pH-dependent receptor binding of specific cargo by the Erv41–Erv46 complex in Golgi compartments identifies escaped ER resident proteins that are then returned to the ER in COPI vesicles.
Abstract
Signal-dependent sorting of proteins in the early secretory pathway is required for dynamic retention of endoplasmic reticulum (ER) and Golgi components. In this study, we identify the Erv41–Erv46 complex as a new retrograde receptor for retrieval of non–HDEL-bearing ER resident proteins. In cells lacking Erv41–Erv46 function, the ER enzyme glucosidase I (Gls1) was mislocalized and degraded in the vacuole. Biochemical experiments demonstrated that the luminal domain of Gls1 bound to the Erv41–Erv46 complex in a pH-dependent manner. Moreover, in vivo disturbance of the pH gradient across membranes by bafilomycin A1 treatment caused Gls1 mislocalization. Whole cell proteomic analyses of deletion strains using stable isotope labeling by amino acids in culture identified other ER resident proteins that depended on the Erv41–Erv46 complex for efficient localization. Our results support a model in which pH-dependent receptor binding of specific cargo by the Erv41–Erv46 complex in Golgi compartments identifies escaped ER resident proteins for retrieval to the ER in coat protein complex I–formed transport carriers.
Introduction
Newly synthesized secretory proteins are folded and modified in the ER before transport to Golgi compartments in a coat protein complex II (COPII)–dependent pathway, whereas transport machinery and escaped ER resident proteins are retrieved from Golgi compartments back to the ER through a coat protein complex I (COPI)–dependent retrograde pathway (Brandizzi and Barlowe, 2013). Transported cargo proteins can be selectively incorporated into COPI- and COPII-coated carrier vesicles through direct and indirect binding to subunits of these coat complexes (Cosson and Letourneur, 1994; Kuehn et al., 1998) or can traffic in a passive bulk-flow manner (Thor et al., 2009). For selective incorporation of transmembrane proteins, cytoplasmically exposed sorting signals have been identified that bind to defined recognition sites in COPI and COPII subunits (Mossessova et al., 2003; Jackson et al., 2012). However, not all transmembrane proteins that traffic through the early secretory pathway display known COPI or COPII sorting signals, and a large number of soluble cargos cannot be directly recognized by coat subunits because they are luminally disposed. To efficiently transport these proteins, a diverse family of sorting receptors is required to link specific cargo to COPI and COPII coat subunits (Dancourt and Barlowe, 2010). For example, ERGIC53 and Erv14 link soluble and transmembrane secretory cargo to COPII adaptor subunits for forward transport (Appenzeller et al., 1999; Powers and Barlowe, 2002), whereas the KDEL receptor and Rer1 bind soluble and transmembrane cargo to the COPI coat for retrograde transport from Golgi compartments (Lewis and Pelham, 1990; Semenza et al., 1990; Sato et al., 1997). Understanding how coat complexes and cargo receptors manage the broad spectrum of proteins that must be sorted in the early secretory pathway and how cargo binding is regulated remain challenging questions.
Proteomic analyses of purified COPII vesicles identified the heteromeric Erv41–Erv46 complex as efficiently packaged ER vesicle proteins that localized to ER and Golgi membranes (Otte et al., 2001). Erv41 and Erv46 are related integral membrane proteins that each contains two transmembrane segments, short cytosolic N- and C-terminal regions, and large luminal domains. Expression of Erv41 and Erv46 are interdependent such that the level of Erv46 was reduced in an erv41Δ strain and Erv41 was not detected in an erv46Δ strain. Both proteins contain COPII sorting motifs on their C termini, and Erv46 contains a conserved COPI binding dilysine motif on its C terminus, which cycles the Erv41–Erv46 complex between the ER and Golgi complex (Otte and Barlowe, 2002). In mammalian cells, the Erv41–Erv46 complex is distributed between the ER, ER–Golgi intermediate compartment, and cis-Golgi compartments (Orci et al., 2003; Breuza et al., 2004). Although it has been shown that yeast strains lacking the Erv41–Erv46 complex are viable and display cold sensitivity, the precise biological function of the Erv41–Erv46 complex is unknown. A study following in vitro translocation and transport of glyco-pro–α factor in yeast revealed that ER microsomes from an erv41Δ strain displayed a mild defect in glucose trimming of the attached N-linked core oligosaccharide and produced a similarly sized product as observed in gls1Δ microsomes (Welsh et al., 2006). Glucosidase I (Gls1) cleaves the terminal α-1,2–linked glucose from the newly attached N-linked core glycan and is thought to function in folding and quality control of nascent glycoproteins (Moremen et al., 1994; Hitt and Wolf, 2004). In this study, we investigated the localization of the Gls1 protein in erv41Δ strains and observed reduced ER levels coincident with mislocalization to the vacuole. Whole cell stable isotope labeling by amino acids in culture (SILAC) proteomics identified other ER proteins that depended on the Erv41–Erv46 complex for wild-type (WT) expression levels. In combination with biochemical experiments, our findings support a model in which the Erv41–Erv46 complex functions as a retrograde receptor for a new class of ER resident proteins.
