Abstract
Whole-genome sequencing of Cryptosporidium spp. is hampered by difficulties in obtaining sufficient, highly pure genomic DNA from clinical specimens. In this study, we developed procedures for the isolation and enrichment of Cryptosporidium genomic DNA from fecal specimens and verification of DNA purity for whole-genome sequencing. The isolation and enrichment of genomic DNA were achieved by a combination of three oocyst purification steps and whole-genome amplification (WGA) of DNA from purified oocysts. Quantitative PCR (qPCR) analysis of WGA products was used as an initial quality assessment of amplified genomic DNA. The purity of WGA products was assessed by Sanger sequencing of cloned products. Next-generation sequencing tools were used in final evaluations of genome coverage and of the extent of contamination. Altogether, 24 fecal specimens of Cryptosporidium parvum, C. hominis, C. andersoni, C. ubiquitum, C. tyzzeri, and Cryptosporidium chipmunk genotype I were processed with the procedures. As expected, WGA products with low (<16.0) threshold cycle (CT) values yielded mostly Cryptosporidium sequences in Sanger sequencing. The cloning-sequencing analysis, however, showed significant contamination in 5 WGA products (proportion of positive colonies derived from Cryptosporidium genomic DNA, ≤25%). Following this strategy, 20 WGA products from six Cryptosporidium species or genotypes with low (mostly <14.0) CT values were submitted to whole-genome sequencing, generating sequence data covering 94.5% to 99.7% of Cryptosporidium genomes, with mostly minor contamination from bacterial, fungal, and host DNA. These results suggest that the described strategy can be used effectively for the isolation and enrichment of Cryptosporidium DNA from fecal specimens for whole-genome sequencing.
INTRODUCTION
Cryptosporidium spp. are an important cause of moderate to severe diarrhea in humans and various animals (1, 2). Over the past decade, great efforts have been made to understand the interaction between Cryptosporidium spp. and their hosts. Thus far, at least 26 species and more than 60 genotypes have been described (3), most with some host specificity. In contrast to the expanding knowledge of the taxonomic complexity of the genus Cryptosporidium, little progress has been made in understanding the molecular basis of phenotypic traits such as the host specificity. This is mainly due to the lack of whole-genome characterization of most Cryptosporidium species and genotypes.
Thus far, the genomes of only four Cryptosporidium isolates have been sequenced. Sequence data of the whole genomes of C. parvum and C. hominis, the two species responsible for most human infections (4), were first published in 2004 (5, 6). Several years later, the genome of C. muris was sequenced, and data from the project are available on CryptoDB (http://cryptodb.org). More recently, the genome of anthroponotic subtype family IIc of C. parvum has been sequenced using the next-generation sequencing (NGS) tools (7). The availability of the whole-genome sequence data from Cryptosporidium spp. has greatly improved our understanding of the basic biology of Cryptosporidium spp. and the development of new intervention strategies (8). The data have also facilitated the development of high-resolution molecular typing tools (4, 9). Population genetic characterizations of C. parvum and C. hominis using these advanced molecular detection tools have started to improve our understanding of the genetic determinants for host specificity and virulence (10–12).
One major factor hindering the NGS analysis of genomes of Cryptosporidium spp. is the difficulty in acquiring sufficient numbers of highly purified oocysts because of the lack of effective animal models and in vitro culture systems. This presents a major obstacle in obtaining DNA in suitable concentrations and purity for NGS analysis. Currently, the diagnostic procedures used in the concentration of Cryptosporidium oocysts from fecal specimens frequently lead to the copurification of contaminating bacteria, food particles, and even host cells.
Several methods have been developed to recover oocysts from fecal and environmental specimens, such as sucrose flotation (13), discontinuous sucrose gradient centrifugation (14), cesium chloride (CsCl) gradient centrifugation (15), and immunomagnetic separation (IMS) (16). These methods usually result in significant oocyst losses, thus generating an insufficient amount of DNA for NGS analysis. In addition, when used alone, these methods frequently lead to the copurification of objects with similar buoyant-density characteristics or adhered to the surface of Cryptosporidium oocysts. As all DNA fragments present in a DNA extraction are sequenced by the shotgun-based NGS technologies, an abundance of contaminating DNA will reduce the sequence coverage of the Cryptosporidium genome. This is especially important for the whole-genome sequencing of Cryptosporidium spp., which have small (∼9-Mb) genomes in comparison to the genomes of the host cells (>3 Gb) and food particles (mostly >300 Mb). Therefore, the isolation and enrichment of parasite DNA free of significant contamination by nontarget organisms are imperative for successful sequencing of Cryptosporidium genomes.
