetmic.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Netherwood T, Wood JL, Townsend HG, Mumford JA, Chanter N. 1996. Foal diarrhoea between 1991 and 1994 in the United Kingdom associated with Clostridium perfringens, rotavirus, Strongyloides westeri and Cryptosporidium spp. Epidemiol Infect 117:375–383. doi: 10.1017/S0950268800001564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukhopadhya I, Sarkar R, Menon VK, Babji S, Paul A, Rajendran P, Sowmyanarayanan TV, Moses PD, Iturriza-Gomara M, Gray JJ, Kang G. 2013. Rotavirus shedding in symptomatic and asymptomatic children using reverse transcription-quantitative PCR. J Med Virol 85:1661–1668. doi: 10.1002/jmv.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Archambault D, Morin G, Elazhary Y, Roy RS. 1990. Study of virus excretion in feces of diarrheic and asymptomatic calves infected with rotavirus. Zentralbl Veterinarmed B 37:73–76. [DOI] [PubMed] [Google Scholar]
- 41.Bridger JC, Oldham G. 1987. Avirulent rotavirus infections protect calves from disease with and without inducing high levels of neutralizing antibody. J Gen Virol 68:2311–2317. doi: 10.1099/0022-1317-68-9-2311. [DOI] [PubMed] [Google Scholar]
- 42.Debouck P, Pensaert M. 1983. Rotavirus excretion in suckling pigs followed under field circumstances. Ann Rech Vet 14:447–448. [PubMed] [Google Scholar]
- 43.Marshall JA, Healey DS, Studdert MJ, Scott PC, Kennett ML, Ward BK, Gust ID. 1984. Viruses and virus-like particles in the faeces of dogs with and without diarrhoea. Aust Vet J 61:33–38. doi: 10.1111/j.1751-0813.1984.tb07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz VLA, Brandão PE, Gregori F, Rodriguez CAR, Souza SLP, Jerez JA. 2009. Isolation of rotavirus from asymptomatic dogs in Brazil. Arq Bras Med Vet Zootec 61:996–999. doi: 10.1590/S0102-09352009000400031. [DOI] [Google Scholar]
- 45.Gelberg HB, Woode GN, Kniffen TS, Hardy M, Hall WF. 1991. The shedding of group A rotavirus antigen in a newly established closed specific pathogen-free swine herd. Vet Microbiol 28:213–229. doi: 10.1016/0378-1135(91)90077-S. [DOI] [PubMed] [Google Scholar]
- 46.Estes MK, Kapikian AZ. 2007. Rotaviruses, p 1917–1974. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 47.Atchison CJ, Tam CC, Hajat S, van Pelt W, Cowden JM, Lopman BA. 2010. Temperature-dependent transmission of rotavirus in Great Britain and The Netherlands. Proc Biol Sci 277:933–942. doi: 10.1098/rspb.2009.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Korsun N, Kroneman A, Lappalainen M, László B, Maunula L, Matthinjnssens J, Midgley S, Mladenova Z, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Schreier E, Steyer A, Sidaraviciute I, Tran AN, Usonis V, Van Ranst M, de Rougemont A, Gray J. 2009. Rotavirus surveillance in Europe, 2005–2008: Web-enabled reporting and real-time analysis of genotyping and epidemiological data. J Infect Dis 200(Suppl 1):S215–S221. doi: 10.1086/605049. [DOI] [PubMed] [Google Scholar]