Results
Deletion of ERV41 mislocalizes Gls1
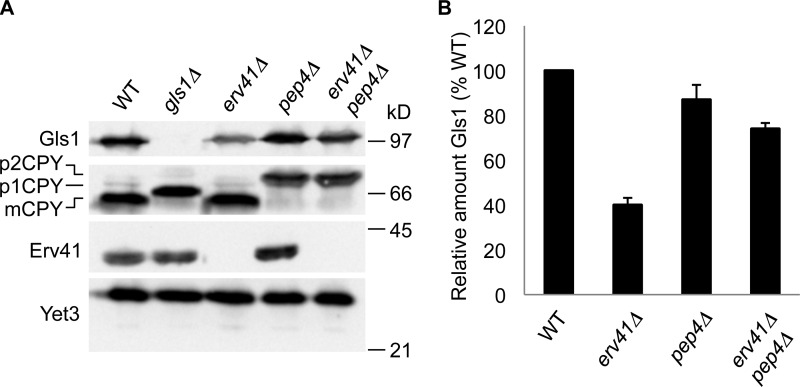
A previous study indicated a deficiency in Gls1 trimming of the N-linked core oligosaccharide when the Erv41–Erv46 complex was depleted from cells (Welsh et al., 2006). To further investigate Erv41–Erv46 function in these cells, we prepared polyclonal antibodies specific for the Gls1 protein. Gls1 is predicted to have a short cytosolic N terminus (residues 1–10), a hydrophobic transmembrane domain (residues 11–28), and a large C-terminal domain (residues 29–833) in the lumen of the ER. We purified the luminal domain of Gls1 fused to the C terminus of GST (GST-Gls1) for production of polyclonal serum against Gls1, and specificity was confirmed by immunoblotting for Gls1 in WT and gls1Δ strains (Fig. 1 A). Using anti-Gls1 antiserum, we found that deletion of ERV41 reduced the steady-state cellular levels of Gls1 by ∼60% compared with the WT strain (Fig. 1, A and B). Moreover, when pep4Δ was combined with erv41Δ, the cellular levels of Gls1 were restored to near WT levels (Fig. 1, A and B). Because PEP4 encodes proteinase A, a vacuolar aspartyl protease required for processing of vacuolar precursors, PEP4 deletion leads to the inhibition of vacuolar protease activity (Ammerer et al., 1986). Carboxypeptidase Y (CPY) is a well-characterized yeast vacuolar enzyme, which is synthesized and glycosylated at the ER as the proenzyme p1CPY form (67 kD), transported to the Golgi for further glycosylation to the p2CPY form (69 kD), and then proteolytically processed at the vacuole in a Pep4-dependent manner to produce the mature CPY (mCPY) form (61 kD; Hemmings et al., 1981; Stevens et al., 1982). We used CPY maturation as an indicator for pep4Δ and note that gls1Δ shifted mCPY to a larger size because of the presence of untrimmed glucose residues on the mCPY N-linked carbohydrate. Because Erv41 and Erv46 form a heteromeric complex and their expression levels are interdependent (Otte et al., 2001), erv41Δ and erv46Δ displayed similar effects on the steady-state cellular levels of Gls1 (Fig. S1), indicating that both Erv41 and Erv46 are required to maintain cellular levels of Gls1.
Figure 1.
Deletion of ERV41 reduces the cellular levels of Gls1 caused by vacuolar degradation. (A) Cells grown to mid–log phase were lysed, resolved on a 10.5% polyacrylamide gel, and immunoblotted for Gls1, CPY, Erv41, and Yet3 (loading control). WT (CBY740), gls1Δ (CBY1086), erv41Δ (CBY1168), pep4Δ (CBY2732), and erv41Δ pep4Δ (CBY3306) strains were compared. (B) Relative amounts of Gls1 with standard error bars (n = 3). Gls1 levels were normalized with Yet3 as the loading control and plotted as a percentage relative to WT.
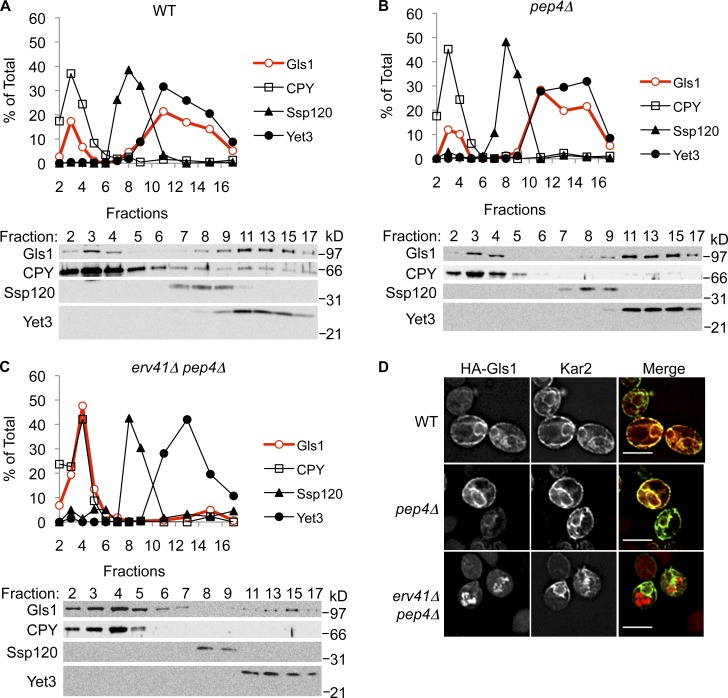
Stabilization of Gls1 in erv41Δ pep4Δ cells suggested that Gls1 was transported to vacuoles in the absence of the Erv41–Erv46 complex. To test for mislocalization of Gls1 to vacuoles in erv41Δ cells, we examined the distribution of Gls1 by subcellular fractionation of membranes on sucrose density gradients (Fig. 2, A–C). In WT and pep4Δ cells, most Gls1 cosedimented with the ER marker Yet3 and a smaller fraction with the vacuole marker CPY (Fig. 2, A and B). In contrast, almost all Gls1 was colocalized with vacuolar membranes in pep4Δ erv41Δ cells (Fig. 2 C), suggesting that the absence of Erv41 caused mislocalization of Gls1 to vacuoles. We observed that little Gls1 co-migrated with the Golgi marker Ssp120 (Huh et al., 2003) in both WT and erv41Δ cells. In addition, we performed immunofluorescence microscopy to examine the localization of Gls1 (Fig. 2 D and Fig. S2). 3HA-tagged Gls1 (HA-Gls1) colocalized with the ER marker Kar2 in WT and pep4Δ strains, whereas HA-Gls1 colocalized with the vacuolar marker alkaline phosphatase (ALP) in the pep4Δ erv41Δ strain. From these results, we concluded that the Erv41–Erv46 complex was required for Gls1 localization to the ER and that Gls1 trimming deficiencies observed in erv41Δ membranes (Welsh et al., 2006) were caused by sharp reductions in ER levels of Gls1.
Figure 2.
