Abstract
Episomal HIV-1 two-long terminal repeat (2-LTR) circles are considered markers for ongoing viral replication. Two sample processing procedures were compared to accurately quantify 2-LTR in patients by using droplet digital PCR (ddPCR). Here, we show that plasmid isolation with a spiked non-HIV plasmid for normalization enables more accurate 2-LTR quantification than genomic DNA isolation.
TEXT
With the persistence of a viral HIV-1 reservoir despite the use of combination antiretroviral therapy (cART) (1), there is a strong need for sensitive tools to characterize the viral reservoir and its dynamics. Recent data indicate that episomal unintegrated circular HIV-1 DNA (two-long terminal repeat [2-LTR] circles) serves as a marker for viral replication (2–4).
DNA isolation procedures for 2-LTR recovery were recently compared (5). Although 2-LTR circles were recovered more efficiently by total DNA isolation than by plasmid DNA isolation in vitro, the small number of patients (n = 3) assessed impeded an in vivo evaluation at the lower end of detection. This is especially important, because of the small abundance of 2-LTR circles in patients on cART (6, 7). PCR inhibition by excessive DNA load limits the input material per reaction of total genomic DNA (gDNA) (8), while plasmid DNA isolation which specifically isolates episomal DNA from the bulk chromosomal DNA allows input from larger amounts of cells.
Here, two sample processing procedures were compared to quantify 2-LTR episomal HIV-1 DNA by using droplet digital PCR (ddPCR) in peripheral blood mononuclear cells (PBMCs) from a large number of HIV-1-infected patients. This comparison was made by ddPCR, which provides direct absolute quantification with a sensitivity similar to or better than that of real-time quantitative PCR (qPCR), as shown in studies assessing low levels of plasma HIV-1 RNA (9), cell-associated HIV-1 RNA (10), total viral DNA (8, 11–13), and 2-LTR circles (8, 13). In addition, absolute quantification by ddPCR provides a direct measure to assess template loss in pre-PCR processing steps.
A modified plasmid DNA isolation method was optimized, using a reference plasmid (pSIF1-H1) (see Fig. S1 in the supplemental material), further referred to as pSIF, spiked in the samples before DNA isolation for normalization to cell equivalents (see Methods and Results in the supplemental material for in-depth optimization and Fig. S2 in the supplemental material for the workflow). The pSIF plasmid was quantified by detection of the Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (see Methods and Table S1 in the supplemental material), further referred to as the pSIF assay.
Quantification of pSIF in plasmid DNA and RPP30 in gDNA isolates revealed a higher number of cell equivalents per ddPCR replicate in plasmid DNA than in genomic DNA isolates, i.e., 6.1-fold and 12.7-fold more cell equivalents in the dilution series and in the patient-derived samples (n = 59), respectively (see Results in the supplemental material). 2-LTR quantities were normalized to cell equivalents by using the pSIF reference plasmid in plasmid DNA isolates or to the chromosomal gene RPP30 in genomic DNA isolates. Of note, a high variation in plasmid DNA isolation efficiency was observed by comparing absolute numbers of pSIF copies/μl by plasmid DNA isolation (median = 258, interquartile range [IQR] = 62.8 to 485). This high variation in isolation efficiency supports the necessity of using the spiked reference plasmid for normalization.
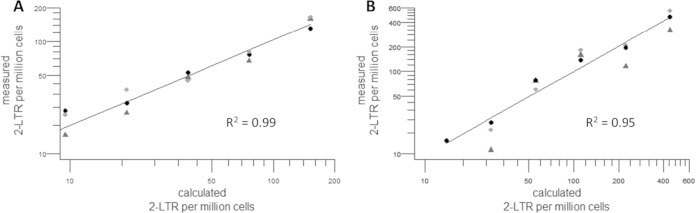
A standard 2-fold dilution series of in vitro-infected SupT1 cells was used to compare 2-LTR quantification in isolates obtained by both DNA isolation methods. A slightly higher variation between replicates was observed in genomic DNA isolates (R2 = 0.95) than in plasmid DNA isolates (R2 = 0.99), especially for the lowest dilutions (Fig. 1A and B).
FIG 1.
Quantification of 2-LTR circles in 2-fold dilution series of infected cells in both DNA isolation methods. (A and B) Linear correlations are observed with both dilution curves, but a higher variation within the three replicates was observed in genomic DNA isolates (B) than in plasmid DNA isolates (A).
