Abstract
Conventional identification of Aeromonas species based on biochemical methods is challenged by the heterogeneous nature of the species. Here, we present a new multiplex PCR method directed toward the gyrB and rpoB genes that identifies four Aeromonas species, A. hydrophila, A. media, A. veronii, and A. caviae, and we describe the application of this method on a Danish strain collection.
TEXT
Aeromonas spp. are highly adapted to aquatic environments and have been described as pathogenic to humans and animals. The genus Aeromonas consists of more than 20 valid species, of which A. hydrophila, A. caviae (synonymous with A. punctata), A. media, A. veronii bv. sobria, and A. veronii bv. veronii are of particular clinical significance, because they can cause gastroenteritis, wound and soft tissue infections, and septicemia (1). Aeromonas spp. may produce an array of virulence factors (e.g., cytolytic toxins with hemolytic activity and enterotoxins). Recent reviews suggested that only a subset of Aeromonas spp. are truly pathogenic and may be transmitted by hitherto unknown routes, and they proposed that further epidemiological and molecular studies are needed (2, 3). Aeromonas species identification has traditionally been performed by a combination of different biochemical tests. However, these are not always conclusive, since some Aeromonas species display heterogeneous biochemical properties; compared to molecular methods, the correct identification rate with biochemical tests has been shown to be very low (4, 5). Molecular species identification has been exploited by the 16S rRNA gene, either by restriction fragment length polymorphism (RFLP) (6–9) or direct sequencing (10, 11). However, due to insufficient interspecies sequence variation and heterogeneity among copies of ribosomal RNA operons in the same bacteria (10, 12, 13), this gene may not be an optimal target. A number of studies have shown that the sequences of several different housekeeping genes are able to differentiate this tight taxonomic group of organisms. These genes include an RNA polymerase B subunit (rpoB), an RNA polymerase D subunit (rpoD), and a DNA gyrase B subunit (gyrB) (10, 14–16). The objective of the present study was to identify species of Aeromonas by partial gyrB and rpoB sequencing in order to develop a multiplex PCR (mPCR) that targets the four most prevalent and clinically relevant species identified by sequence analysis on a Danish strain collection.
The strain collection used in this study was composed of 51 Aeromonas spp. collected from diarrheagenic patients during the period of 2005 to 2010. Each stool specimen was grown on enteric medium (Statens Serum Institut, Hillerød, Denmark) (17), and Aeromonas was identified by its distinct colony morphology, while further delineation of clinically relevant species was performed manually according to their biochemical characteristics, including Voges-Proskauer test results and lysine decarboxylase, ornithine decarboxylase, arginine dihydrolase, glucose (gas), esculin, and acid production from the fermentation of carbohydrates (4). Bacterial colonies were prepared for PCR and for gyrB and rpoB sequencing by 10 min of boiling in 10% Chelex 100 (Bio-Rad, Hercules, CA, USA) in 10 mM Tris-HCl and 1 mM EDTA (pH 8), followed by centrifugation and 10-fold dilution of the supernatant in PCR-grade water. PCR was performed in a total reaction volume of 25 μl containing the following reagents: 5 μl of