Abstract
Bacterial grain rot (BGR), caused by the bacterial pathogen Burkholderia glumae, is a destructive disease of rice. At anthesis, rice panicles are attacked by the pathogen, and the infection causes unfilled or aborted grains, reducing grain yield and quality. Thus, increasing the level of BGR resistance is an important objective for rice breeding. A quantitative trait locus (QTL) on rice chromosome 1 that controls BGR resistance was previously detected in backcross inbred lines (BILs) derived from a cross between Kele, a resistant traditional lowland cultivar (indica) that originated in India, and Hitomebore, a susceptible modern lowland cultivar (temperate japonica) from Japan. Further genetic analyses using a BC3F6 population derived from a cross between a resistant BIL (BC2F5) and Hitomebore confirmed that a QTL for BGR resistance was located on the long arm of chromosome 1. To define more precisely the chromosomal region underlying this QTL, we identified nine BC2F6 plants in which recombination occurred near the QTL. Substitution mapping using homozygous recombinant and nonrecombinant plants demonstrated that the QTL, here designated as Resistance to Burkholderia glumae 2 (RBG2), was located in a 502-kb interval defined by simple sequence repeat markers RM1216 and RM11727.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-015-0192-x) contains supplementary material, which is available to authorized users.
Keywords: Oryza sativa L., Disease resistance, Bacterial grain rot, QTL, Linkage analysis, Panicle blight
Burkholderia glumae causes bacterial grain rot (BGR) and seedling rot in rice (Oryza sativa L.), both of which are highly destructive to rice production (Ham et al. 2011). Until now, there have been two reports of quantitative trait loci (QTLs) for BGR resistance (Mizobuchi et al. 2013a; Pinson et al. 2010). As described in Mizobuchi et al. (2013a), we detected a QTL for BGR resistance on the long arm of chromosome 1 by using backcross inbred lines (BILs) derived from a cross between the traditional lowland indica cultivar Kele (JP13287) and the modern lowland temperate japonica cultivar Hitomebore. The Kele allele at the QTL decreased the ratio of diseased spikelets (RDS).
To validate the effect of the Kele allele at this QTL, we used a resistant BIL (BC2F5) line (HK19; Fig. 1). Most of the chromosome regions of HK19 originated from the susceptible cultivar Hitomebore, but HK19 also contains a large segment of chromosome 1 and small segments of chromosomes 2, 5, 8, 10, 11, and 12 derived from Kele. Twenty-nine F2 plants (BC3F6) were produced by crossing Hitomebore with HK19. Plants were grown in paddy fields in the summer of 2013 at the National Institute of Agrobiological Sciences (NIAS) in Tsukuba, Japan. Thirty-day-old seedlings were transplanted at a density of one seedling per hill and planted in a single row of 10 hills at a spacing of 18 cm between hills and 30 cm between rows. Basal fertilizer was applied at a rate of 56 kg N, 56 kg P, and 56 kg K ha−1. Days from sowing to heading for Kele and Hitomebore, which were transplanted on May 15, were 89 and 94 days, respectively. In contrast, days to heading of the F2 plants ranged from 99 to 114 days. Therefore, the F2 plants were categorized by heading date and inoculated on different dates (from July 26 to August 7). The Kele and Hitomebore controls were seeded and transplanted on several dates after the F2 seeding and transplanting dates to better match the heading dates of the F2 plants. We measured resistance to BGR by the modified cut-panicle inoculation method in which panicles containing only spikelets at 1 day after anthesis were harvested and inoculated as previously described (Mizobuchi et al. 2013a). Inoculation and measurement were conducted as previously described (Mizobuchi et al. 2013a). Simple sequence repeat (SSR) markers in the target chromosome regions were screened to identify those detecting polymorphism between Hitomebore and HK19 (IRGSP 2005). The F2 plants were then genotyped with 28 SSR markers (Supplemental Table 1). PCR analysis was performed as previously described (Mizobuchi et al. 2013b). Linkage mapping was performed using version 3.0 of MAPMAKER/EXP software (Lander et al. 1987), and the Kosambi map function was used to calculate genetic distances.
Fig. 1.