- 49.Patel MM, Pitzer VE, Alonso WJ, Vera D, Lopman B, Tate J, Viboud C, Parashar UD. 2013. Global seasonality of rotavirus disease. Pediatr Infect Dis J 32:e134–e147. doi: 10.1097/INF.0b013e31827d3b68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pitzer VE, Viboud C, Simonsen L, Steiner C, Panozzo CA, Alonso WJ, Miller MA, Glass RI, Glasser JW, Parashar UD, Grenfell BT. 2009. Demographic variability, vaccination, and the spatiotemporal dynamics of rotavirus epidemics. Science 325:290–294. doi: 10.1126/science.1172330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pitzer VE, Viboud C, Lopman BA, Patel MM, Parashar UD, Grenfell BT. 2011. Influence of birth rates and transmission rates on the global seasonality of rotavirus incidence. J R Soc Interface 8:1584–1593. doi: 10.1098/rsif.2011.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ergler CR, Kearns RA, Witten K. 2013. Seasonal and locational variations in children's play: implications for wellbeing. Soc Sci Med 91:178–185. doi: 10.1016/j.socscimed.2012.11.034. [DOI] [PubMed] [Google Scholar]
- 53.Wastlhuber J. 1991. History of domestic cats and cat breeds, p 1–59. In Pedersen N. (ed), Feline husbandry: diseases and management in the multiple-cat environment, 1st ed American Veterinary Publications, Inc, Goleta, CA. [Google Scholar]
- 54.Jöchle W, Jöchle M. 1993. Reproduction in a feral cat population and its control with a prolactin inhibitor, cabergoline. J Reprod Fertil 47:419–424. [PubMed] [Google Scholar]
- 55.Martella V, Potgieter AC, Lorusso E, De Grazia S, Giammanco GM, Matthijnssens J, Bányai K, Ciarlet M, Lavazza A, Decaro N, Buonavoglia C. 2011. A feline rotavirus G3P[9] carries traces of multiple reassortment events and resembles rare human G3P[9] rotaviruses. J Gen Virol 92:1214–1221. doi: 10.1099/vir.0.027425-0. [DOI] [PubMed] [Google Scholar]
- 56.Silberstein I, Shulman LM, Mendelson E, Shif I. 1995. Distribution of both rotavirus VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J Clin Microbiol 33:1421–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos N, Volotão EM, Soares CC, Albuquerque MC, da Silva FM, de Carvalho TRB, Pereira CF, Chizhikov V, Hoshino Y. 2001. Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J Clin Microbiol 39:1157–1160. doi: 10.1128/JCM.39.3.1157-1160.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khamrin P, Maneekarn N, Peerakome S, Tonusin S, Phan TG, Okitsu S, Ushijima H. 2007. Molecular characterization of rare G3P[9] rotavirus strains isolated from children hospitalized with acute gastroenteritis. J Med Virol 79:843–851. doi: 10.1002/jmv.20840. [DOI] [PubMed] [Google Scholar]
- 59.Novikova NA, Ponomareva NV, Novikov DV, Prilipov AG, Epifanova NV, Golitsyna LN. 2008. [Nucleotide sequence analysis of the NSP4 gene from group A rotaviruses isolated in Nizhni Novgorod]. Vopr Virusol 53:35–39. (In Russian.) [PubMed] [Google Scholar]
- 60.Bányai K, Bogdán A, Domonkos G, Kisfali P, Molnár P, Tóth A, Melegh B, Martella V, Gentsch JR, Szucs G. 2009. Genetic diversity and zoonotic potential of human rotavirus strains, 2003–2006, Hungary. J Med Virol 81:362–370. doi: 10.1002/jmv.21375. [DOI] [PubMed] [Google Scholar]
- 61.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brussow H, Hoshino Y, Flores J. 1992. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol 30:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffin DD, Nakagomi T, Hoshino Y, Nakagomi O, Kirkwood CD, Parashar UD, Glass RI, Gentsch JR, National Rotavirus Surveillance System . 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6],G12) and rare P3[9] strains related to bovine rotaviruses. Virol 294:256–269. doi: 10.1006/viro.2001.1333. [DOI] [PubMed] [Google Scholar]
