Abstract
An IMP-4-producing Acinetobacter pittii strain coproducing oxacillinases was isolated from a leg wound of a 67-year-old female patient. Identification to the species level by rpoB and gyrB sequencing and multiplex-PCR-based analysis revealed that the isolate was A. pittii. Whole-genome sequencing of this A. pittii isolate determined the presence of blaOXA-96, blaCARB-2, and a novel blaOXA-421 gene. The position of this novel blaOXA-421 gene was similar to that of blaOXA-51 in A. baumannii, downstream of the phosphinothricin N-acetyltransferase gene and upstream of fxsA in the chromosome. This A. pittii isolate was found to belong to sequence type 119 (ST119). Here, we report the first isolation of IMP-4-producing A. pittii ST119 with a novel blaOXA-421 gene from a patient in Australia and characterize its draft genome.
CASE REPORT
A 67-year-old diabetic woman suffered a fall leading to a displaced distal spiral tibial plateau fracture. In the weeks prior to the fall, she had received multiple antimicrobials (clindamycin, lincomycin, cephalexin, ciprofloxacin, and ceftazidime) for an infected hematoma of the breast and a series of lower respiratory tract infections. The patient underwent definitive repair of the fracture but postoperatively developed osteomyelitis. Debridement of the leg wound was performed. Acinetobacter species and vancomycin-resistant Enterococcus strains were isolated from the tissue removed. This Acinetobacter species (CR12-42) was carbapenem resistant. Despite ongoing antibiotic treatment, the patient's leg required amputation in March 2013, after continuous inflammation, infections for more than 5 months, and an episode of severe Clostridium difficile infection resulting in colectomy. The leg infection was resolved by the amputation.
The initial identification of this Acinetobacter species was done by Vitek 2. Antimicrobial susceptibility testing by Vitek 2 (bioMérieux) showed resistance to carbapenems, ceftazidime, ceftriaxone, cefepime, gentamicin, tobramycin, trimethoprim-sulfamethoxazole, ticarcillin-clavulanic acid, and ciprofloxacin according to the EUCAST standard (1). The isolate was referred to our laboratory at the University of Queensland Centre for Clinical Research. The Acinetobacter isolate was identified to the species level by a gyrB multiplex PCR, which revealed that CR12-42 was Acinetobacter pittii (2). Partial rpoB sequencing (3) confirmed that CR12-42 was A. pittii.
Phenotypic characterization to determine the class of carbapenemase was performed as previously described (4–6). The A. pittii isolate showed a metallo-β-lactamase phenotype by producing a larger inhibition zone around carbapenem disks with EDTA than around carbapenem disks alone (>5-mm breakpoint increase in the size of the inhibition zone). The isolate also produced a positive result in the modified Hodge and Carba NP tests for carbapenemase production. MICs were determined with Etest (bioMérieux). The isolate was resistant to all of the carbapenems tested, i.e., ceftazidime, cefotaxime, cefepime, cefoxitin, ticarcillin-clavulanic acid, trimethoprim-sulfamethoxazole, and ciprofloxacin (Table 1). Interestingly, this A. pittii isolate was susceptible to tetracycline, minocycline, colistin, and tigecycline (Table 1).
TABLE 1.