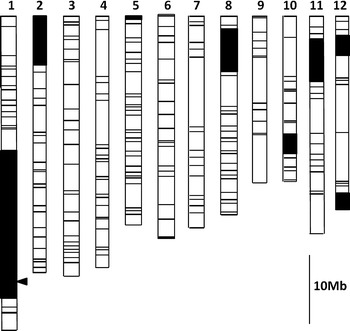
Graphical genotype of a resistant BC2F5 line (HK19) used for fine mapping of QTLs. Chromosome numbers are indicated above each linkage map. Positions of marker loci used for genotyping are shown as horizontal lines and were obtained from the linkage map of BILs derived from a cross between Kele and Hitomebore (Mizobuchi et al. 2013a). The arrowhead next to the long arm of chromosome 1 shows the putative position of the QTL for resistance to bacterial grain rot (BGR) (Mizobuchi et al. 2013a) examined in the present study. White boxes indicate regions homozygous for Hitomebore marker alleles; black boxes indicate regions homozygous for Kele marker alleles
We performed QTL analyses by using composite interval mapping, as implemented by the Zmapqtl program (model 6) provided in version 2.5 of the QTL Cartographer software (Wang et al. 2005). By QTL analysis, we detected one QTL between RM11725 and RM11727 on the long arm of chromosome 1 (Fig. 2a). The QTL accounted for 35.4 % of the total phenotypic variance in the F2 plants, and the Kele allele decreased RDS by 10.4 %. The F2 plants derived from the cross of Hitomebore and HK19 showed a wide range of variation (20.6–84.7 %) in RDS (Fig. 2b). The correlation between heading date and RDS was not significant (R 2 = 0.0562). On the basis of the genotype at RM11727, the SSR marker nearest to LOD peak, F2 plants were classified into three genotypic classes; homozygous for the Kele allele, homozygous for the Hitomebore allele, or heterozygous (Fig. 2b). F2 plants homozygous for the Kele allele (n = 8) showed a low mean RDS (34.8 %), ranging from 20.8 to 66.8 %. Heterozygous plants (n = 9) also had a low mean RDS (32.8 %), ranging from 20.6 to 46.4 %. In contrast, the mean RDS was 55.7 %, ranging from 27.9 to 84.7 %, in plants homozygous for the Hitomebore allele (n = 12). Thus, plants homozygous for the Kele allele tended to show lower RDS values than those homozygous for the Hitomebore allele. These results clearly support the existence of the previously detected QTL on the long arm of chromosome 1 and show that the Kele allele at the QTL decreases the RDS.
Fig. 2.
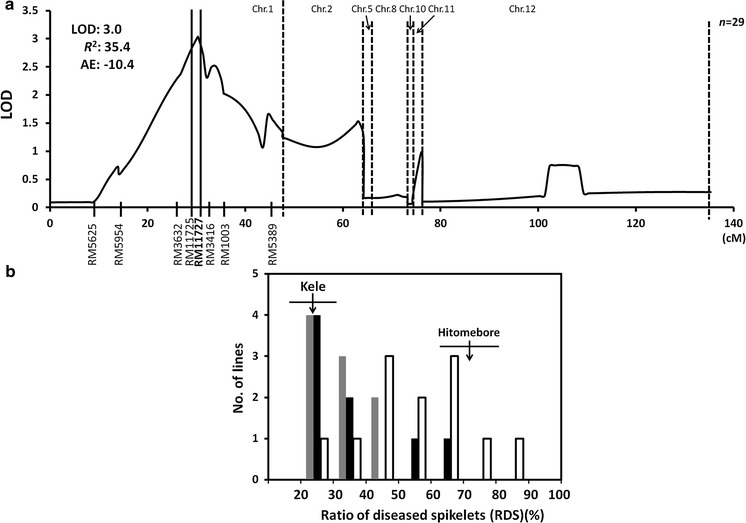
Chromosomal location of a QTL for resistance to bacterial grain rot (BGR) on the long arm of chromosome 1 and effects of allelic differences at linked marker RM11727. a The log-likelihood curve indicates a putative QTL position on chromosome 1 in an F2 population derived from Hitomebore × HK19 (a resistant BC2F5 line). We used genome-wide threshold values (α = 0.05) to detect putative QTLs on the basis of the results of 1,000 permutations. LOD logarithm of odds, R 2 percentage of phenotypic variance explained, and AE additive effect of the allele from Kele relative to that from Hitomebore. b Frequency distribution of the ratio of diseased spikelets (RDS) in F2 plants showing the three genotype classes of SSR marker RM11727, which was found to be nearest to LOD peak. The x-axis labels indicate the maximum RDS in each bin. Genotypes at RM11727 are represented as white bars (homozygous for Hitomebore allele), gray bars (heterozygous), and black bars (homozygous for Kele allele). The RDS values of the F2 plants were scored 5 days after inoculation. Arrows indicate the mean values for Kele and Hitomebore; horizontal lines across the arrows indicate the standard deviations