Absence of Erv41 mislocalizes Gls1 to vacuoles. (A–C) Lysates from WT (CBY740; A), pep4Δ (CBY2732; B), and erv41Δ pep4Δ (CBY3306; C) cells were separated on 18–60% sucrose density gradients. Fractions were collected starting with fraction 1 from the top of the gradient, resolved on 10.5% polyacrylamide gels, and immunoblotted for Gls1, CPY (vacuole marker), Ssp120 (Golgi marker), and Yet3 (ER marker). The relative amounts of each protein were determined by densitometry and plotted. The sucrose gradients shown are from a single representative experiment out of two independent repeats of the experiment. (D) Triple HA-tagged Gls1 was visualized by immunofluorescence microscopy using anti-HA monoclonal antibody and anti–mouse IgG Texas red–conjugated secondary antibody in WT (CBY3841), pep4Δ (CBY3864), and erv41Δ pep4Δ (CBY3867) strains. The same cells were also stained for Kar2 as an ER marker using polyclonal antibody against Kar2 and anti–rabbit IgG FITC-conjugated secondary antibody. Bars, 5 µm.
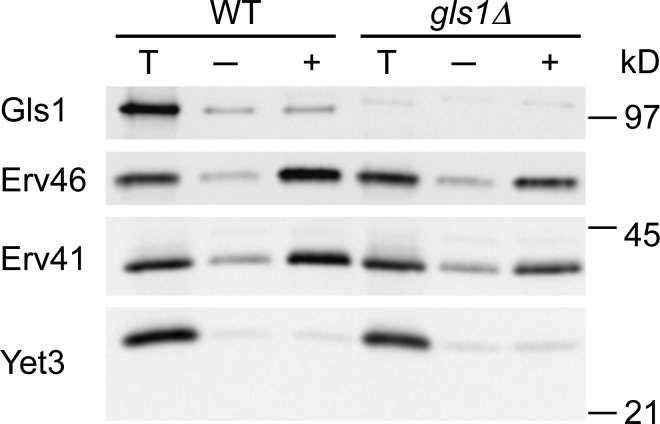
To gain further insight into potential ER retention or retrieval mechanisms that could underlie Gls1 mislocalization in erv41Δ cells, we performed in vitro budding assays with ER microsomes from WT and gls1Δ strains in the absence or presence of COPII components (Fig. 3). Erv41 and Erv46 were efficiently packaged into COPII vesicles as previously reported (Otte et al., 2001), and gls1Δ did not influence these packaging efficiencies. In WT membranes, Gls1 was not efficiently packaged into COPII vesicles as observed for the ER resident protein Yet3 (Wilson and Barlowe, 2010). These results indicated that Gls1 is not actively incorporated into COPII transport vesicles, although low levels of Gls1 are likely to leak into ER-derived vesicles. Gls1 dependence on the actively cycling Erv41–Erv46 complex for ER localization is more consistent with a retrieval mechanism as observed for the dependence of other ER resident proteins on the cycling KDEL receptor and Rer1 (Lewis and Pelham, 1990; Sato et al., 1997). Thus, we next explored the hypothesis that Erv41–Erv46 functions as a retrograde cargo receptor to retrieve escaped Gls1 back to the ER. In this model, strains that lack Erv41–Erv46 complex would not retrieve Gls1 from post-ER compartments, resulting in trafficking to the vacuole and degradation.
Figure 3.
Gls1 is not efficiently packaged into COPII vesicles. In vitro budding reactions were performed with microsomes from WT (CBY740) and gls1Δ (CBY1086) strains in the absence (−) or presence (+) of purified COPII components. Membranes from one tenth of the total reaction (T) and vesicle fractions were collected by centrifugation, resolved on 10.5% polyacrylamide gels, and immunoblotted for Gls1, Erv41, and Erv46 as well as the ER resident protein, Yet3, as a negative control.
Erv41–Erv46 complex binds to Gls1 in a pH-dependent manner
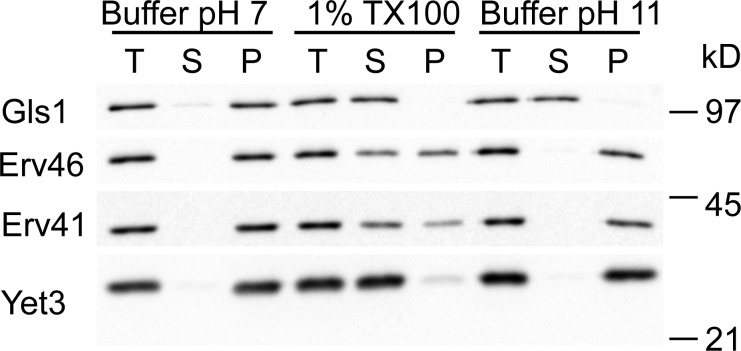
Although GLS1 is reported to encode a type II integral membrane protein (Jiang et al., 1996), additional studies show that a soluble form of Gls1 results from proteolytic cleavage between residues Ala24 and Thr25 near the end of the predicted transmembrane domain during overexpression and purification of Gls1 (Dhanawansa et al., 2002). Moreover, this cleavage site matches a predicted signal sequence cleavage site in Gls1 (Nielsen et al., 1997; Faridmoayer and Scaman, 2004). To examine whether Gls1 is indeed an integral membrane protein or not, we tested its solubilization properties from ER microsomes under various conditions (Fig. 4). Gls1 was solubilized and shifted to the supernatant fraction after treatment with 0.1 M sodium carbonate, pH 11, or detergent (1% Triton X-100), indicating that Gls1 is an ER luminal protein but does not contain a transmembrane anchor. In contrast, only detergent treatment (1% Triton X-100) solubilized the integral membrane proteins Erv41, Erv46, and Yet3. These results indicate that Gls1 has its signal sequence cleaved to produce a soluble protein and that potential sorting information would be contained within this luminal domain.
Figure 4.