MICs of antimicrobials for A. pittii CR12-42 as determined by Etest
| Antimicrobial(s) | MIC (mg/liter) | Interpretationa |
|---|---|---|
| Ertapenem | >32 | Resistant |
| Imipenem | 24 | Resistant |
| Meropenem | 12 | Resistant |
| Doripenem | >32 | Resistant |
| Cefepime | 64 | Resistant |
| Ceftazidime | >256 | Resistant |
| Cefotaxime | >32 | Resistant |
| Ceftriaxone | >32 | Resistant |
| Cefuroxime | >256 | Resistant |
| Cefoxitin | >256 | Resistant |
| Piperacillin-tazobactam | 12 | Resistant |
| Ampicillin-sulbactam | 2 | Susceptibleb |
| Ticarcillin-clavulanic acid | 256 | Resistant |
| Piperacillin | >256 | Resistant |
| Amikacin | 12 | Intermediate |
| Gentamicin | >256 | Resistant |
| Netilmicin | 24 | Resistant |
| Ciprofloxacin | 3 | Resistant |
| Tetracycline | 0.75 | Susceptibleb |
| Minocycline | 0.023 | Susceptibleb |
| Trimethoprim-sulfamethoxazole | >32 | Resistant |
| Colistin | 0.094 | Susceptible |
| Tigecycline | 0.094 | Susceptible |
Carbapenem resistance in Acinetobacter species is commonly associated with the presence of carbapenem-hydrolyzing class D β-lactamase- or oxacillinase-encoding genes such as blaOXA-23 and blaOXA-51 in Acinetobacter baumannii (7, 8). A PCR assay and sequencing for all of the blaOXA genes frequently present in Acinetobacter species, i.e., blaOXA-23-like, blaOXA-51-like, blaOXA-40-like, and blaOXA-58-like, were performed (7–9). The isolate was positive for the blaOXA-58-like subclass and negative for other subclasses of blaOXA. A PCR assay for ISAba1, the common insertion element in A. baumannii, was also negative. A PCR assay and sequencing for other carbapenemase-encoding genes (10, 11), i.e., blaIMP, blaNDM, blaKPC, and blaVIM, were positive for blaIMP-4. A prepared pair-ended library of the whole genomic DNA was sequenced via Illumina MiSeq to further characterize the resistance mechanisms of A. pittii CR12-42 and to analyze its genome.
Whole-genome DNA sequencing produced a total of 138,932,382 paired-end reads with 30× average coverage. We used the CLC genomic workbench version 7.5 (CLC Bio, Aarhus, Denmark) for de novo assembly with a 500-bp minimum threshold resulting in 127 contigs. The draft genome consisted of 4,372,178 nucleotides and was annotated by rapid annotations using subsystems technology (RAST) (12). RAST annotation showed that Acinetobacter calcoaceticus PHEA-2 (score, 503) and Acinetobacter sp. strain SH024 (score, 436) are the two closest neighbors of A. pittii CR12-42. Our isolate was related to only one other A. pittii strain, TG6411, but with a lower score of 221. A total of 13 A. pittii draft genomes have been described in the BioProject (http://www.ncbi.nlm.nih.gov/bioproject/); however, draft genomes of only three isolates were published, including one draft genome of an NDM-1-producing A. pittii strain from China (13).
In silico identification of CR12-42 to the species level by using rpoB and gyrB showed it to be 100% identical to A. pittii. A. pittii belongs, together with Acinetobacter nosocomialis, within the A. calcoaceticus-baumannii complex and was formerly named Acinetobacter genomic species 3 (14). In silico analysis of A. baumannii multilocus sequence typing (MLST) by the Pasteur scheme (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html) identified A. pittii CR12-42 as being of sequence type 119 (ST119). The alleles found were cpn-60 (n = 36), fusA (n = 20), gltA (n = 38), pyrG (n = 16), recA (n = 38), rplB (n = 18), and rpoB (n = 20). It has been reported that MLST by the Pasteur scheme is capable of providing the ST of A. pittii (15). The clinical significance of A. pittii ST119 is indicated by the fact that it has been reported to be the predominant clone among the A. pittii strains (18 out of 25) isolated in four hospitals in Japan (16). Interestingly, these Japanese A. pittii isolates possessed a different blaIMP variant, blaIMP-19 (16). Of note, A. pittii ST119 has not been reported previously in Australia.