In this study, we developed a strategy for the isolation and enrichment of Cryptosporidium DNA and verification of DNA purity for whole-genome sequencing. The method uses combined sucrose and cesium chloride density gradient separation and IMS for purification of oocysts from fecal specimens, DNA extraction using a commercial kit, whole-genome amplification (WGA) to increase the quantity of the extracted genomic DNA, quantitative PCR (qPCR) analysis of WGA products for initial evaluation of Cryptosporidium DNA quality, cloning-sequencing of WGA products for initial assessment of DNA purity, and NGS analysis of WGA products for the final evaluation of genome coverage and of the extent of nontarget contamination. The procedures developed in this study should make routine whole-genome sequencing of Cryptosporidium spp. feasible.
MATERIALS AND METHODS
Specimens.
Twenty-four fecal specimens of six Cryptosporidium species or genotypes from mostly naturally infected humans and animals were used in this study (Table 1). All human specimens were from patients with diarrhea, whereas the animal specimens were mostly from healthy animals. All fecal specimens were previously confirmed as being positive for Cryptosporidium spp. by sequence analysis of the small-subunit rRNA (ssrRNA) gene (17). They were stored in 2.5% potassium dichromate (K2Cr2O7) for less than 1 year before being used in oocyst purification.
TABLE 1.
Cryptosporidium specimens used in this study and results of the assessment of the quality of whole-genome amplification products by qPCR, Sanger sequencing, and next-generation sequencing
| Species | Specimen no. | Origin | Source | Concn of purified WGA product (ng/μl) |
CT value |
% positive colonies derived from Cryptosporidium (no. of positive colonies/total no. of colonies) | Length (bp) of assembly | Length (bp) of mapped contigs (% genome coverage)c | % contamination in NGS analysis | Major contaminant(s) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Purified WGA product | 1:100 dilution | 1:1,000 dilution | ||||||||||
| C. andersoni | 31636a | Ethiopia | Cattle | 262.9 | 15.79 | 21.58 | 24.77 | 3.70 (1/27) | ||||
| 31729a | China | Cattle | 214.8 | 12.27 | 21.14 | 21.74 | 85.71 (18/21) | 10,027,502 | 9,072,248 (98.13) | 9.53 | Serratia liquefaciens, Delftia acidovorans, Ralstonia pickettii, unknown | |
| 33583a | Japan | Cattle via SCID mouse | 242.6 | 12.80 | 18.56 | 22.14 | 21.74 (5/23) | 18,347,829 | 9,086,745 (98.29) | 50.48 | Pseudomonas fluorescens | |
| 30847a | Canada | Cattle | 212.8 | 12.88 | 20.10 | 23.28 | 80.95 (17/21) | 9,578,724 | 9,078,261 (98.19) | 5.22 | Penicillium chrysogenum, Serratia liquefaciens | |
| 30972a,b | Japan | Cattle | 147.5 | 29.12 | 36.09 | 37.59 | 0 (0/26) | 2,153,082 | 3,984 (0.04) | 99.81 | Serratia liquefaciens, Penicillium chrysogenum, Bos taurus, Acanthamoeba sp., Delftia acidovorans | |
| 37034a | Egypt | Cattle | 160.1 | 15.65 | 22.49 | 26.16 | 62.50 (20/32) | 13,720,950 | 8,902,966 (96.30) | 35.11 | Unknown | |
| 38986a | Japan | Cattle via SCID mouse | 332.8 | 11.57 | 15.98 | 19.07 | 90.91 (20/22) | 9,060,784 | 9,034,514 (97.72) | 0.29 | Lactobacillus sakei plasmid, Homo sapiens | |
| C. parvum | 35090 | Egypt | Cattle | 169.7 | 10.58 | 16.71 | 19.47 | 96.42 (27/28) | 8,851,552 | 8,820,515 (96.90) | 0.35 | Acidovorax sp. |
| 35102 | Egypt | Cattle | 205.3 | 24.01 | 31.50 | 33.16 | 0 (0/19) | |||||
| 39011 | United States | Human | 199.1 | 12.44 | 18.83 | 22.69 | 64.71 (11/17) | 9,233,306 | 9,075,221 (99.70) | 1.71 | Cloning vector pFD288, enterobacterial phage, unknown | |
| 39187 | United States | Human | 42.5 | 13.74 | 20.71 | 24.84 | 70 (7/10) | 9,104,201 | 9,075,601 (99.71) | 0.31 | Sphingobacterium sp., Homo sapiens | |