To further delimit the candidate genomic region of the QTL for BGR resistance, we used a BIL (BC2F5) line (HK114) in which the region of interest on the long arm of chromosome 1 was heterozygous (Supplemental Fig. 1). We identified nine recombinants (BC2F6) from the BIL line and then selected homozygous recombinant and nonrecombinant plants from the BC2F7 progeny of each one. Thus, we evaluated nine pairs of lines in the inoculation test. Significant difference about RDS was detected among seven pairs (BC2F7-12W-1, -2, -3, -5, -6, -7, and -8), whereas two pairs (BC2F7-12W-4 and -9) had high RDS values that were not significantly different between those of the recombinant and nonrecombinant lines (Fig. 3). Together, the genotype and phenotype information clearly delimit the QTL for BGR resistance between SSR markers RM1216 and RM11727 (a 502-kb interval in the Nipponbare genome reference sequence) on chromosome 1 (Fig. 3). Because we have already identified and named RBG1 (Resistance to Burkholderia glumae 1; formerly named qRBS1), a QTL on chromosome 10 involved in resistance to bacterial seedling rot (Mizobuchi et al. 2013b), we have designated this QTL for BGR resistance as Resistance to Burkholderia glumae 2 (RBG2), following the nomenclature recommended by McCouch and CGSNL (Committee on Gene Symbolization 2008).
Fig. 3.
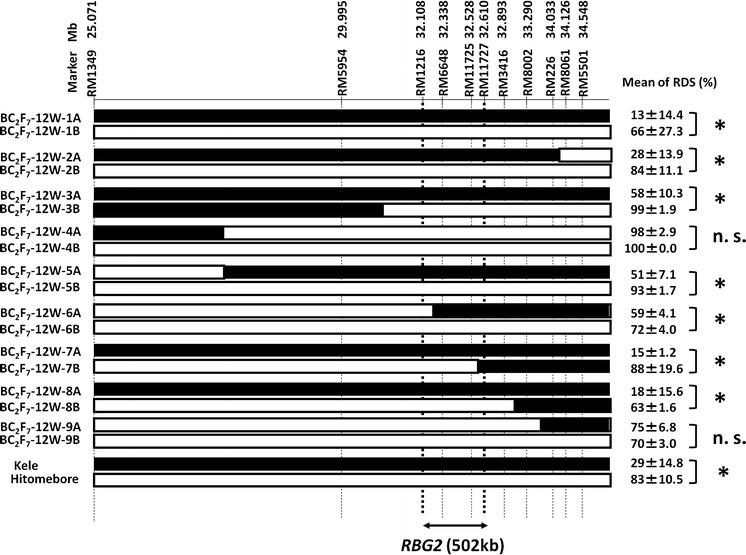
Substitution mapping of a QTL controlling resistance to bacterial grain rot (BGR) on the long arm of chromosome 1 in recombinant BC2F7 lines. Each pair of lines (e.g., 1A and 1B) was identified from the progeny of a recombinant BC2F6 plant. Black bars indicate chromosome regions derived from Kele (resistant); white bars indicate chromosome regions derived from Hitomebore (susceptible). Positions are based on the International Rice Genome Sequencing Project (IRGSP) 1.0 pseudomolecules of the Nipponbare genome. The location of the candidate QTL (RBG2), indicated at the bottom, is based on the phenotypic data obtained in an inoculation test, tabulated on the right. The ratio of diseased spikelets (RDS) scores of the two lines in each pair was compared by using Student’s test. *P < 0.05; ns not significant, P > 0.05
We surveyed the candidate genomic region of RBG2 using the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/ (Ohyanagi et al. 2006)) to nominate candidate genes. Among the predicted genes, there are none known to be related to disease resistance such as nucleotide-binding-site–leucine-rich repeat (NBS-LRR) genes. It is hard to predict which of the genes might be related to BGR resistance because there have been no reports of genes associated with BGR resistance and because the morphological and physiological functions of RBG2 are not yet known. Thus, further delimitation of the candidate genomic region of RBG2 will be necessary to identify the gene corresponding to RBG2.