To assess the applicability of the proposed assay in patient samples, 59 samples were collected from HIV-1-infected patients with undetectable viral loads on cART (n = 44) or with detectable viral loads who either recently initiated cART (n = 6) (median = 31 days, IQR = 5.5 to 125.3) or were off therapy (n = 9). The comparison of 2-LTR circles between the groups is shown in Results and Fig. S3 in the supplemental material. Samples were derived from two studies which were approved by the ethical committee of Ghent University Hospital (reference numbers B67020071646 [14] and B670201111928). Both studies included 2-LTR quantification for comparison with clinical characteristics.
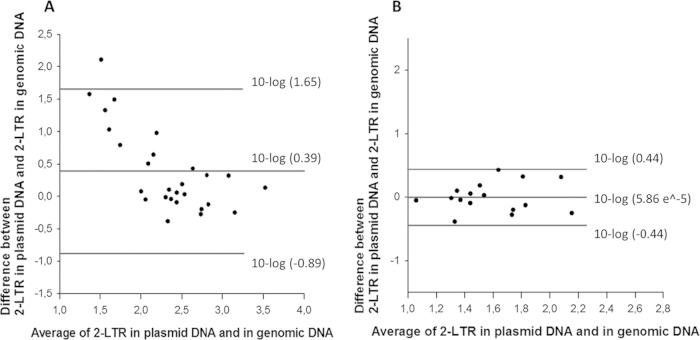
Out of 59 HIV-1-infected patients, 27 (45.8%) patients had detectable 2-LTR counts with both methods. Bland-Altman analysis revealed an average agreement between both methods, with a mean difference (bias) ± standard deviation (SD) of 0.39 ± 1.27 log10 and with 95% limits of agreement (LoA) from −0.89 to 1.65 log10 (Fig. 2A). Since sampling error or infrequent false-positive calls may influence these data (8, 10), a more stringent comparison was made using only samples in which values of more than 10 2-LTR copies/million PBMCs were recorded with both DNA isolation methods (n = 17 samples). Bland-Altman analysis of these samples revealed a mean difference (bias) of 5.86e−5 ± 0.44 log10, with 95% LoA between −0.44 and 0.44 log10 (Fig. 2B). 2-LTR copies were undetectable in 14 plasmid DNA samples and in 26 genomic DNA samples, indicating a higher sensitivity of the plasmid DNA isolation than of the genomic DNA isolation. This difference was significant by Fisher's exact test (P = 0.03), indicating a low probability that the difference is caused by random variability of false positives. Interestingly, the mean fluorescent amplitude of 2-LTR-positive droplets was higher in plasmid DNA isolates than in genomic DNA isolates (see Results and Fig. S4 and S5 in the supplemental material). This increases the sensitivity and the ease of discerning true-positive from false-positive events.
FIG 2.
Agreement evaluation between the plasmid and genomic DNA isolation methods for the 2-LTR circle quantification in patient-derived samples. Bland-Altman plots comparing the 2-LTR quantification in both plasmid and genomic DNA isolates of 27 detectable samples (A) and of 17 samples with 2-LTR copies above 10 per million PBMCs (B). Mean differences and the 95% limits of agreement are shown on the graph.
To compare the concentration of 2-LTR templates after DNA isolation in both methods, the amounts of positive droplets per sample were compared by Wilcoxon signed-rank test on the raw positive-droplet counts of the 27 detectable samples. A significantly higher number of positive droplets was observed in plasmid DNA isolates, with detection of 2.4-fold more positive droplets than in genomic DNA isolates (P = <0.001) (see Results and Fig. S4 to S6 in the supplemental material).
The higher number of template molecules found in the plasmid DNA isolates can be explained by the higher concentrations of sample that can be loaded in ddPCR than by using genomic DNA isolates, which suffer from inhibition by bulk chromosomal DNA and the restriction buffer (see Results and Fig. S7 in the supplemental material). In the patient-derived samples, an average of 610,256 cell equivalents were loaded in the ddPCR well from the plasmid DNA isolates, whereas only 47,900 cell equivalents could be loaded for genomic DNA isolates (see Fig. S2 in the supplemental material).