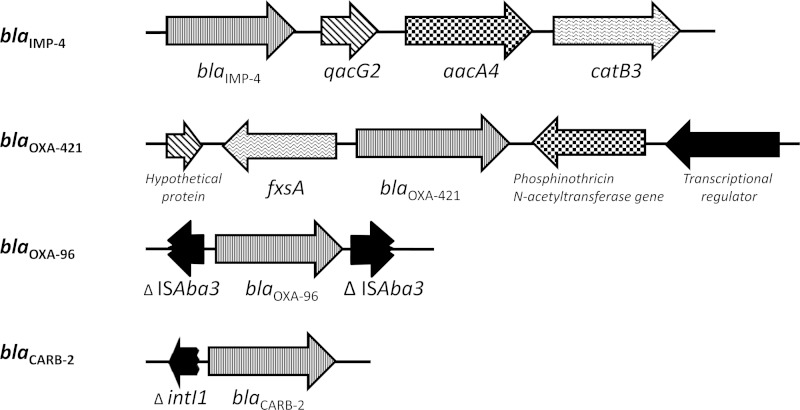
The resistance genes were screened with ResFinder (17). The β-lactamase-encoding genes blaIMP-4, blaOXA-96, and blaCARB-2 were identified. blaOXA-96 has a single nucleotide difference (a guanine-for-adenine substitution at position 483) from blaOXA-58. blaOXA-96 had been reported within an A. baumannii isolate from Singapore that also harbored blaOXA-23 and blaOXA-64 (18). In our isolate, blaOXA-96 had a genetic context similar to that of blaOXA-58, which was bracketed by ISAba3 (GenBank accession number JX968506) (Fig. 1).
FIG 1.
Genetic contexts of the four β-lactamase-encoding genes in A. pittii CR12-42.
In addition, a novel blaOXA gene, blaOXA-421, was identified (Fig. 1). This gene had a genetic environment identical to that of the chromosomal blaOXA-51 gene in A. baumannii (19), which includes two genes that are usually present upstream and downstream of blaOXA-51 in A. baumannii, the phosphinothricin N-acetyltransferase-encoding gene and fxsA, respectively. blaOXA-421 has 95% identity with the previously reported blaOXA gene (GenBank accession number CP002177, locus tag BDGL_000903) from the genome of A. calcoaceticus PHEA-2 (20), which is the closest neighbor of our CR12-42 isolate, as previously mentioned. The second closest relative of blaOXA-421 was blaOXA of Acinetobacter oleivorans, with 89% similarity (GenBank accession number CP002080, locus tag AOLE_1170) (21). The other blaOXA genes similar to blaOXA-421 were blaOXA-324, blaOXA-325, blaOXA-326, blaOXA-332, and blaOXA-354 (88 to 89% similarity), which were recently identified in A. calcoaceticus (22). The carbapenemase activity of OXA-421 warrants further investigation.
The blaIMP-4 gene in A. pittii CR12-42 was located inside a class 1 integron. Downstream from blaIMP-4 were qacG2 and the aminoglycoside and chloramphenicol resistance genes aacA4 and catB2 (Fig. 1). This genetic context of blaIMP-4 in CR12-42 was found to be identical to that in an IMP-4-producing A. baumannii strain from Singapore (GenBank accession number DQ532122) (18). blaIMP-4 has also been reported in Acinetobacter junii from Australia; however, the genetic context was not characterized (23). Our genetic context was also similar to that of blaIMP-4 in the IncHI2-type plasmid from Enterobacter cloacae and Escherichia coli and the IncL/M plasmid carrying blaIMP-4 in an E. cloacae strain from Australia (24, 25). However, the plasmid backbone of these sequences could not be identified within our draft genome. Further investigation is needed to determine if blaIMP-4 is located on a plasmid or the chromosome of CR12-42.
A carbenicillinase gene, blaCARB-2 was identified with ResFinder. blaCARB-2, which was also designated blaPSE-1, was first reported in Pseudomonas aeruginosa (26). The genetic context of blaCARB-2 in CR12-42 was also potentially a class 1 integron with a truncated integrase (intI1) located upstream of blaCARB-2 (Fig. 1). Other resistance genes found in this strain included sul1 (sulfonamide resistance), msr(E) and mph(E) (macrolide resistance), and aac-3-IId (aminoglycoside resistance). Consistent with this, the A. pittii strain was resistant to gentamicin and tobramycin but susceptible to amikacin. Of note, no 16S rRNA methylase was found in this isolate.