| 31727a | China | Human via gerbil | 209.1 | 13.00 | 21.13 | 24.28 | 41.67 (10/24) | 9,158,968 | 9,041,928 (99.34) | 1.28 | Delftia acidovorans, Stenotrophomonas maltophilia, Homo sapiens, Uncultured marine virus | |
| 34902 | Egypt | Cattle | 137.1 | 13.94 | 19.67 | 22.70 | 96.67 (29/30) | 9,075,198 | 9,003,222 (98.91) | 0.79 | Cloning vector pUC19c, Stenotrophomonas maltophilia, Delftia acidovorans, Pinus taeda | |
| 37266a | Egypt | Cattle | 240.5 | 19.29 | 23.69 | 25.33 | 0 (0/20) | |||||
| C. hominisc | 33537a,b | United States | Human | 233.2 | 13.93 | 20.68 | 24.11 | 66.67 (21/31) | 14,161,808 | 8,665,098 (95.20) | 38.81 | Serratia liquefaciens |
| 30976a | United States | Human | 254.3 | 12.61 | 19.72 | 23.07 | 22,133,082 | 9,054,314 (99.47) | 59.09 | Escherichia coli, Hafnia alvei, unknown Enterobacteriaceae | ||
| 37999a | United States | Human | 318.0 | 12.82 | 19.61 | 22.00 | 86.21 (25/29) | 9,054,010 | 9,041,990 (99.34) | 0.13 | Stenotrophomonas maltophilia | |
| 30974a,b | United States | Human | 207.5 | 11.39 | 16.19 | 22.19 | 96.15 (25/26) | 8,871,639 | 8,826,344 (96.97) | 0.51 | Bacteroides fragilis | |
| C. ubiquitum | 33496a,b | United States | Sifaka | 261.9 | 9.81 | 19.14 | 24.52 | 82.35 (28/34) | 11,431,018 | 8,605,875 (94.55) | 24.71 | Akkermansia muciniphila, Pseudomonas fluorescens |
| 39668 | United States | Human | 142.2 | 13.17 | 20.21 | 23.88 | 91.30 (21/23) | 9,456,213 | 8,957,037 (98.40) | 5.28 | Uncultured bacterium plasmid, Escherichia coli-Bacteroides shuttle vector, Bifidobacterium bifidum plasmid, enterobacteria phage, Bacteroides dorei, Homo sapiens | |
| 39725 | United States | Human | 172.5 | 12.91 | 19.89 | 23.31 | 65.22 (15/23) | 11,526,951 | 9,030,621 (99.21) | 21.66 | Homo sapiens, Propionibacterium acnes, Meyerozyma guilliermondii | |
| 39726 | United States | Human | 222.8 | 12.19 | 18.04 | 22.70 | 96.77 (30/31) | 9,057,158 | 8,962,965 (98.47) | 1.04 | Acinetobacter baumannii, unknown fungi, Homo sapiens | |
| C. tyzzeri | 37035b | Czech Republic | House mouse | 180.9 | 11.07 | 19.29 | 22.88 | 100 (32/32) | 8,666,741 | 8,606,333 (94.55) | 0.70 | Pseudomonas pseudoalcaligenes |
| Cryptosporidium chipmunk genotype I | 37763a | United States | Human | 306.6 | 10.95 | 17.52 | 21.26 | 100 (11/11) | 9,509,783 | 9,009,492 (98.98) | 5.26 | Cryptosporidium hominis |
DNA extracted from IMS-purified oocysts without bleach treatment.
Sequenced by 454 FLX; all others sequenced by Illumina.
For C. andersoni, the reference genome is the published C. muris RN 66 whole-genome sequence (DDBJ accession no. AAZY02000000), whereas for the rest, the reference genome is the C. parvum Iowa whole-genome sequence (DDBJ accession no. AAEE01000000).
Oocyst purification.
Approximately 5 ml of fecal suspension from each specimen was transferred to a 20-mesh stainless steel sieve to remove large debris and washed with 45 ml of 0.85% saline solution. The sieved fecal suspension was centrifuged at 1,500 × g for 10 min. The pellet was resuspended in 0.85% saline solution and applied to a discontinuous sucrose gradient as previously described (14). The oocysts harvested were purified with a cesium chloride (CsCl) gradient technique (18). The purified secondary oocysts were further separated from residual contaminants by IMS using a Dynabeads anti-Cryptosporidium kit (Invitrogen, Oslo, Norway) per manufacturer-recommended procedures, except that twice the recommended volume of beads was used. Finally, the bead-oocyst suspension was treated on ice with 10% Clorox Regular Bleach (Oakland, CA) for 10 min to dissolve any bacteria or fungi attached to oocysts.