In conclusion, these data show that plasmid DNA isolation combined with a reference plasmid for normalization provides an optimal method for 2-LTR circle quantification in patient-derived samples.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by internal funding without pharmaceutical sponsoring. E.M. and P.B. are supported by the Agency for Innovation by Science and Technology of the Flemish Government (IWT; grant no. 111286 and 111393). M.K. is supported by a Special Research Grant—BOF Grant of Ghent University (grant no. 01N02712). J.V. is supported by an FWO Aspirant Fellowship Research Foundation—Flanders (FWO). L.V and B.V. are senior clinical investigators supported by FWO (grant no. 1.8.020.09.N.00 and grant no. 007414N). B.V. is supported by Ghent University grant BOF11/GOA/013. Additionally, this work was performed by the support of The Foundation for AIDS Research (AmfAR) (grant no. 108314-51-RGRL), King Baudouin Foundation (grant no. 2010-R20640-003), and HIV-ERA (IRIFCURE) and HIV-ERA (130442 SBO; EURECA).
We acknowledge D. N. Levy, New York University College of Dentistry, New York, NY, for the kind donation of the NLENG1-IRES HIV construct. We thank the study participants for their involvement.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03087-14.
REFERENCES
- 1.Eisele E, Siliciano RF. 2012. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37:377–388. doi: 10.1016/j.immuni.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharkey M, Babic DZ, Greenough T, Gulick R, Kuritzkes DR, Stevenson M. 2011. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog 7:e1001303. doi: 10.1371/journal.ppat.1001303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu W, Jiao Y, Lei R, Hua W, Wang R, Ji Y, Liu Z, Wei F, Zhang T, Shi X, Wu H, Zhang L. 2011. Rapid turnover of 2-LTR HIV-1 DNA during early stage of highly active antiretroviral therapy. PLoS One 6:e21081. doi: 10.1371/journal.pone.0021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray JM, McBride K, Boesecke C, Bailey M, Amin J, Suzuki K, Baker D, Zaunders JJ, Emery S, Cooper DA, Koelsch KK, Kelleher AD, Pint Study Team . 2012. Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards. AIDS 26:543–550. doi: 10.1097/QAD.0b013e328350fb3c. [DOI] [PubMed] [Google Scholar]
- 5.Badralmaa Y, Natarajan V. 2013. Impact of the DNA extraction method on 2-LTR DNA circle recovery from HIV-1 infected cells. J Virol Methods 193:184–189. doi: 10.1016/j.jviromet.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graf EH, Mexas AM, Yu JJ, Shaheen F, Liszewski MK, Di Mascio M, Migueles SA, Connors M, O'Doherty U. 2011. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog 7:e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzon MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. 2010. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med 16:460–465. doi: 10.1038/nm.2111. [DOI] [PubMed] [Google Scholar]
- 8.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson E, Wiegand A, Boltz VF, Spindler J, Dueppen P, Troppman R, Maldarelli F, Mellors JW, Kearney M, Coffin JM. 2012. Single-copy detection of plasma HIV-1 RNA using droplet digital PCR technology, abstr 679. Abstr 19th Conf Retroviruses Opportun Infect. [Google Scholar]
- 10.Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. 2014. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 9:e85999. doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Spiegelaere W, Malatinkova E, Kiselinova M, Bonczkowski P, Verhofstede C, Vogelaers D, Vandekerckhove L. 2013. Touchdown digital polymerase chain reaction for quantification of highly conserved sequences in the HIV-1 genome. Anal Biochem 439:201–203. doi: 10.1016/j.ab.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 12.Strain MC, Richman DD. 2013. New assays for monitoring residual HIV burden in effectively treated individuals. Curr Opin HIV AIDS 8:106–110. doi: 10.1097/COH.0b013e32835d811b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henrich TJ, Gallien S, Li JZ, Pereyra F, Kuritzkes DR. 2012. Low-level detection and quantitation of cellular HIV-1 DNA and 2-LTR circles using droplet digital PCR. J Virol Methods 186:68–72. doi: 10.1016/j.jviromet.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messiaen P, De Spiegelaere W, Alcami J, Vervisch K, Van Acker P, Verhasselt B, Meuwissen P, Calonge E, Gonzalez N, Gutierrez-Rodero F, Rodriguez-Martin C, Sermijn E, Poppe B, Vogelaers D, Verhofstede C, Vandekerckhove L. 2012. Characterization of LEDGF/p75 genetic variants and association with HIV-1 disease progression. PLoS One 7:e50204. doi: 10.1371/journal.pone.0050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.