Gls1 is a soluble ER-luminal protein. Microsomes from WT (CBY740) cells were incubated in buffer pH 7.0 (20 mM Hepes, pH 7, 150 mM potassium acetate, and 2 mM EDTA), in 1% TX100 (buffer pH 7.0 with 1% Triton X-100), or in buffer pH 11.0 (0.1 M sodium carbonate, pH 11, and 2 mM EDTA). Total lysates (T) were separated into supernatant (S) and pellet (P) fractions and resolved on 10.5% polyacrylamide gels and immunoblotted for Gls1, Erv41, Erv46, and Yet3.
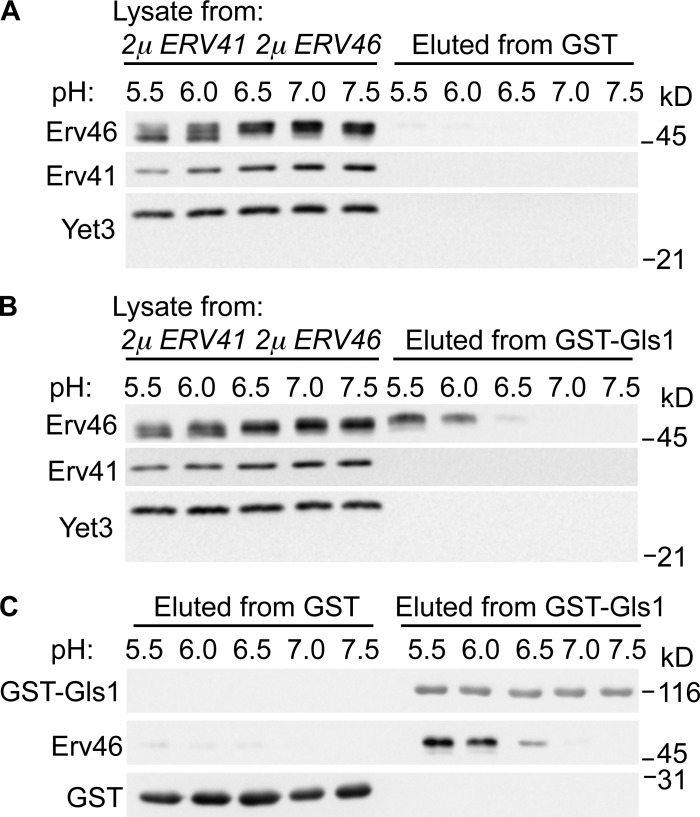
To test whether the Erv41–Erv46 complex can bind Gls1 as a cargo, we immobilized the soluble domain of Gls1 fused to GST (GST-Gls1) on glutathione agarose beads and mixed with detergent solubilized extracts from microsomes overexpressing the Erv41–Erv46 complex. Binding interactions for other cargo receptors in the early secretory pathway are thought to be regulated by compartmental pH (Wilson et al., 1993; Appenzeller-Herzog et al., 2004), and reported pH gradients between the ER and Golgi range from 7.4 to 6.2 (Paroutis et al., 2004). Therefore, we monitored binding interactions between Gls1 and the Erv41–Erv46 complex under different pH conditions ranging from 5.5 to 7.5 (Fig. 5). For these experiments, membrane proteins were solubilized in buffers at the indicated pH followed by binding to GST-Gls1 and multiple washes at the constant pH. Bound proteins were eluted from the beads for immunoblot analysis and revealed that Erv46 bound to GST-Gls1 under acidic pH buffer conditions (Fig. 5 B). Erv46 did not bind immobilized GST alone (Fig. 5 A) to support specificity of the interaction, and Erv41 was not detected in the eluted fractions under these conditions, suggesting that Gls1 interaction with the complex was mediated through the Erv46 subunit. These results are consistent with a model in which the Erv41–Erv46 complex binds to Gls1 in the reduced pH environment of luminal Golgi compartments.
Figure 5.
Erv41–Erv46 complex binds to Gls1 in a pH-dependent manner in vitro. (A and B) GST (A)- or GST-Gls1 (B)–bound glutathione agarose beads were incubated with cell lysate from microsomes overexpressing Erv41–Erv46 complex (CBY978) in different pH buffers (pH 5.5, 6.0, 6.5, 7.0, and 7.5) at 4°C. After washing, proteins bound to the beads were eluted and resolved on 10.5% polyacrylamide gels and immunoblotted for Gls1, Erv41, Erv46, Yet3, and GST. (C) Eluted samples were compared on a single blot for levels of GST-Gls1, Erv46, and GST. Figure panels were assembled from single immunoblots, and the molecular mass markers shown indicate relative positions across neighboring strips.