DNA extraction and whole-genome amplification.
DNA was extracted from purified oocysts using a QIAamp DNA minikit (Qiagen Sciences, Hilden, Germany). Briefly, 180 μl of the ATL buffer from the kit was transferred to the bleached treated bead-oocyst complex in a 1.5-ml tube, and the suspension was subjected to 5 freeze-thaw (−70°C and 56°C) cycles. The suspension was then digested with 20 μl of proteinase K at 56°C overnight. Genomic DNA was extracted from the oocysts following the manufacturer-recommended procedures. WGA was performed on 5 μl of the extracted DNA using a REPLI-g Midi kit (Qiagen Sciences). After WGA, 5 μl of each amplified product was analyzed by electrophoresis on a 1.5% agarose gel. The remaining 45 μl of WGA product was washed with 60 μl Tris-EDTA (TE) buffer (0.01 M, pH 8.0) by vacuum filtration using MultiScreen plates (EMD Millipore, Billerica, MA). The cleaned WGA product was resuspended in 100 μl TE buffer, and the DNA concentration was measured using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Cryptosporidium DNA analysis by qPCR.
The WGA products and 1:100 and 1:1,000 dilutions of them were analyzed by ssrRNA gene-based qPCR. Each PCR reaction mixture had a final volume of 50 μl and contained 200 μM (each) deoxynucleoside triphosphates (dNTPs), 3 mM MgCl2, 500 nM (each) primer (primer 18S-qF2, 5′-AAG TAT AAA CCC CTT TAC AAG TA-3′; primer 18S-qR2, 5′-TAT TAT TCC ATG CTG GAG TAT TC-3′), 400 ng/μl of nonacetylated bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 1× GeneAmp PCR buffer (Applied Biosystems, Foster City, CA), 2.5 U of GoTaq DNA polymerase (Promega, Madison, WI), 1× EvaGreen (Biotium, Hayward, CA), and 1 μl of the DNA template. All reactions were performed on a LightCycler 480 system (Roche, Mannheim, Germany) for 50 cycles of amplification (95°C for 5 s, 55°C for 10 s, and 72°C for 40 s), with an initial denaturation step (95°C for 3 min) and a final cooling step (40°C for 30 s). The threshold cycle (CT) value from the qPCR was used as an indicator of the yield of Cryptosporidium genomic DNA in WGA products.
Assessment of purity of Cryptosporidium genomic DNA in WGA products by cloning and Sanger sequencing.
The purified WGA products were digested with BamHI (Thermo Fisher Scientific, Waltham, MA) in a 60-μl reaction volume containing 20 μl of purified WGA product. The digested products were loaded onto a 1.5% agarose gel, and restriction products of between 1,000 and 1,500 bp were cut out and purified with a QIAquick gel extraction kit (Qiagen Sciences). These products were cloned into BamHI-linearized pUC19 vectors (Thermo Fisher Scientific) using a Rapid DNA ligation kit (Thermo Fisher Scientific) in a 20-μl volume containing 1.5 μl of linear pUC19 vectors, 10 μl of gel-purified insertion DNA, 4 μl of 5× Rapid ligation buffer, and 1 μl T4 DNA ligase. Two microliters of ligation products was used to transform JM109 competent cells (Promega). Transformed Escherichia coli cells were plated on LB plates containing ampicillin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl-β-d-thiogalactopyranoside). After 16 h of incubation at 37°C, up to 40 white colonies were picked and boiled in 5 μl of nuclease-free water for 5 min to release plasmid DNA. They were screened for positive colonies by PCR using universal vector primers (Forward, 5′-GCCAGGGTTTTCCCAGTCACGA-3′; Reverse, 5′-GAGCGGATAACAATTTCACACAGG-3′). Positive PCR products were sequenced on an ABI 3130 Genetic Analyzer (Applied Biosystems) using the forward primer and a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems). The obtained sequences were subjected to BLAST analysis using the NCBI nucleotide database to determine the source of the cloned WGA product. The percentage of positive colonies from Cryptosporidium was determined to assess the extent of contamination by nontarget organisms.
Assessment of contamination from nontargets and coverage of Cryptosporidium genomes by NGS.