template, 0.2 mM each dATP, dCTP, dGTP, and dTTP (GeneAmp; Applied Biosystems), 1× PCR buffer, 2.0 mM MgCl2, 1.25 U Platinum Taq DNA polymerase (Invitrogen), and 0.5 μM each of primers up1 and up2r (18) or Pasrpob-L and Rpob-R (19) when gyrB or rpoB, respectively, was amplified. The amplicon size of the partial gyrB gene was 1,273 bp, and that of rpoB was 560 bp. In order to obtain reliable sequence quality in both directions, sequencing was performed by a combination of the PCR primers mentioned above and additional primers designed here for this purpose for gyrB (gyrB-F2[493], 5′-GAGGACTACAGCAAGAAGGCCA-3′; gyrB-R2[516], 5′-GACTTGGCCTTCTTGCTGTAGTC-3′) and for rpoB (rpoB-F2, 5′-CAACTTCGTCGGTGATCACA-3′; rpoB-R2, 5′-TGTGATCACCGACGAAGTGG-3′). The obtained partial gyrB and rpoB gene sequences were deposited in GenBank (see Table S1 in the supplemental material). Based on the sequences obtained in this study combined with GenBank sequences from Küpfer et al. (10), alignments of the rpoB and gyrB sequences were constructed, and species-specific primers were designed for mPCR toward four Aeromonas species, A. hydrophila, A. media, A. veronii, and A. caviae. All sequence analyses were done by the CLC DNA Workbench software, version 6.5 (CLC bio, Århus, Denmark). The resulting mPCR procedure was performed in a 25-μl reaction mixture containing the following reagents: 1.5 μl of template, 0.2 mM each dATP, dCTP, dGTP, and dTTP (GeneAmp, Applied Biosystems), 1.2× PCR buffer, 2.0 mM MgCl2, and 1.25 U Platinum Taq DNA polymerase (Invitrogen); the primer concentrations are listed in Table 1, and the thermocycler conditions were as follows: 2-min hot start at 95°C, 6 cycles of 94°C for 40 s, 67°C for 50 s, and 72°C for 40 s, and 30 cycles of 94°C for 40 s, 65°C for 50 s, and 72°C for 40 s.
TABLE 1.
Primers used in the mPCR method for Aeromonas species identification
| Primer | Target organism (gene) | Sequence | Product (bp) | Primer concn (μM) |
|---|---|---|---|---|
| A-cav F(4) | A. caviae (gyrB) | 5′-TGCTGCTGACCATCCGC-3′ | 0.5 | |
| A-cav R(74) | 5′-GGTGCCTGCGGCTCG-3′ | 70 | 0.5 | |
| A-med F(176) | A. media (gyrB) | 5′-GGCCAAGCGTCTGCGT-3′ | 0.2 | |
| A-med R(275) | 5′-CGCCCTCGTAGCAGAAGTGA-3′ | 99 | 0.2 | |
| A-hyd F(533) | A. hydrophila (gyrB) | 5′-AGTCTGCCGCCAGTGGC-3′ | 0.45 | |
| A-hyd R(677) | 5′-CRCCCATCGCCTGTTCG-3′ | 144 | 0.45 | |
| A-Ver F(b1) | A. veronii (rpoB) | 5′-CGTGCCGGCTTTGAAGTC-3′ | 0.15 | |
| A-Ver R(b1) | 5′-GATCACGTACTTGCCTTCTTCAATA-3′ | 224 | 0.15 | |
| A-16S F(270) | Aeromonas | 5′-CGACGATCCCTAGCTGGTCT-3′ | 0.05 | |
| A-16S R(731) | Universal (16S rRNA gene) | 5′-GCCTTCGCCACCGGTAT-3′ | 461 | 0.05 |
In the present study, we used rpoB and gyrB sequencing to determine the species identity of clinical Aeromonas isolates derived from patients suffering from gastroenteritis. In order to do so, we took advantage of previous findings where internal variations of the genes rpoB and gyrB were shown to be linked to phylogeny and, hence, correlated to species identity (10, 11, 14–16). However, due to variations in sequence quality and length, we decided to accept partial rpoB and gyrB sequences of 390 bp and 1,113 bp, respectively. With respect to the rpoB sequence, 390 bp is shorter than what was reported in previous studies, where a 560-bp sequence was used (10, 19), but phylogenetic analyses on reference strain sequences showed the same topologies with trees built from short (390-bp) and long (560-bp) sequences (data not shown).