- 63.Bányai K, Gentsch JR, Griffin DD, Holmes JL, Glass RI, Szücs G. 2003. Genetic variability among serotype G6 human rotaviruses: identification of a novel lineage isolated in Hungary. J Med Virol 71:124–134. doi: 10.1002/jmv.10462. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto D, Kawaguchiya M, Ghosh S, Ichikawa M, Numazaki K, Kobayashi N. 2011. Detection and full genomic analysis of G6P[9] human rotavirus in Japan. Virus Genes 43:215–223. doi: 10.1007/s11262-011-0624-6. [DOI] [PubMed] [Google Scholar]
- 65.Cooney MA, Gorrell RJ, Palombo EA. 2001. Characterisation and phylogenetic analysis of the VP7 proteins of serotype G6 and G8 human rotaviruses. J Med Microbiol 50:462–467. [DOI] [PubMed] [Google Scholar]
- 66.Diwakarla S, Clark R, Palombo EA. 2002. Expanding distribution of human serotype G6 rotaviruses in Australia. Microbiol Immunol 46:499–502. doi: 10.1111/j.1348-0421.2002.tb02726.x. [DOI] [PubMed] [Google Scholar]
- 67.Chang KO, Parwani AV, Saif LJ. 2000. Comparative sequence analysis of the VP7 genes of G6, G8 and G10 bovine group A rotaviruses and further characterization of G6 subtypes. Arch Virol 145:725–737. doi: 10.1007/s007050050666. [DOI] [PubMed] [Google Scholar]
- 68.Collins PJ, Mulherin E, Cashman O, Lennon G, Gunn L, O'Shea H, Fanning S. 2014. Detection and characterisation of bovine rotavirus in Ireland from 2006–2008. Ir Vet J 67:13. doi: 10.1186/2046-0481-67-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okada N, Matsumoto Y. 2002. Bovine rotavirus G and P types and sequence analysis of the VP7 gene of two G8 bovine rotaviruses from Japan. Vet Microbiol 84:297–305. doi: 10.1016/S0378-1135(01)00445-X. [DOI] [PubMed] [Google Scholar]
- 70.Pisanelli G, Martella V, Pagnini U, De Martino L, Lorusso E, Iovane G, Buonavoglia C. 2005. Distribution of G (VP7) and P (VP4) genotypes in buffalo group A rotaviruses isolated in Southern Italy. Vet Microbiol 110:1–6. doi: 10.1016/j.vetmic.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 71.Gazal S, Taku AK, Kumar B. 2012. Predominance of rotavirus genotype G6P[11] in diarrhoeic lambs. Vet J 193:299–300. doi: 10.1016/j.tvjl.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 72.Muñoz M, Lanza I, Alvarez M, Cármenes P. 1995. Prevalence of neutralizing antibodies to 9 rotavirus strains representing 7 G-serotypes in sheep sera. Vet Microbiol 45:351–361. doi: 10.1016/0378-1135(95)00002-R. [DOI] [PubMed] [Google Scholar]
- 73.Fitzgerald TA, Muñoz M, Wood AR, Snodgrass DR. 1995. Serological and genomic characterization of group-A rotaviruses from lambs. Arch Virol 140:1541–1548. doi: 10.1007/BF01322528. [DOI] [PubMed] [Google Scholar]
- 74.Ghosh S, Alam MM, Ahmed MU, Talukdar RI, Paul SK, Kobayashi N. 2010. Complete genome constellation of a caprine group A rotavirus strain reveals common evolution with ruminant and human rotavirus strains. J Gen Virol 91:2367–2373. doi: 10.1099/vir.0.022244-0. [DOI] [PubMed] [Google Scholar]
- 75.Pratelli A, Martella V, Tempesta M, Buonavoglia C. 1999. Characterization by polymerase chain reaction of ruminant rotaviruses isolated in Italy. Microbiologica 22:105–109. [PubMed] [Google Scholar]