WGA products from five specimens (30972, 30974, 33496, 33537, and 37035) were sequenced with 454 technology on a GS-FLX Titanium System (Roche, Branford, CT), using the standard Roche library protocol, generating 500,000 (specimens 30972, 33496, and 37035) to 1 million (specimens 30974 and 33537) reads of ∼350 bp for each specimen, giving an estimated 20- to 40-fold coverage of the Cryptosporidium genome. The remaining WGA products, except for three with high CT values, were sequenced using the Illumina TruSeq (v3) library protocol on an Illumina Genome Analyzer IIx or Illumina HiSeq 2500 system (Illumina, San Diego, CA), giving an estimated 140- to 200-fold coverage of the Cryptosporidium genome. For Illumina sequencing, 100-by-100-bp paired-end sequencing was used for most WGA products, except for C. andersoni specimen 38986, C. parvum specimens 34902 and 35090, and Cryptosporidium chipmunk genotype I specimen 37763, for which only single-end reads were available because of premature termination of the sequencing run. The sequence reads from each specimen were assembled using the CLC Genomics Workbench (CLC Bio, Boston, MA). The contigs generated were mapped to published whole-genome sequences of C. muris (for C. andersoni genomes) and C. parvum (for other Cryptosporidium genomes) using Mauve (http://gel.ahabs.wisc.edu/mauve/).
Nucleotide sequence accession number.
The sequences determined in this work have been deposited in the Whole Genome Shotgun Project for C. hominis at DDBJ/EMBL/GenBank under BioProject accession no. PRJNA252787. Raw sequence reads and assembled contigs for other Cryptosporidium species will be submitted once data analyses are complete.
RESULTS
Efficiency of WGA.
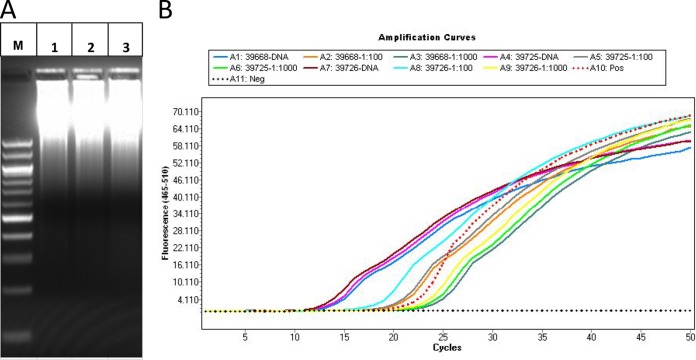
After WGA, electrophoresis of the product showed the presence of a range of DNA amplicons for all specimens, indicating that the amplification was successful. Most of the WGA products were large molecules located in the top of the gel (Fig. 1A). The concentrations of purified WGA DNA ranged between 42.5 ng/μl and 332.8 ng/μl, but most of them were in the range of 180.9 ng/μl to 332.8 ng/μl (Table 1). To estimate the abundance of Cryptosporidium genomic DNA within the WGA products, qPCR analysis was carried out on purified WGA products and their 1:100 and 1:1,000 dilutions using Cryptosporidium-specific primers (Fig. 1B). Of the 24 WGA products, 19 generated relatively low CT values ranging from 9.81 to 13.94 for undiluted WGA products (Table 1 and Fig. 1B), indicating that Cryptosporidium genomic DNA was present in these specimens in high concentrations. The CT values of the remaining WGA products from five specimens (30972, 31636, 35102, 37034, and 37266) were higher, with two having extremely high CT values (24.01 and 29.12) (Table 1).
FIG 1.
Whole-genome amplification (WGA) of Cryptosporidium ubiquitum DNA extracted from oocysts purified from human fecal specimens 39668 (second lane), 39725 (third lane), and 39726 (fourth lane) (A) and assessment of the quality of WGA products by qPCR analysis of the Cryptosporidium small-subunit rRNA gene in undiluted and 1:100 and 1:1,000 dilutions of WGA products (B). See Table 1 for CT values of the qPCR for the three specimens under analysis. Lane M, molecular marker; Pos, positive; Neg, negative.
Purity of WGA products by Sanger sequencing.