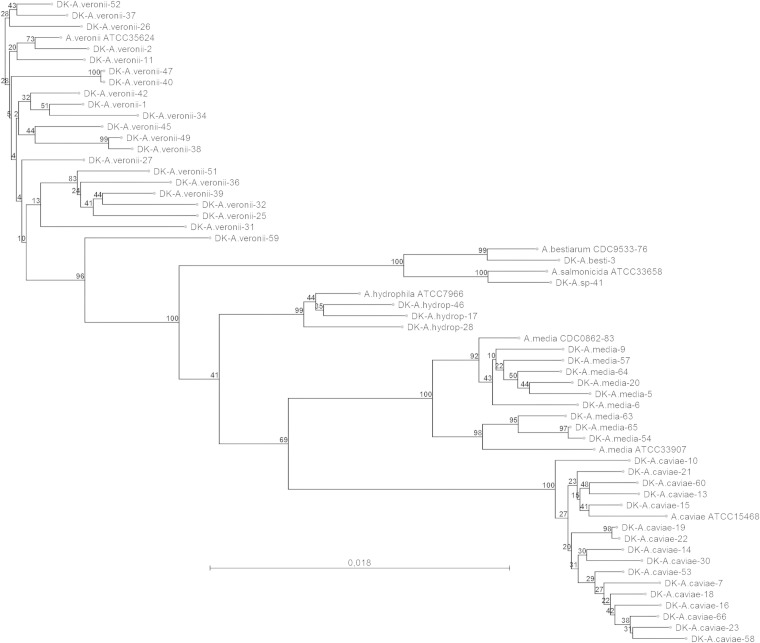
Species identities of the clinical isolates were investigated by a combination of individual GenBank BLAST searches and alignment and phylogenetic tree construction for the individual sequences (data not shown) and for the concatenated sequences of gyrB plus rpoB (Fig. 1). With one exception among the 51 clinical isolates, the individual gyrB and rpoB gene sequences and the concatenated sequences resulted in the same species identifications. The one exception was a gyrB sequence (GenBank accession no. KJ747141) that was identical to A. salmonicida, while the rpoB sequence (KJ747178) may have originated from either A. salmonicida or A. encheleia. The remaining isolates were identified as A. veronii (n = 21, 41.2%), A. caviae (n = 16, 31.4%), A. media (n = 9, 17.6%), A. hydrophila (n = 3, 5.9%), and A. bestiarum (n = 1, 2%). The maximum interspecies divergences of gyrB and rpoB were found to be 10% (111 out of 1,113 nucleotides) and 9% (35 out of 390 nucleotides), respectively. The maximum intraspecies divergences of the gyrB gene were 5% (A. veronii, 53 nucleotides), 2.3% (A. hydrophila, 26 nucleotides), 2.5% (A. caviae, 28 nucleotides), and 3.1% (A. media, 35 nucleotides), and those of rpoB were 2.3% (A. veronii, 9 nucleotides), 1.5% (A. hydrophila, 6 nucleotides), 1.3% (A. caviae, 5 nucleotides), and 2.8% (A. media, 11 nucleotides).
FIG 1.
Neighbor-joining tree constructed from concatenated partial gyrB (1,113-bp) and rpoB (390-bp) sequences obtained from clinical strains in this study (accession numbers listed in Table S1 in the supplemental material) compared to reference sequences obtained from GenBank. Numbers at the nodes indicate bootstrap values as the percentage of 1,000 replicates. The scale bar indicates 1.8% sequence divergence.
Only 19/51 (37.3%) Aeromonas isolates were correctly identified by the phenotypic method. Of the 18 Aeromonas isolates correctly identified by the phenotypic method, 2 (66.7%) were A. hydrophila, 2 (9.5%) were A. veronii, and 15 (93.8%) were A. caviae (for further details, see Table S1 in the supplemental material). In particular, high discrepancies were observed for A. veronii and A. media, which may be explained by the phenotypic resemblance between A. veronii bv. sobria and A. hydrophila (both species negative for ornithine decarboxylase) and A. media and A. caviae (almost identical biochemically), respectively (4, 5). In order to subclassify A. veronii strains, tests for ornithine decarboxylase, arginine dihydrolase, salicin, esculin, and tartrate, for example, can be applied. The 21 molecularly identified strains of A. veronii belonged to A. veronii bv. sobria when biochemically tested. A. veronii bv. sobria and A. caviae have been shown to be the most prevalent Aeromonas spp. associated with traveler's diarrhea, which includes symptoms of watery diarrhea, abdominal cramps, and fever (20).