- 76.Schoondermark-van de Ven E, Van Ranst M, de Bruin W, van den Hurk P, Zeller M, Matthijnssens J, Heylen E. 2013. Rabbit colony infected with a bovine-like G6P[11] rotavirus strain. Vet Microbiol 166:154–164. doi: 10.1016/j.vetmic.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 77.Gouvea V, Santos N, Timenetsky Mdo C. 1994. Identification of bovine and porcine rotavirus-G types by PCR. J Clin Microbiol 32:1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Parra GI, Vidales G, Gomez JA, Fernandez FM, Parreño V, Bok K. 2008. Phylogenetic analysis of porcine rotavirus in Argentina: increasing diversity of G4 strains and evidence of interspecies transmission. Vet Microbiol 126:243–250. doi: 10.1016/j.vetmic.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 79.Doan YH, Nakagomi T, Aboudy Y, Silberstein I, Behar-Novat E, Nakagomi O, Shulman LM. 2013. Identification by full-genome analysis of a bovine rotavirus transmitted directly to and causing diarrhea in a human child. J Clin Microbiol 51:182–189. doi: 10.1128/JCM.02062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steyer A, Sagadin M, Kolenc M, Poljšak-Prijatelj M. 2013. Whole genome sequence analysis of bovine G6P[11] rotavirus strain found in a child with gastroenteritis. Infect Genet Evol 13:89–95. doi: 10.1016/j.meegid.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 81.Afrad MH, Matthijnssens J, Moni S, Kabir F, Ashrafi A, Rahman MZ, Faruque ASG, Azim T, Rahman M. 2013. Genetic characterization of a rare bovine-like human VP4 mono-reassortant G6P[8] rotavirus strain detected from an infant in Bangladesh. Infect Genet Evol 19:120–126. doi: 10.1016/j.meegid.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 82.De Grazia S, Martella V, Rotolo V, Bonura F, Matthijnssens J, Bányai K, Ciarlet M, Giammanco GM. 2011. Molecular characterization of genotype G6 human rotavirus strains detected in Italy from 1986 to 2009. Infect Genet Evol 11:1449–1455. doi: 10.1016/j.meegid.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 83.El Sherif M, Esona MD, Wang Y, Gentsch JR, Jiang B, Glass RI, Abou Baker S, Klena JD. 2011. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect Genet Evol 11:1436–1442. doi: 10.1016/j.meegid.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 84.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PPC, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Iturriza-Gómara M, German AC, Dove W, Morgan KL, Nawaz S, Nakagomi T, Nakagomi O, Radford A, Sandrasegaram M, Cunliffe NA. 2013. Molecular epidemiology of feline rotaviruses in the UK. Proceedings of the 5th European Rotavirus Biology Meeting, 6 to 9 October 2013, Valencia, Spain. [Google Scholar]
- 86.Oka T, Nakagomi T, Nakagomi O. 2001. A lack of consistent amino acid substitutions in NSP4 between rotaviruses derived from diarrheal and asymptomatically-infected kittens. Microbiol Immunol 45:173–177. doi: 10.1111/j.1348-0421.2001.tb01277.x. [DOI] [PubMed] [Google Scholar]
- 87.De Grazia S, Martella V, Giammanco GM, Iturriza-Gómara M, Ramirez S, Cascio A, Colomba C, Arista S. 2007. Canine-origin G3P[3] rotavirus strain in child with acute gastroenteritis. Emerg Infect Dis 13:1091–1093. doi: 10.3201/eid1307.070239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li B, Clark HF, Gouvea V. 1993. Nucleotide sequence of the VP4-encoding gene of an unusual human rotavirus (HCR3). Virology 196:825–830. doi: 10.1006/viro.1993.1540. [DOI] [PubMed] [Google Scholar]
- 89.Santos N, Clark HF, Hoshino Y, Gouvea V. 1998. Relationship among serotype G3P5A rotavirus strains isolated from different host species. Mol Cell Probes 12:379–386. doi: 10.1006/mcpr.1998.0198. [DOI] [PubMed] [Google Scholar]