The purity of WGA products was estimated initially by cloning and Sanger sequencing of the products. BLAST analysis of 10 to 34 sequences obtained from each WGA product indicated that most specimens had low levels of contamination (Table 1). Five specimens, including 30972 (C. andersoni), 31636 (C. andersoni), 33583 (C. andersoni), 35102 (C. parvum), and 37266 (C. parvum), however, had very high levels of contamination, with only 0/26, 1/27, 5/23, 0/19, and 0/20 colonies deriving from Cryptosporidium genomic DNA, respectively. WGA products of four of the specimens (31636, 33583, 30972, and 37266), however, were generated from IMS-purified oocysts without bleach treatment prior to DNA extraction during the early phase of the study. For the remaining specimens, the percentages of positive colonies derived from Cryptosporidium ranged from 41.7% to 100% (Table 1).
Purity of WGA products by NGS.
The purity of the WGA products was further assessed by NGS analysis. All WGA products with low (<16.0) CT values in qPCR and a significant proportion (>20%) of Cryptosporidium sequences in cloning-Sanger sequencing were sequenced by NGS. As a control, one WGA product from specimen 30972 with a high CT value (29.12) and no Cryptosporidium sequence in Sanger sequencing (0/26) was also sequenced by NGS. Mapping of the assembled NGS contigs to reference genomes of Cryptosporidium indicated that 10 of the 21 sequenced WGA products had minimal (<2% of the overall contig sequences) contamination by nontarget organisms (Table 1). Among the remaining 11 WGA products sequenced, 4 had <10% and 6 had 21.7% to 59.1% contamination with sequences from nontarget organisms. In contrast, 454 sequencing of the WGA product from specimen 30972 produced a poor genome assembly (2,153,082 bp in 1,384 contigs; N50 = 3,922 bp) because of the generation of mostly (57.25%) nonaligned reads. The assembly contained only 0.04% Cryptosporidium sequences. Thus, there was a fairly good agreement in contamination assessments between Sanger sequencing of cloned WGA products and direct NGS sequencing; most WGA products showing low levels of contaminations in Sanger sequencing produced mostly Cryptosporidium contigs in NGS analysis (Table 1).
As expected, host (especially human) DNA and enteric bacteria such as Serratia liquefaciens, Escherichia coli, and Delftia acidovorans and their mobile elements (phages and plasmids) were major contaminants. However, some other Gram-negative bacterial species such as Stenotrophomonas maltophilia and Pseudomonas spp. and, occasionally, fungi were also present in WGA products from some specimens (Table 1). These sequences generally had more balanced G/C content (∼50%) than the sequences from Cryptosporidium spp. (∼30%).
Genome coverage by WGA.
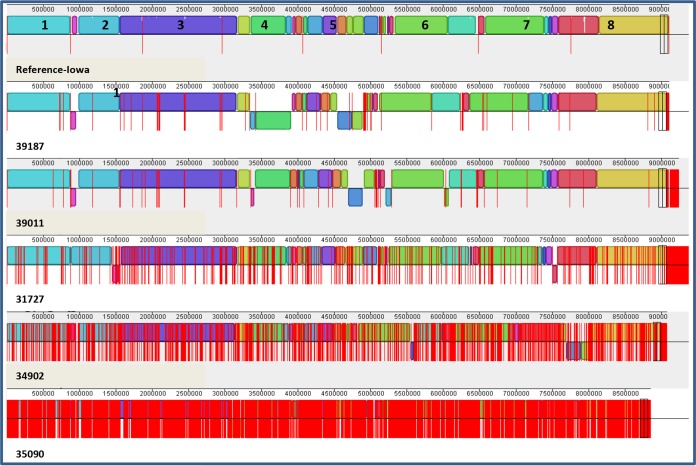
Mapping of the assembled NGS contigs to reference genomes from C. muris (for C. andersoni genomes) and C. parvum (for genomes of remaining species in the study) indicated that 94.6% to 99.7% of the Cryptosporidium genome was covered by NGS analysis of the WGA products from Cryptosporidium oocysts purified from fecal materials (Table 1; see Fig. 2 for mapping results for C. parvum specimens). As expected, the genome coverage of WGA products by 454 sequencing was lower than that by Illumina, at 94.6% to 97.0% (mean ± standard deviation, 95.32% ± 1.14%) versus 96.3% to 99.7% (98.57% ± 0.98%), because of the differences in the depth of sequencing efforts (∼20-fold to ∼40-fold coverage by 454 sequencing versus ∼140-fold to ∼200-fold coverage by Illumina). The highest coverage came from WGA of C. parvum and C. hominis genomes by Illumina paired-end sequencing, with most WGA products yielding sequences covering 98.9% to 99.7% of the genome. Thus, 77 and 79 contigs covered the entire genomes of C. parvum specimens 39187 and 39011 (Fig. 2), and 47 and 64 contigs covered the genomes of C. hominis specimens 30976 and 37999. Similarly, 35 and 38 contigs covered all genomic sequences of C. ubiquitum specimens 39726 and 39668 that were mapped to the complete reference C. parvum genome (data not shown). There were no obvious differences in genome coverage between human and animal specimens.