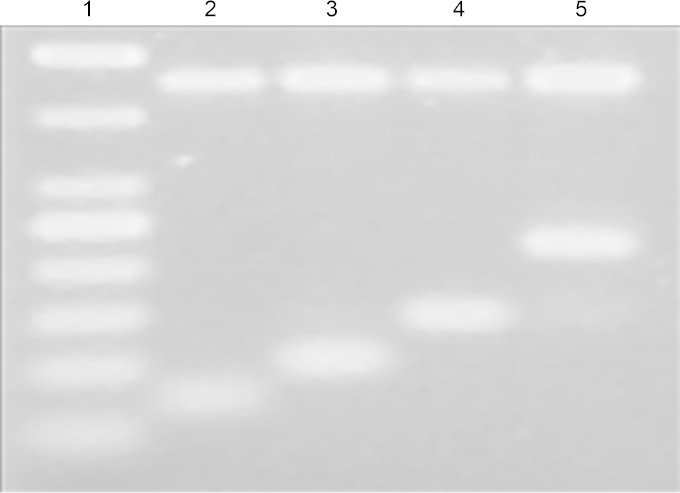
The mPCR method was designed toward four species, A. hydrophila, A. media, A. veronii, and A. caviae, and all 49 isolates in the present study were correctly identified. Figure 2 shows an example of four strains analyzed by the mPCR method. A. bestiarum and A. salmonicida or A. encheleia were not included in the analysis because they had little or no clinical significance and few sequences available with relatively high sequence variation; hence, no optimal primer design was possible. Therefore, if isolates investigated by the present mPCR method do not generate an amplicon, they should be subjected to, e.g., rpoB and/or gyrB sequencing in order to obtain correct species identification. In conclusion, the present mPCR method offers easy and precise identification of Aeromonas species that may improve routine diagnostics and shed new light on the controversial clinical and epidemiological aspects of this genus. Especially significant is the identification of A. media and A. veronii, which, according to this study, are underdiagnosed when phenotypic methods for species determination are applied.
FIG 2.
Four clinical isolates analyzed by mPCR for Aeromonas species identifications. Each primer set was designed to be specific for the particular species and to have a unique amplicon size when analyzed by agarose gel electrophoresis. Also included in the mPCR was a primer set targeting 16S rRNA genes, which served as an internal PCR control (461 bp). Lane 1, 50-bp DNA marker; lane 2, A. caviae (70 bp); lane 3, A. media (99 bp); lane 4, A. hydrophila (144 bp); and lane 5, A. veronii (244 bp).
Nucleotide sequence accession numbers.
New sequences determined in this study were deposited in GenBank under accession numbers KJ747109 to KJ747187, KJ775031, and KJ775032 (for further details, see Table S1 in the supplemental material).
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01963-14.