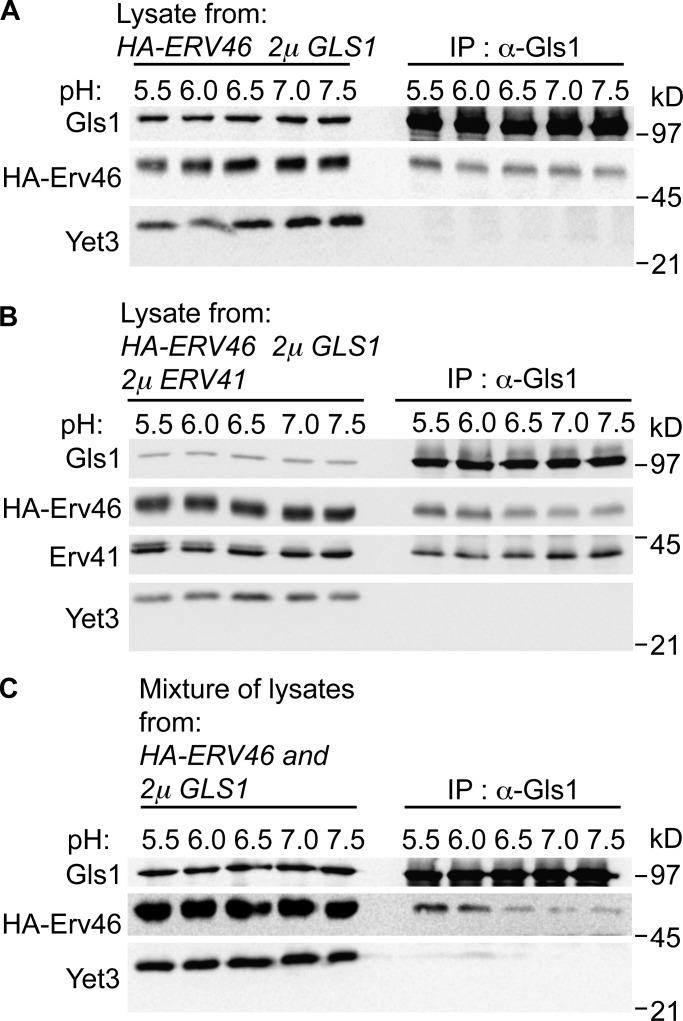
To further explore interactions between the Erv41–Erv46 complex and Gls1, coimmunoprecipitation (IP; co-IP) experiments were performed using detergent-solubilized membrane extracts from cells expressing different levels and tagged versions of these proteins. In cells overexpressing HA-Erv46 and Gls1 together, co-IP of Gls1 from Triton X-100–solubilized membranes across a pH range of 5.5 to 7.5 showed that HA-Erv46 was recovered in complex with Gls1 (Fig. 6 A). However, equivalent levels of HA-Erv46 were recovered over this pH range, suggesting that stability of preexisting Gls1-Erv46 complexes was independent of pH. Similarly, in cells overexpressing Erv41, HA-Erv46, and Gls1, co-IP of Gls1 from these detergent-solubilized membranes recovered both HA-Erv46 and Erv41 across the pH range (Fig. 6 B). These results imply that a fraction of Gls1 is assembled into complexes with Erv41–Erv46 under overexpression conditions and that these complexes are insensitive to pH during the co-IP procedure. To examine why the co-IP assay did not show a pH-dependent interaction, mixing IP assays were performed in which HA-Erv46 and Gls1 were individually overexpressed in cells, membranes from these cells were solubilized with Triton X-100 in the indicated pH buffers, and then membrane extracts were combined before adding anti-Gls1 antibodies. Under these conditions, HA-Erv46 coprecipitated with Gls1 in a pH-dependent manner (Fig. 6 C) and displayed a similar pH profile as observed for the GST-Gls1 pull-down assay. Based on these collective findings, we speculate that a reduced pH environment promotes binding of Gls1 to the Erv41–Erv46 complex, although reversal of this interaction is not trigged by pH alone and may depend on additional factors for efficient dissociation.
Figure 6.
Erv41–Erv46 complex interacts with Gls1 in vivo. (A and B) Semi-intact cells from strains overexpressing HA-Erv46 and Gls1 (CBY3728; A) or HA-Erv46, Gls1, and Erv41 (CBY3785; B) were solubilized with 1% Triton X-100 and immunoprecipitated with anti-Gls1 antibody at 4°C in different pH buffers (pH 5.5, 6.0, 6.5, 7.0, and 7.5). (C) Mixing-IP assay in which Triton X-100–solubilized lysates from semi-intact cells overexpressing HA-Erv46 (CBY770) or Gls1 (CBY3832) were mixed and immunoprecipitated with the anti-Gls1 antibody. Immunoprecipitated proteins were resolved on 10.5% polyacrylamide gels and immunoblotted for Gls1, HA, Erv41, and Yet3.
Proteomic analysis of erv41Δ and erv46Δ mutants identifies additional Erv41–Erv46 cargo
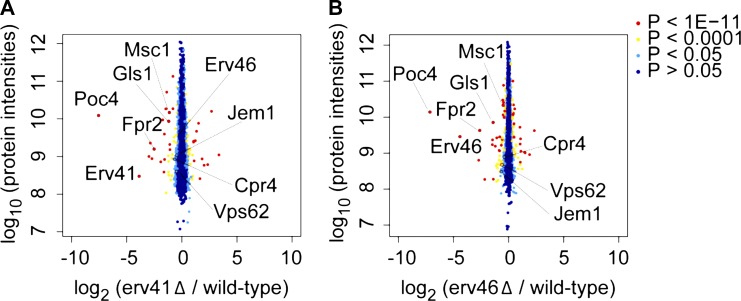
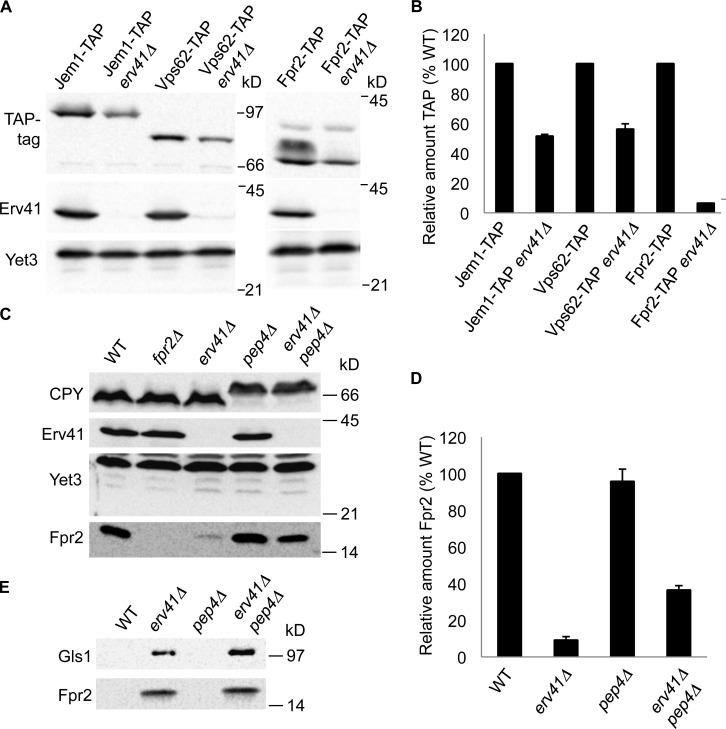
Because other characterized retrograde and anterograde cargo receptors in the early secretory pathway are known to recognize multiple cargo proteins (Dancourt and Barlowe, 2010), we investigated whether other ER resident proteins might depend on the Erv41–Erv46 complex for retrograde retrieval. Based on the reduced steady-state protein levels of Gls1 in the erv41Δ strain, we hypothesized that additional Erv41–Erv46-dependent cargo proteins would be mistargeted to vacuoles and present at reduced levels. Advances in mass spectrometry–based quantitative proteomics using SILAC can provide comparative protein levels for most endogenously expressed proteins in dividing yeast cells (Fröhlich et al., 2013). Through this approach, we quantified yeast proteome changes in the erv41Δ and erv46Δ strains against a WT standard. In total, 3,905 proteins were identified with relative abundance measurements for 3,494 and 3,486 proteins in the erv41Δ and the erv46Δ strain, respectively (Table S3). In comparing protein abundance changes expressed as the log2 ratio of deletion strain/WT, we focused on 20 proteins that were significantly reduced (log2 < −0.4) in both the erv41Δ and erv46Δ strains. Importantly, Gls1 was detectably reduced in the erv41Δ strain (log2 = −1.19) and the erv46Δ strain (log2 = −1.40) in accord with our initial observations. Moreover, Erv41 (log2 = −3.87) and Erv46 (log2 = −4.41) protein levels were markedly reduced in the respective deletion strains as expected. A plot of protein intensities against log2 ratios is shown in Fig. 7, with proteins of interest highlighted including Fpr2, Msc1, Vps62, Jem1, and Cpr4. Next, we focused our attention on potential ER resident proteins that were detected at reduced steady-state levels as candidate cargo for the Erv41–Erv46 complex. We obtained strains containing C-terminally tandem affinity purification (TAP)–tagged Jem1, Vps62, and Fpr2 (Ghaemmaghami et al., 2003), generated erv41::kanR deletions in the TAP-tagged strain background, and monitored their steady-state expression levels to confirm the proteomic findings. In our analysis of these strains, significant decreases were detected in Jem1-TAP, Vps62-TAP, and Fpr2-TAP levels in erv41Δ strains of 49%, 44%, and 94%, respectively, compared with the WT background (Fig. 8, A and B). These decreases are in good agreement with the proteomic results.
Figure 7.
Whole cell proteome changes in erv41Δ and erv46Δ strains using SILAC. Plot of protein intensities normalized against heavy/light SILAC ratios of the WT strain (CBY740). (A and B) Proteome changes in erv41Δ (CBY1168) cells with 3,494 proteins quantified (A) and in erv46Δ (CBY3612) cells with 3,486 proteins quantified (B). Proteins of interest that were detected at reduced levels in both deletion strains are indicated.
Figure 8.
Deletion of ERV41 reduces cellular levels of Fpr2 caused by vacuolar degradation and extracellular secretion. (A) Fpr2-TAP (CBY3793), Fpr2-TAP erv41Δ (CBY3813), Vps62-TAP (CBY3897), Vps62-TAP erv41Δ (CBY3908), Jem1-TAP (CBY3791), and Jem1-TAP erv41Δ (CBY3811) cells grown to mid–log phase were lysed, resolved on a 10.5% polyacrylamide gel, and immunoblotted for the TAP-tag, Erv41, and Yet3. (B) Relative amounts of Fpr2-TAP, Vps62-TAP, and Jem1-TAP with standard error bars (n = 3). TAP-tagged protein levels were normalized with Yet3 as the loading control and plotted as a percentage relative to WT. (C) Cells grown to mid–log phase were lysed, resolved on 15% polyacrylamide gels, and immunoblotted for Fpr2, CPY, Erv41, and Yet3 (loading control). WT (CBY740), fpr2Δ (CBY3758), erv41Δ (CBY1168), pep4Δ (CBY2732), and erv41Δ pep4Δ (CBY3306) strains were compared. (D) Relative amounts of Fpr2 with standard error bars (n = 3). Fpr2 levels were normalized with Yet3 as a loading control and plotted as a percentage relative to WT. (E) Proteins secreted to the extracellular medium were precipitated by TCA, resolved on a 15% polyacrylamide gel, and immunoblotted for Gls1 and Fpr2.
The yeast FPR2 gene encodes a 15-kD membrane-associated peptidyl-prolyl cis–trans-isomerase that localizes to the ER and has a predicted N-terminal signal sequence (Nielsen et al., 1992; Huh et al., 2003). To further examine endogenous Fpr2 protein levels in WT and erv41Δ strains, we prepared polyclonal antiserum specific for Fpr2. Immunoblot analysis of whole cell extracts demonstrated that erv41Δ reduced steady-state levels of Fpr2 by ∼91% compared with the WT strain (Fig. 8, C and D). Moreover, combining pep4Δ with erv41Δ produced a recovery of Fpr2 cellular levels consistent with mislocalization of Fpr2 to vacuolar compartments in the absence of a functional Erv41–Erv46 complex. Finally, we assessed extracellular secretion of Fpr2 into the growth media because inactivation of Pep4-dependent vacuolar proteases only restored Fpr2 levels to ∼36% of normal WT cellular levels (Fig. 8, D and E). Strikingly, both Fpr2 and Gls1 were detected in the extracellular medium of erv41Δ and erv41Δ pep4Δ strains, indicating that failure to retain these ER resident proteins in the early secretory pathway resulted in missorting into both vacuolar-targeted and exocytic transport vesicles. Based on these results, we conclude that the Erv41–Erv46 complex is required for localization of multiple ER resident proteins and propose that the complex acts as a retrograde receptor to retrieve a subset of escaped ER proteins.