Since B. glumae was first discovered in Japan (Goto and Ohata 1956; Goto et al. 1987; Kurita and Tabei 1967; Uematsu et al. 1976), it has also been reported in other countries in East Asia (Chien and Chang 1987; Cottyn et al. 1996a, b; Jeong et al. 2003; Luo et al. 2007; Trung et al. 1993) and Latin America (Nandakumar et al. 2007b; Zeigler and Alvarez 1989). Although several cultivars show partial resistance to BGR (Goto and Watanabe 1975; Groth et al. 2007; Imbe et al. 1986; Mogi and Tsushima 1985; Nandakumar et al. 2007a; Nandakumar and Rush 2008; Pinson et al. 2010; Prabhu and Bedendo 1988; Sayler et al. 2006; Sha et al. 2006; Takita et al. 1988; Wasano and Okuda 1994; Yasunaga et al. 2002), only one report of QTL analysis of BGR resistance has been published other than our previous report (Mizobuchi et al. 2013a; Pinson et al. 2010). This may be because the level of resistance is highly influenced by environmental conditions, making genetic analysis of BGR resistance very difficult (Tsushima 1996; Tsushima et al. 1985). Pinson et al. (2010) found a major QTL on chromosome 3 for BGR resistance colocated with a QTL for heading date. Because late-flowering panicles are subjected to cooler temperatures that are less conductive to disease development during grain fill, it is possible that the genetic effects of the heading-related QTLs affected the disease scoring. On the other hand, by selecting parental cultivars with similar heading dates and using a method (cut-panicle inoculation) that minimizes the effect of heading date variation, we successfully detected a major QTL for BGR resistance on chromosome 1, and no QTL for heading date was detected near this BGR resistance QTL (Mizobuchi et al. 2013a). In the QTL analysis of this study, the correlation between heading date and RDS was not significant (R 2 = 0.0562). The nine pairs of BC2F7 lines used for substitution mapping had similar heading dates between homozygous recombinant and nonrecombinant plants. Therefore, we suppose that the disease resistance derived from RBG2 is not a pleiotropic effect of the QTL for heading date. To enhance our understanding of the genetic control of BGR resistance, we undertook fine mapping of the QTL and successfully defined a candidate genomic region for the QTL, RBG2. The RBG2 map information obtained in this study opens the way not only for the use of RBG2 in breeding programs, but also for gene isolation that will enable us to elucidate the genetic mechanism of BGR resistance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Table 1 Primer pairs used in this study (XLS 32 kb)
Supplemental Fig. 1 Graphical genotype of a BC2F5 line (HK114) used for substitution mapping of RBG2. Chromosome numbers are indicated above each linkage map. Positions of marker loci used for genotyping are shown as horizontal lines and were obtained from the linkage map of BILs derived from a cross between Kele and Hitomebore (Mizobuchi et al. 2013a). The arrowhead shows RM11727, the nearest marker detected by QTL analysis of an F2 population derived from Hitomebore × HK19 (described in Fig. 2). White boxes indicate regions homozygous for Hitomebore marker alleles, black boxes indicate regions homozygous for Kele marker alleles, and the gray box indicates a heterozygous region. (TIFF 76 kb)
Acknowledgments
We thank Dr. T. Imbe (NARO) for valuable advice. We also thank the staff of the technical support section of the NIAS for their field management and experimental support. This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Project for Climate Change, Rice-2006 and Rice-3003) and the NIAS Strategic Research Fund.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Chien CC, Chang YC. The susceptibility of rice plants at different growth stages and 21 commercial rice varieties to Pseudomonas glumae. J Agric Res China. 1987;36:302–310. [Google Scholar]