REFERENCES
- 1.Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Graevenitz A. 2007. The role of Aeromonas in diarrhea: a review. Infection 35:59–64. doi: 10.1007/s15010-007-6243-4. [DOI] [PubMed] [Google Scholar]
- 3.Parker JL, Shaw JG. 2011. Aeromonas spp. clinical microbiology and disease. J Infect 62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Abbott SL, Cheung WK, Janda JM. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J Clin Microbiol 41:2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ormen O, Granum PE, Lassen J, Figueras MJ. 2005. Lack of agreement between biochemical and genetic identification of Aeromonas spp. APMIS 113:203–207. doi: 10.1111/j.1600-0463.2005.apm1130308.x. [DOI] [PubMed] [Google Scholar]
- 6.Graf J. 1999. Diverse restriction fragment length polymorphism patterns of the PCR-amplified 16S rRNA genes in Aeromonas veronii strains and possible misidentification of Aeromonas species. J Clin Microbiol 37:3194–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueras MJ, Soler L, Chacon MR, Guarro J, Martinez-Murcia AJ. 2000. Extended method for discrimination of Aeromonas spp. by 16S rDNA RFLP analysis. Int J Syst Evol Microbiol 50:2069–2073. doi: 10.1099/00207713-50-6-2069. [DOI] [PubMed] [Google Scholar]
- 8.Borrell N, Acinas SG, Figueras MJ, Martinez-Murcia AJ. 1997. Identification of Aeromonas clinical isolates by restriction fragment length polymorphism of PCR-amplified 16S rRNA genes. J Clin Microbiol 35:1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghatak S, Agarwal RK, Bhilegaonkar KN. 2007. Species identification of clinically important Aeromonas spp. by restriction fragment length polymorphism of 16S rDNA. Lett Appl Microbiol 44:550–554. doi: 10.1111/j.1472-765X.2006.02104.x. [DOI] [PubMed] [Google Scholar]
- 10.Küpfer M, Kuhnert P, Korczak BM, Peduzzi R, Demarta A. 2006. Genetic relationships of Aeromonas strains inferred from 16S rRNA, gyrB and rpoB gene sequences. Int J Syst Evol Microbiol 56:2743–2751. doi: 10.1099/ijs.0.63650-0. [DOI] [PubMed] [Google Scholar]
- 11.Soler L, Yanez MA, Chacon MR, Aguilera-Arreola MG, Catalan V, Figueras MJ, Martinez-Murcia AJ. 2004. Phylogenetic analysis of the genus Aeromonas based on two housekeeping genes. Int J Syst Evol Microbiol 54:1511–1519. doi: 10.1099/ijs.0.03048-0. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Murcia AJ, Benlloch S, Collins MD. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int J Syst Bacteriol 42:412–421. doi: 10.1099/00207713-42-3-412. [DOI] [PubMed] [Google Scholar]
- 13.Morandi A, Zhaxybayeva O, Gogarten JP, Graf J. 2005. Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains. J Bacteriol 187:6561–6564. doi: 10.1128/JB.187.18.6561-6564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yáñez MA, Catalan V, Apraiz D, Figueras MJ, Martinez-Murcia AJ. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int J Syst Evol Microbiol 53:875–883. doi: 10.1099/ijs.0.02443-0. [DOI] [PubMed] [Google Scholar]
- 15.Lamy B, Laurent F, Kodjo A. 2010. Validation of a partial rpoB gene sequence as a tool for phylogenetic identification of aeromonads isolated from environmental sources. Can J Microbiol 56:217–228. doi: 10.1139/W10-006. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Murcia AJ, Monera A, Saavedra MJ, Oncina R, Lopez-Alvarez M, Lara E, Figueras MJ. 2011. Multilocus phylogenetic analysis of the genus Aeromonas. Syst Appl Microbiol 34:189–199. doi: 10.1016/j.syapm.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Blom M, Meyer A, Gerner-Smidt P, Gaarslev K, Espersen F. 1999. Evaluation of Statens Serum Institut enteric medium for detection of enteric pathogens. J Clin Microbiol 37:2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto S, Harayama S. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol 61:3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korczak B, Christensen H, Emler S, Frey J, Kuhnert P. 2004. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int J Syst Evol Microbiol 54:1393–1399. doi: 10.1099/ijs.0.03043-0. [DOI] [PubMed] [Google Scholar]
- 20.Vila J, Ruiz J, Gallardo F, Vargas M, Soler L, Figueras MJ, Gascon J. 2003. Aeromonas spp. and traveler's diarrhea: clinical features and antimicrobial resistance. Emerg Infect Dis 9:552–555. doi: 10.3201/eid0905.020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.