Mutation of a COPI binding motif in Erv46 mislocalizes Gls1
A previous study has revealed COPI and COPII sorting signals within the cytoplasmic tail sequences of the Erv41–Erv46 complex that are required to cycle the complex between ER and Golgi compartments (Otte and Barlowe, 2002). More specifically, Erv46 contains a conserved dilysine motif on its C-terminal tail, which is important for packaging into retrograde-directed COPI vesicles (Cosson and Letourneur, 1994; Otte and Barlowe, 2002). It has been reported that mutation of dilysine residues in ER resident proteins to diarginines disrupts ER retention because of loss of binding to the COPI complex (Cosson and Letourneur, 1994). To test the model that the Erv41–Erv46 complex functions in retrieval of escaped Gls1, the dilysine motif in Erv46 was mutated to diarginines (Erv46 KK/RR) to block retrograde transport of the Erv41–Erv46 complex and to monitor the influence on Gls1 localization. By comparing WT Erv46 and Erv46 KK/RR mutant strains (Fig. S3), we found that mutation of the COPI-binding motif in the Erv46 tail reduced the steady-state cellular levels of Gls1 by ∼30% compared with the WT strain. Moreover, Gls1 was secreted to the extracellular medium in the Erv46 KK/RR mutant strain. These results indicate that retrograde trafficking of the Erv41–Erv46 complex is important for retrieval of leaked Gls1. Although mutation of the Erv46 dilysine motif caused Gls1 mislocalization, we noted that this phenotype was not as severe as the erv41Δ-null mutation, suggesting that partially redundant COPI sorting information remained in the point mutant. Regardless, the impact of this Erv46 dilysine mutation on Gls1 localization supports a retrograde receptor function for the Erv41–Erv46 complex.
Disruption of intracellular pH gradients causes Gls1 mislocalization
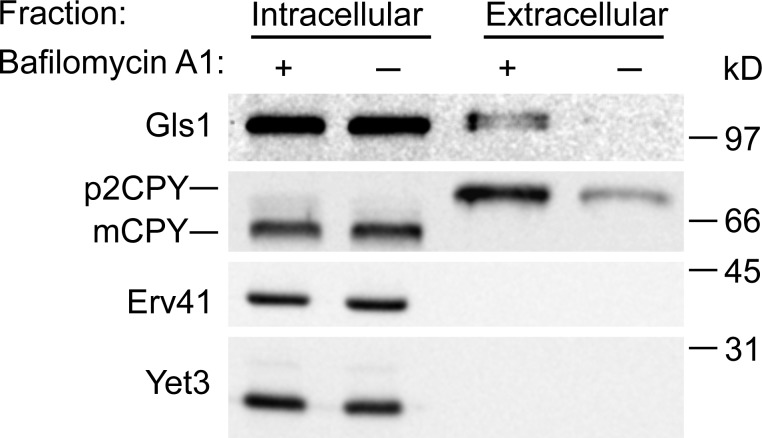
The in vitro pH-dependent interaction between Gls1 and the Erv41–Erv46 complex led us to investigate its physiological significance in vivo. The vacuolar ATPase plays an important role in acidification of vacuoles/lysosomes, endosomes, and Golgi compartments in eukaryotic cells (Forgac, 2007). The drug bafilomycin A1 has been shown to be a specific inhibitor of vacuolar ATPases (Bowman et al., 1988) and to increase the pH in Golgi compartments (Llopis et al., 1998). To examine the role of this pH gradient across Golgi membranes in Gls1 localization, we tested the effect of bafilomycin A1 on Gls1 sorting in vivo. WT yeast cells were treated with 20 µM bafilomycin A1 for 2 h and separated into intracellular and extracellular fractions by centrifugation. We used CPY as a positive control because a previous study demonstrated that bafilomycin A1 caused missorting and secretion of the vacuolar hydrolase CPY (Banta et al., 1988). As shown in Fig. 9, Gls1 was secreted into the extracellular fraction, and the level of secreted p2CPY (Golgi form of CPY) was increased in the presence of bafilomycin A1, indicating that the pH gradient across Golgi membranes plays a role in Gls1 localization. These results provide in vivo evidence that an intracellular pH gradient regulates binding between escaped ER resident proteins and the Erv41–Erv46 complex for retrieval back to the ER.
Figure 9.
In vivo disturbance of pH gradients by bafilomycin A1 causes secretion of Gls1. WT (CBY740) cells were incubated for 2 h in the presence or absence of 20 µM bafilomycin A1 (in DMSO). Cultures were separated into intracellular and extracellular fractions. Pellets in the intracellular fraction were lysed, proteins secreted to the extracellular fraction were precipitated by TCA, and samples were resolved on a polyacrylamide gel then immunoblotted for Gls1, CPY, Erv41, and Yet3.
Discussion
Organelle composition in the secretory pathway is maintained under dynamic cellular conditions through protein targeting, retention, and retrieval mechanisms. In this study, we describe a new retrieval activity in which specific ER resident proteins that have trafficked to Golgi compartments are recognized and returned to the ER by the cycling Erv41–Erv46 retrograde receptor. Our results show that ER resident proteins Gls1 and Fpr2 are secreted or mislocalized in Erv41–Erv46-deficient cells and that the luminal domain of Gls1 binds directly to the Erv41–Erv46 complex under mildly acidic conditions. The Erv41–Erv46 complex is efficiently packaged into ER-derived COPII vesicles and actively recycles from Golgi compartments to the ER in COPI vesicles (Otte and Barlowe, 2002). In contrast, ER resident proteins, such as Gls1, are not efficiently packaged into COPII vesicles, and we speculate that a low level of bulk-flow exit of ER resident proteins must be countered by retrograde retrieval from early Golgi compartments. If not, escaped ER resident proteins traffic through the process of Golgi maturation (Losev et al., 2006) followed by Golgi-to-cell surface and Golgi-to-vacuole transport, which results in protein mislocalization. Pull-down and co-IP experiments indicated that interactions between the Erv41–Erv46 complex and Gls1 are regulated by pH. Moreover, disruption of intracellular pH gradients by bafilomycin A1 treatment led to secretion of Gls1. Together, these findings support a model in which escaped Gls1 binds to Erv41–Erv46 in the reduced pH environment of early Golgi compartments for retrograde transport and release in the neutral pH of the ER lumen.