Regardless of its resistance to multiple antimicrobials, A. pittii CR12-42 remained susceptible to tetracycline and minocycline, which was consistent with the absence of a tetracycline resistance gene within the draft genome. In addition, the MIC of ampicillin-sulbactam remained low (2 mg/liter), despite the presence of multiple carbapenemase-encoding genes. Further, sulbactam is known to have activity against A. baumannii (27). In a study by Higgins et al., the ampicillin-sulbactam MIC50 of 115 A. baumannii strains was 2 mg/liter (27). Ampicillin-sulbactam susceptibility was also shown in the majority of the previously reported A. pittii ST119 strains harboring blaIMP-19 (94%) in Japan (16). In addition, 94% of these were susceptible to minocycline, similar to the antimicrobial phenotype of CR12-42 (16). Apart from the difference in blaIMP variants, CR12-42 has an antimicrobial phenotype and genotype identical to those of A. pittii ST119 from Japan.
IMP-producing Enterobacteriaceae strains have been frequently reported in Australia. Although OXA-23-like is the main subclass of carbapenemases identified in A. baumannii, IMP-4 is occasionally identified in A. pittii in locations such as Hong Kong and Singapore (18, 28). Other variants of blaIMP, such as blaIMP-1, blaIMP-8, blaIMP-11, and blaIMP-19, have been described in A. pittii in Southeast Asia (16, 29, 30). A. pittii has also recently been reported to produce NDM (31, 32).
Generally, A. baumannii is considered the most important and the most prevalent Acinetobacter species causing infections. However, A. pittii has caused hospital outbreaks in The Netherland and China (32, 33) and was reported as the most common Acinetobacter species causing nosocomial infections in Germany (34). Our study illustrates the emergence of a multidrug-resistant A. pittii strain in Australia. Therefore, accurate identification to the species level and characterization of the prevalence of A. pittii among the Acinetobacter species isolated in our region and its antibiotic resistance warrant further investigation.
This work was approved by the Royal Brisbane and Women's Hospital Human Research Ethics Committee (HREC/13/QRBW/391: epidemiology, clinical significance, treatment, and outcome of infections by carbapenem-resistant Enterobacteriaceae and Acinetobacter species in Queensland). This project is registered as BioProject PRJNA255268 and BioSample SAMN03003652.
Nucleotide sequence accession numbers.
The GenBank accession number of blaOXA-421 is KM401566. The GenBank accession number of the draft genome of A. pittii CR12-42 is JQNT00000000.
ACKNOWLEDGMENTS
We thank the microbiology staff at the Gold Coast Hospital microbiology laboratory for the study isolate.
We thank the Pathology Queensland—Study, Education, and Research Trust Fund (4177). W.K. has received a research high degree scholarship from Siriraj Hospital, Mahidol University, Bangkok, Thailand.