- 90.Khamrin P, Maneekarn N, Peerakome S, Yagyu F, Okitsu S, Ushijima H. 2006. Molecular characterization of a rare G3P[3] human rotavirus reassortant strain reveals evidence for multiple human-animal interspecies transmissions. J Med Virol 78:986–994. doi: 10.1002/jmv.20651. [DOI] [PubMed] [Google Scholar]
- 91.Banerjee I, Iturriza-Gomara M, Rajendran P, Primrose B, Ramani S, Gray JJ, Brown DW, Kang G. 2007. Molecular characterization of G11P[25] and G3P[3] human rotavirus strains associated with asymptomatic infection in South India. J Med Virol 79:1768–1774. doi: 10.1002/jmv.20988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fulton RW, Johnson CA, Pearson NJ, Woode GN. 1981. Isolation of a rotavirus from a newborn dog with diarrhea. Am J Vet Res 42:841–843. [PubMed] [Google Scholar]
- 93.Hoshino Y, Wyatt RG, Scott FW, Appel MJ. 1982. Isolation and characterization of a canine rotavirus. Arch Virol 72:113–125. doi: 10.1007/BF01314456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoshino Y, Wyatt RG, Greenberg HB, Kalica AR, Flores J, Kapikian AZ. 1983. Serological comparison of canine rotavirus with various simian and human rotaviruses by plaque reduction neutralization and hemagglutination inhibition tests. Infect Immun 41:169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang BK, Song DS, Jung KI, Lee CS, Park SJ, Oh JS, An DJ, Yang JS, Moon HJ, Lee SS, Yoon YD, Park BK. 2007. Genetic characterization of canine rotavirus isolated from a puppy in Korea and experimental reproduction of disease. J Vet Diagn Invest 19:78–83. doi: 10.1177/104063870701900112. [DOI] [PubMed] [Google Scholar]
- 96.Martella V, Pratelli A, Elia G, Decaro N, Tempesta M, Buonavoglia C. 2001. Isolation and genetic characterization of two G3P5A[3] canine rotavirus strains in Italy. J Virol Methods 96:43–49. doi: 10.1016/S0166-0934(01)00312-3. [DOI] [PubMed] [Google Scholar]
- 97.Mochizuki M, Hsüan S. 1984. Isolation of a rotavirus from canine diarrheal feces. Nippon Juigaku Zasshi 46:905–908. doi: 10.1292/jvms1939.46.905. [DOI] [PubMed] [Google Scholar]
- 98.He B, Yang F, Yang W, Zhang Y, Feng Y, Zhou J, Xie J, Feng Y, Bao X, Guo H, Li Y, Xia L, Li N, Matthijnssens J, Zhang H, Tu C. 2013. Characterization of a novel G3P[3] rotavirus isolated from a lesser horseshoe bat: a distant relative of feline/canine rotaviruses. J Virol 87:12357–12366. doi: 10.1128/JVI.02013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miño S, Matthijnssens J, Badaracco A, Garaicoechea L, Zeller M, Heylen E, Van Ranst M, Barrandeguy M, Parreño V. 2013. Equine G3P[3] rotavirus strain E3198 related to simian RRV and feline/canine-like rotaviruses based on complete genome analyses. Vet Microbiol 161:239–246. doi: 10.1016/j.vetmic.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 100.Westerman LE, Jiang B, McClure HM, Snipes-Magaldi LJ, Griffin DD, Shin G, Gentsch JR, Glass RI. 2006. Isolation and characterization of a new simian rotavirus, YK-1. Virol J 3:40. doi: 10.1186/1743-422X-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JB, Youn SJ, Nakagomi T, Park SY, Kim TJ, Song CS, Jang HK, Kim BS, Nakagomi O. 2003. Isolation, serologic and molecular characterization of the first G3 caprine rotavirus. Arch Virol 148:643–657. doi: 10.1007/s00705-002-0963-7. [DOI] [PubMed] [Google Scholar]
- 102.Ghosh S, Varghese V, Samajdar S, Sinha M, Kobayashi N, Naik TN. 2007. Molecular characterization of bovine group A rotavirus G3P[3] strains. Arch Virol 152:1935–1940. doi: 10.1007/s00705-007-1009-y. [DOI] [PubMed] [Google Scholar]