- Cottyn B, Cerez MT, VanOutryve MF, Barroga J, Swings J, Mew TW. Bacterial diseases of rice. 1. Pathogenic bacteria associated with sheath rot complex and grain discoloration of rice in the Philippines. Plant Dis. 1996;80:429–437. doi: 10.1094/PD-80-0429. [DOI] [Google Scholar]
- Cottyn B, VanOutryve MF, Cerez MT, DeCleene M, Swings J, Mew TW. Bacterial diseases of rice. 2. Characterization of pathogenic bacteria associated with sheath rot complex and grain discoloration of rice in the Philippines. Plant Dis. 1996;80:438–445. doi: 10.1094/PD-80-0438. [DOI] [Google Scholar]
- Goto K, Ohata K. New bacterial diseases of rice (brown stripe and grain rot) Ann Phytopathol Soc Jpn. 1956;21:46–47. [Google Scholar]
- Goto T, Watanabe B. Varietal resistance to bacterial grain rot of rice, caused by Pseudomonas glumae. Proc Assoc Plant Prot Kyushu. 1975;21:141–143. doi: 10.4241/kyubyochu.21.141. [DOI] [Google Scholar]
- Goto T, Nishiyama K, Ohata K. Bacteria causing grain rot of rice. Ann Phytopathol Soc Jpn. 1987;53:141–149. doi: 10.3186/jjphytopath.53.141. [DOI] [Google Scholar]
- Groth DE, Linscombe SD, Sha X. Registration of two disease-resistant germplasm lines of rice. J Plant Regist. 2007;1:63–64. doi: 10.3198/jpr2006.10.0677crg. [DOI] [Google Scholar]
- Ham J, et al. Molecular genetic and genomic studies on bacterial panicle blight of rice and its causative agent Burkholderia glumae. Phytopathology. 2011;101:S266–S266. [Google Scholar]
- Imbe T, Tsushima S, Nishiyama H. Varietal resistance of rice to bacterial grain rot and screening method. Proc Assoc Plant Prot Kyushu. 1986;32:17–19. doi: 10.4241/kyubyochu.32.17. [DOI] [Google Scholar]
- IRGSP The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T, Hwang I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003;87:890–895. doi: 10.1094/PDIS.2003.87.8.890. [DOI] [PubMed] [Google Scholar]
- Kurita T, Tabei H. On the pathogenic bacterium of bacterial grain rot of rice. Ann Phytopathol Soc Jpn. 1967;33:111. [Google Scholar]
- Lander E, Green P, Abrahamson J, Barlow A, Daley M, Lincoln S, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Luo J, Xie G, Li B, Lihui X. First report of Burkholderia glumae isolated from symptomless rice seeds in China. Plant Dis. 2007;91:1363. doi: 10.1094/PDIS-91-10-1363B. [DOI] [PubMed] [Google Scholar]
- McCouch S, CGSNL (Committee on Gene Symbolization. Nomenclature and Linkage, Rice Genetics Cooperative) Gene nomenclature system for rice. Rice. 2008;1:72–84. doi: 10.1007/s12284-008-9004-9. [DOI] [Google Scholar]
- Mizobuchi R, Sato H, Fukuoka S, Tanabata T, Tsushima S, Imbe T, Yano M. Mapping a quantitative trait locus for resistance to bacterial grain rot in rice. Rice. 2013;6:13. doi: 10.1186/1939-8433-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizobuchi R, Sato H, Fukuoka S, Tsushima S, Imbe T, Yano M. Identification of qRBS1, a QTL involved in resistance to bacterial seedling rot in rice. Theor Appl Genet. 2013;126:2417–2425. doi: 10.1007/s00122-013-2145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi S, Tsushima S. Varietal resistance to bacterial grain rot in rice, caused by Pseudomonas glumae. Kyushu Agric Res. 1985;47:103. [Google Scholar]
- Nandakumar R, Rush MC. Analysis of gene expression in Jupiter rice showing partial resistance to rice panicle blight caused by Burkholderia glumae. Phytopathology. 2008;98:S112. [Google Scholar]