REFERENCES
- 1.EUCAST. 2013. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST, Basel, Switzerland: http://www.eucast.org/clinical_breakpoints/ Accessed 1 May. [Google Scholar]
- 2.Higgins PG, Lehmann M, Wisplinghoff H, Seifert H. 2010. gyrB multiplex PCR to differentiate between Acinetobacter calcoaceticus and Acinetobacter genomic species 3. J Clin Microbiol 48:4592–4594. doi: 10.1128/JCM.01765-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gundi VA, Dijkshoorn L, Burignat S, Raoult D, La Scola B. 2009. Validation of partial rpoB gene sequence analysis for the identification of clinically important and emerging Acinetobacter species. Microbiology 155:2333–2341. doi: 10.1099/mic.0.026054-0. [DOI] [PubMed] [Google Scholar]
- 4.Doi Y, Potoski BA, Adams-Haduch JM, Sidjabat HE, Pasculle AW, Paterson DL. 2008. Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type beta-lactamase by use of a boronic acid compound. J Clin Microbiol 46:4083–4086. doi: 10.1128/JCM.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picão RC, Andrade SS, Nicoletti AG, Campana EH, Moraes GC, Mendes RE, Gales AC. 2008. Metallo-beta-lactamase detection: comparative evaluation of double-disk synergy versus combined disk tests for IMP-, GIM-, SIM-, SPM-, or VIM-producing isolates. J Clin Microbiol 46:2028–2037. doi: 10.1128/JCM.00818-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins PG, Perez-Llarena FJ, Zander E, Fernandez A, Bou G, Seifert H. 2013. OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother 57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Runnegar N, Sidjabat H, Goh HM, Nimmo GR, Schembri MA, Paterson DL. 2010. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a single institution over a 10-year period. J Clin Microbiol 48:4051–4056. doi: 10.1128/JCM.01208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang HY, Lee HJ, Suh JT, Lee KM. 2009. Outbreaks of imipenem resistant Acinetobacter baumannii producing OXA-23 beta-lactamase in a tertiary care hospital in Korea. Yonsei Med J 50:764–770. doi: 10.3349/ymj.2009.50.6.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Sidjabat H, Nimmo GR, Walsh TR, Binotto E, Htin A, Hayashi Y, Li J, Nation RL, George N, Paterson DL. 2011. Carbapenem resistance in Klebsiella pneumoniae due to the New Delhi metallo-beta-lactamase. Clin Infect Dis 52:481–484. doi: 10.1093/cid/ciq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Cui Y, Pu F, Jiang G, Zhao X, Yuan Y, Zhao W, Li D, Liu H, Li Y, Liang T, Xu L, Wang Y, Song Q, Yang J, Liang L, Yang R, Han L, Song Y. 2012. Draft genome sequence of an Acinetobacter genomic species 3 strain harboring a bla(NDM-1) gene. J Bacteriol 194:204–205. doi: 10.1128/JB.06202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Chen T, Yu R, Lu X, Zong Z. 2013. Acinetobacter pittii and Acinetobacter nosocomialis among clinical isolates of the Acinetobacter calcoaceticus-baumannii complex in Sichuan, China. Diagn Microbiol Infect Dis 76:392–395. doi: 10.1016/j.diagmicrobio.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto M, Nagao M, Matsumura Y, Hotta G, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2013. Regional dissemination of Acinetobacter species harbouring metallo-beta-lactamase genes in Japan. Clin Microbiol Infect 19:729–736. doi: 10.1111/1469-0691.12013. [DOI] [PubMed] [Google Scholar]
- 17.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh TH, Sng LH, Wang GC, Hsu LY, Zhao Y. 2007. IMP-4 and OXA beta-lactamases in Acinetobacter baumannii from Singapore. J Antimicrob Chemother 59:627–632. doi: 10.1093/jac/dkl544. [DOI] [PubMed] [Google Scholar]