FIG 2.
Coverage of the Cryptosporidium genome by assembled contigs generated from Illumina sequencing of whole-genome amplified products from Cryptosporidium parvum oocysts purified from five fecal specimens. The assembled contigs (bordered by vertical red lines) from each specimen were mapped to the reference genome of the C. parvum Iowa isolate to show sequence coverage of eight chromosomes (numbered) and contamination with sequences from nontarget organisms (contigs after the pairs of vertical black boxes). Minimum contamination was seen in WGA from most specimens except for specimen 31727, which has some contigs near the end of the assembly alignment. Better genome coverage is seen with assemblies generated from 100-bp-by-100-bp paired-end sequencing (for specimens 39187, 39011, and 31727) than with those generated from single-end 100-bp reads (for specimens 34902 and 35090). The depths of sequencing for specimens 39187, 39011, 31727, 34902, and 35090 are 190-, 170-, 140-, 170-, and 140-fold genome coverage, respectively.
DISCUSSION
In this study, a strategy was developed for the isolation and enrichment of Cryptosporidium DNA from fecal specimens for NGS analysis of whole genomes. It uses an integrated approach involving oocyst purification, DNA extraction from purified oocysts, and WGA of extracted DNA. Sanger sequencing and NGS analysis of the WGA products indicate that this approach can provide an adequate quantity of Cryptosporidium DNA for whole-genome sequencing. Although some contamination from nontarget microbial and host DNA was seen in WGA products, the high throughput and deep coverage of the NGS analysis enabled the acquisition of sequences covering 94.6% to 99.7% of the genomes of six Cryptosporidium species or genotypes, including four Cryptosporidium species or genotypes sequenced for the first time at the whole-genome level. Previously, because of the difficulty in in vitro and in vivo propagation of Cryptosporidium spp., only four isolates of three Cryptosporidium species had been sequenced.
A critical step in our approach is the reduction of contamination from nontarget organisms. To achieve this, we have combined two density gradient centrifugation steps with IMS to remove food particles and minimize contamination by bacteria, fungi, and host DNA. The effectiveness of this combined oocyst purification procedure was demonstrated by the high percentage of positive colonies derived from Cryptosporidium in traditional sequencing. Among the 24 WGA products generated, 17 produced mostly (from 62.5% to 100%) Cryptosporidium sequences in Sanger sequencing of cloned WGA products. Four of the five WGA products with significant nontarget contamination in Sanger sequencing were from oocysts not subjected to bleach treatment during the early phase of the study, demonstrating the importance of removing residual contaminants attached to isolated oocysts. The assessment of the purity of WGA was supported by successful NGS analysis of the WGA products, which documented the presence of 0.13% to 59.1% contamination by nontargets in 20 WGA products sequenced successfully by 454 or Illumina technology.
Although some residual contamination is inevitably present in most WGA products generated, this contamination can apparently be overcome by the high throughput of NGS analysis. Thus, three WGA products with significant contamination were sequenced successfully by Illumina in this study, yielding sequences that cover 98.3% to 99.5% of the Cryptosporidium genome. It is very easy to filter out sequence contigs from contaminants, as they do not map to reference Cryptosporidium genomes and differ from unmapped species-specific Cryptosporidium sequences by having more balanced (∼50%) instead of low (∼30% for Cryptosporidium spp.) G/C content.
Although the combination of oocyst purification methods can yield highly pure preparations, the number of organisms isolated is small. Consequently, the amount of genomic DNA extracted from the purified oocysts is limited. To increase DNA concentrations for NGS library construction, we have used WGA to amplify extracted DNA, as already done for archiving Cryptosporidium DNA from clinical specimens (19). Although the visualization of genomic DNA of high molecular weight by electrophoresis is a direct demonstration of successes and failures in genome amplification, we used ssrRNA gene-based qPCR to assess the quality and quality of the Cryptosporidium DNA amplified, indicated by the CT values generated (Fig. 1). In the majority of WGA products with low (<16.0) CT values in qPCR, NGS analysis of the WGA products has led to the acquisition of sequences that cover nearly complete genomes of Cryptosporidium spp. In fact, all but one (specimen 37034) of the Cryptosporidium genomes sequenced in this study had WGA products with CT values lower than 14.0. In contrast, WGA products with high CT values all generated a low percentage of Cryptosporidium sequences in Sanger sequencing. Thus, low CT values in qPCR analysis of WGA products can serve as a proxy for high yield of Cryptosporidium genomic DNA. This can potentially eliminate the use of the laborious and time-consuming cloning-sequencing approach in assessing the quantity and purity of Cryptosporidium DNA prior to NGS analysis of the WGA products.