- Nandakumar R, Bollich P, Groth D, Rush MC. Confirmation of the partial resistance of Jupiter rice to bacterial panicle blight caused by Burkholderia glumae through reduced disease and yield loss in inoculated field tests. Phytopathology. 2007;97:S82–S83. [Google Scholar]
- Nandakumar R, Rush MC, Correa F. Association of Burkholderia glumae and B. gladioli with panicle blight symptoms on rice in Panama. Plant Dis. 2007;91:767. doi: 10.1094/PDIS-91-6-0767C. [DOI] [PubMed] [Google Scholar]
- Ohyanagi H, et al. The rice annotation project database (RAP-DB): hub for Oryza sativa ssp japonica genome information. Nucl Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson SRM, Shahjahan AKM, Rush MC, Groth DE. Bacterial panicle blight resistance QTLs in rice and their association with other disease resistance loci and heading date. Crop Sci. 2010;50:1287–1297. doi: 10.2135/cropsci2008.07.0447. [DOI] [Google Scholar]
- Prabhu AS, Bedendo IP. Glume blight of rice in Brazil: etiology, varietal reaction and loss estimates. Trop Pest Manag. 1988;34:85–88. doi: 10.1080/09670878809371215. [DOI] [Google Scholar]
- Sayler RJ, Cartwright RD, Yang YN. Genetic characterization and real-time PCR detection of Burkholderia glumae, a newly emerging bacterial pathogen of rice in the United States. Plant Dis. 2006;90:603–610. doi: 10.1094/PD-90-0603. [DOI] [PubMed] [Google Scholar]
- Sha X, et al. Registration of ‘Jupiter’ rice. Crop Sci. 2006;46:1811–1812. doi: 10.2135/cropsci2005.10-0393. [DOI] [Google Scholar]
- Takita T, Imbe T, Nishiyama H, Tsushima S. Resistance to rice bacterial grain rot in indica and upland rice. Kyushu Agric Res. 1988;50:28. [Google Scholar]
- Trung HM, Van NV, Vien NV, Lam DT, Lien M. Occurrence of rice grain rot disease in Vietnam. Int Rice Res Notes. 1993;18:30. [Google Scholar]
- Tsushima S. Epidemiology of bacterial grain rot of rice caused by Pseudomonas glumae. JARQ. 1996;30:85–89. [Google Scholar]
- Tsushima S, Mogi S, Saito H. Effects of inoculum density, incubation temperature and incubation period on the development of rice bacterial grain rot. Proc Assoc Plant Prot Kyushu. 1985;31:11–12. doi: 10.4241/kyubyochu.31.11. [DOI] [Google Scholar]
- Uematsu T, Yoshimura D, Nishiyama K, Ibaraki T, Fujii H. Occurrence of bacterial seedling rot in nursery flat, caused by grain rot bacterium Pseudomonas glumae. Ann Phytopathol Soc Jpn. 1976;42:310–312. doi: 10.3186/jjphytopath.42.310. [DOI] [Google Scholar]
- Wang S, Basten C, Zeng Z (2005) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
- Wasano K, Okuda S. Evaluation of resistance of rice cultivars to bacterial grain rot by the syringe inoculation method. Breed Sci. 1994;44:1–6. [Google Scholar]
- Yasunaga T, Wada T, Oosata KF, Hamachi Y. Varietal differences in occurrence of bacterial grain rot in rice cultivars with high palatability. Crop Sci Soc Jpn. 2002;68:12–14. [Google Scholar]
- Zeigler RS, Alvarez E. Grain discoloration of rice caused by Pseudomonas glumae in Latin America. Plant Dis. 1989;73:368. doi: 10.1094/PD-73-0368B. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1 Primer pairs used in this study (XLS 32 kb)
Supplemental Fig. 1 Graphical genotype of a BC2F5 line (HK114) used for substitution mapping of RBG2. Chromosome numbers are indicated above each linkage map. Positions of marker loci used for genotyping are shown as horizontal lines and were obtained from the linkage map of BILs derived from a cross between Kele and Hitomebore (Mizobuchi et al. 2013a). The arrowhead shows RM11727, the nearest marker detected by QTL analysis of an F2 population derived from Hitomebore × HK19 (described in Fig. 2). White boxes indicate regions homozygous for Hitomebore marker alleles, black boxes indicate regions homozygous for Kele marker alleles, and the gray box indicates a heterozygous region. (TIFF 76 kb)