- 19.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. 2010. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob Agents Chemother 54:4575–4581. doi: 10.1128/AAC.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Peng Z, Zhan Y, Wang J, Yan Y, Chen M, Lu W, Ping S, Zhang W, Zhao Z, Li S, Takeo M, Lin M. 2011. Novel regulator MphX represses activation of phenol hydroxylase genes caused by a XylR/DmpR-type regulator MphR in Acinetobacter calcoaceticus. PLoS One 6:e17350. doi: 10.1371/journal.pone.0017350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung J, Madsen EL, Jeon CO, Park W. 2011. Comparative genomic analysis of Acinetobacter oleivorans DR1 to determine strain-specific genomic regions and gentisate biodegradation. Appl Environ Microbiol 77:7418–7424. doi: 10.1128/AEM.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamolvit W, Higgins PG, Paterson DL, Seifert H. 2014. Multiplex PCR to detect the genes encoding naturally occurring oxacillinases in Acinetobacter spp. J Antimicrob Chemother 69:959–963. doi: 10.1093/jac/dkt480. [DOI] [PubMed] [Google Scholar]
- 23.Peleg AY, Franklin C, Walters LJ, Bell JM, Spelman DW. 2006. OXA-58 and IMP-4 carbapenem-hydrolyzing beta-lactamases in an Acinetobacter junii blood culture isolate from Australia. Antimicrob Agents Chemother 50:399–400. doi: 10.1128/AAC.50.1.399-400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partridge SR, Ginn AN, Paulsen IT, Iredell JR. 2012. pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob Agents Chemother 56:6029–6032. doi: 10.1128/AAC.01189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidjabat HE, Heney C, George NM, Nimmo GR, Paterson DL. 2014. Interspecies transfer of blaIMP-4 in a patient with prolonged colonization by IMP-4-producing Enterobacteriaceae. J Clin Microbiol 52:3816–3818. doi: 10.1128/JCM.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huovinen P, Jacoby GA. 1991. Sequence of the PSE-1 beta-lactamase gene. Antimicrob Agents Chemother 35:2428–2430. doi: 10.1128/AAC.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins PG, Wisplinghoff H, Stefanik D, Seifert H. 2004. In vitro activities of the beta-lactamase inhibitors clavulanic acid, sulbactam, and tazobactam alone or in combination with beta-lactams against epidemiologically characterized multidrug-resistant Acinetobacter baumannii strains. Antimicrob Agents Chemother 48:1586–1592. doi: 10.1128/AAC.48.5.1586-1592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu YW, Afzal-Shah M, Houang ET, Palepou MI, Lyon DJ, Woodford N, Livermore DM. 2001. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother 45:710–714. doi: 10.1128/AAC.45.3.710-714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang LY, Lu PL, Chen TL, Chang FY, Fung CP, Siu LK. 2010. Molecular characterization of beta-lactamase genes and their genetic structures in Acinetobacter genospecies 3 isolates in Taiwan. Antimicrob Agents Chemother 54:2699–2703. doi: 10.1128/AAC.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim CK, Lee Y, Lee H, Woo GJ, Song W, Kim MN, Lee WG, Jeong SH, Lee K, Chong Y. 2010. Prevalence and diversity of carbapenemases among imipenem-nonsusceptible Acinetobacter isolates in Korea: emergence of a novel OXA-182. Diagn Microbiol Infect Dis 68:432–438. doi: 10.1016/j.diagmicrobio.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 31.Roca I, Mosqueda N, Altun B, Espinal P, Akova M, Vila J. 2014. Molecular characterization of NDM-1-producing Acinetobacter pittii isolated from Turkey in 2006. J Antimicrob Chemother 69:3437–3438. doi: 10.1093/jac/dku306. [DOI] [PubMed] [Google Scholar]
- 32.Yang J, Chen Y, Jia X, Luo Y, Song Q, Zhao W, Wang Y, Liu H, Zheng D, Xia Y, Yu R, Han X, Jiang G, Zhou Y, Zhou W, Hu X, Liang L, Han L. 2012. Dissemination and characterization of NDM-1-producing Acinetobacter pittii in an intensive care unit in China. Clin Microbiol Infect 18:E506–E513. doi: 10.1111/1469-0691.12035. [DOI] [PubMed] [Google Scholar]
- 33.Idzenga D, Schouten MA, van Zanten AR. 2006. Outbreak of Acinetobacter genomic species 3 in a Dutch intensive care unit. J Hosp Infect 63:485–487. doi: 10.1016/j.jhin.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Schleicher X, Higgins PG, Wisplinghoff H, Korber-Irrgang B, Kresken M, Seifert H. 2013. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005–2009). Clin Microbiol Infect 19:737–742. doi: 10.1111/1469-0691.12026. [DOI] [PubMed] [Google Scholar]