In summary, the procedure developed in this study has been shown to be effective for the isolation and enrichment of Cryptosporidium DNA from fecal specimens and verification of DNA purity for whole-genome sequencing. The use of this approach has led to the sequencing of nearly complete genomes of 20 isolates of six Cryptosporidium species. With further refinement, it can potentially lead to the wide use of comparative genomics in epidemiological investigations of cryptosporidiosis in humans and to de novo sequencing of the genomes of other Cryptosporidium species of public health and economic importance.
ACKNOWLEDGMENTS
We thank Lori Rowe and Kristine Knipe for technical assistance.
This work was supported by the National Natural Science Foundation of China (31229005 and 31110103901) and Centers for Disease Control and Prevention, USA.
The findings and conclusions in this report are ours and do not necessarily represent the views of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Cho YI, Han JI, Wang C, Cooper V, Schwartz K, Engelken T, Yoon KJ. 2013. Case-control study of microbiological etiology associated with calf diarrhea. Vet Microbiol 166:375–385. doi: 10.1016/j.vetmic.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan U, Fayer R, Xiao L. 11 August 2014, posting date Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 4.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 6.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, Serrano MG, Puiu D, Manque P, Akiyoshi D, Mackey AJ, Pearson WR, Dear PH, Bankier AT, Peterson DL, Abrahamsen MS, Kapur V, Tzipori S, Buck GA. 2004. The genome of Cryptosporidium hominis. Nature 431:1107–1112. doi: 10.1038/nature02977. [DOI] [PubMed] [Google Scholar]
- 7.Widmer G, Lee Y, Hunt P, Martinelli A, Tolkoff M, Bodi K. 2012. Comparative genome analysis of two Cryptosporidium parvum isolates with different host range. Infect Genet Evol 12:1213–1221. doi: 10.1016/j.meegid.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rider SD Jr, Zhu G. 2010. Cryptosporidium: genomic and biochemical features. Exp Parasitol 124:2–9. doi: 10.1016/j.exppara.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plutzer J, Karanis P. 2009. Genetic polymorphism in Cryptosporidium species: an update. Vet Parasitol 165:187–199. doi: 10.1016/j.vetpar.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Feng Y, Tiao N, Li N, Hlavsa M, Xiao L. 2014. Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J Clin Microbiol 52:524–530. doi: 10.1128/JCM.02973-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. 2013. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int J Parasitol 43:1141–1147. doi: 10.1016/j.ijpara.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, Feng Y. 2013. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis 19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNabb SJ, Hensel DM, Welch DF, Heijbel H, McKee GL, Istre GR. 1985. Comparison of sedimentation and flotation techniques for identification of Cryptosporidium sp. oocysts in a large outbreak of human diarrhea. J Clin Microbiol 22:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrowood MJ, Sterling CR. 1987. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol 73:314–319. doi: 10.2307/3282084. [DOI] [PubMed] [Google Scholar]
- 15.Kilani RT, Sekla L. 1987. Purification of Cryptosporidium oocysts and sporozoites by cesium-chloride and Percoll gradients. Am J Trop Med Hyg 36:505–508. [DOI] [PubMed] [Google Scholar]
- 16.Bukhari Z, McCuin RM, Fricker CR, Clancy JL. 1998. Immunomagnetic separation of Cryptosporidium parvum from source water samples of various turbidities. Appl Environ Microbiol 64:4495–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrowood MJ, Donaldson K. 1996. Improved purification methods for calf-derived Cryptosporidium parvum oocysts using discontinuous sucrose and cesium chloride gradients. J Eukaryot Microbiol 43:89S. doi: 10.1111/j.1550-7408.1996.tb05015.x. [DOI] [PubMed] [Google Scholar]
- 19.Bouzid M, Heavens D, Elwin K, Chalmers RM, Hadfield SJ, Hunter PR, Tyler KM. 2010. Whole genome amplification (WGA) for archiving and genotyping of clinical isolates of Cryptosporidium species. Parasitology 137:27–36. doi: 10.1017/S0031182009991132. [DOI] [PubMed] [Google Scholar]