Our model for Erv41–Erv46 activity in retaining ER resident proteins shares parallels with the well-characterized KDEL receptors that recognize conserved KDEL motifs (HDEL in yeast) found at the C terminus of many soluble ER resident proteins (Munro and Pelham, 1987; Pelham et al., 1988). The purified human KDEL receptor displays pH-dependent affinity for KDEL peptides with increased binding activity at an acidic pH relative to neutral, which is thought to promote KDEL receptor–ligand interactions in the Golgi complex and release KDEL ligands in the neutral pH of the ER (Scheel and Pelham, 1996). Interestingly, the pH gradient between ER and Golgi compartments appears to have an opposite effect on anterograde cargo receptors such as ERGIC53, which binds newly synthesized glycoproteins in the ER for transport and release in more acidic Golgi compartments (Appenzeller et al., 1999; Appenzeller-Herzog et al., 2004). We observed a similar pH profile for Gls1 binding to Erv41–Erv46 as reported for the interaction between KDEL receptor and its ligands (Scheel and Pelham, 1996) in support of the retrograde receptor model. However, we also observed that Gls1 bound to Erv41–Erv46 in detergent-solubilized membranes extracts was not readily dissociated under reduced pH conditions, suggesting that additional factors may be required to facilitate Gls1 release in the ER lumen.
The Erv41–Erv46 proteins are highly conserved across species and localize predominantly to the ER–Golgi intermediate compartment and early Golgi compartments as reported for the KDEL receptors (Orci et al., 2003; Breuza et al., 2004; Raykhel et al., 2007). ER folding machinery components Gls1, or glucosidase I (Romero et al., 1997), and the Fpr2 prolyl-isomerase, or FKBP-13 (Partaledis and Berlin, 1993), are also conserved, suggesting that this retrieval mechanism is likely to operate in many cell types. It is important to note that currently identified Erv41–Erv46 retrieval cargoes are soluble ER-luminal proteins that do not contain KDEL/HDEL signals, and with the bulk of the Erv41–Erv46 mass facing the ER lumen, this retrieval mechanism appears distinct from the seven-transmembrane–containing KDEL receptor. Moreover, a recent structural study on the Erv41 luminal domain reveal an unusual β-sandwich arrangement and a prominent negative electrostatic surface patch thought to promote protein–protein interactions (Biterova et al., 2013), suggesting that the Erv41–Erv46 complex recognizes a class of ER resident proteins through a novel interaction motif. We are currently defining this sorting motif in the 118–amino acid Fpr2 protein, which should permit bioinformatic searches for other Erv41–Erv46 retrieval cargo. Because certain microbes are known to express KDEL-bearing pathogenic factors to gain access to the ER (Sandvig and van Deurs, 2002), identification of analogous sorting signals for Erv41–Erv46 retrieval may help explain additional host–pathogen relationships.
Our whole cell SILAC proteomic analysis of deletion strains provides powerful insights into the consequences of loss-of-function Erv41–Erv46. Reduced intracellular protein levels of non-HDEL–bearing ER resident proteins are likely a result of failures in retrieval that lead to secretion of these proteins from cells or mislocalization and degradation in the vacuole. Several other notable changes in steady-state levels of secretory cargo and cytosolic proteins may reflect secondary consequences of, or cellular responses to, loss of Erv41–Erv46 function. For example, decreased levels of the plasma membrane transporters Zrt1/2 and Pdr12 may be caused by loss of specific ER folding machinery needed for their biogenesis and result in precocious ER-associated degradation (Kota et al., 2007). Strong reductions in the proteasome assembly factor Poc4 (Le Tallec et al., 2007) were observed in both the erv41Δ and erv46Δ proteomic datasets. However, we detected only modest reductions through analysis of TAP-tagged Poc4 in the erv41Δ background, indicating that further studies will be required to understand this result.
In our initial characterization of the erv41Δ and erv46Δ mutants, we observed a modest reduction in apparent fusion of COPII vesicles with Golgi membrane in cell-free transport reactions (Otte et al., 2001). Based on the current evidence, we suspect this effect is caused by indirect consequences of the erv41Δ/erv46Δ mutations. Reduced levels of ER resident proteins in the mutant cells may result in inefficient ER export of proteins necessary for fusion at Golgi membranes or for glycosylation of glyco-pro–α-factor that serves as the readout for fusion in cell-free assays (Baker et al., 1988). Interestingly, it was recently reported that the Golgi-localized mannosyltransferase Ktr4 accumulates in the ER of erv41Δ and erv46Δ strains (Noda et al., 2014), which could explain reduced levels of Golgi-modified glyco-pro–α-factor in our fusion assays. It is also possible that some cellular changes are caused by Erv41–Erv46 function as an anterograde cargo receptor for specific proteins as proposed for Golgi localization of Ktr4 (Noda et al., 2014). However, we note that the erv41Δ and erv46Δ strains do not display an activated unfolded protein response as reported for other anterograde cargo receptor deletion strains (Jonikas et al., 2009).
Finally, observed increases in cell surface proteins (e.g., Tos1 and Ecm25) and transcription regulatory machinery (e.g., Ngg1 and Sap30) may reflect cellular responses to perturbation of the ER luminal environment. Further analyses will be required to confirm the magnitude of these increases. Regardless, the global snapshot provided by whole cell proteomics suggests clear models that can be tested through SILAC and experimental analyses of available yeast mutant strains to more fully define the cellular function of Erv41–Erv46.
Materials and methods
Yeast strains and growth media
Yeast strains used in this study are listed in Table 1. Strains were grown at 30°C in rich medium (YPD [1% Bacto yeast extract, 2% Bacto-peptone, and 2% glucose]). Growth of cell cultures was monitored by absorbance (OD) at 600 nm (OD600). Precultures were grown overnight in YPD to stationary phase, and fresh YPD was inoculated with precultures to an OD600 of 0.1. Cells were grown at 30°C and harvested in mid–log phase between 0.8 and 1.2 OD600. Yeast strains containing plasmids were grown overnight in minimal medium (YMD [0.7% yeast nitrogen base without amino acids, 2% glucose, and the appropriate supplement mixture (complete supplement mixture; MP Biomedicals)]) and back diluted into YPD medium. For overexpression from the GAL1 promoter, cells were grown overnight in YPD or YMD containing 1% galactose and 1% glucose and back diluted to YPD containing 1.5% galactose and 0.5% glucose. For overexpression from the Cu2+-inducible CUP1 promoter in plasmid pYEX4T-1-Gls1, GST-Gls1 expression was induced by the addition of 0.5 mM copper sulfate.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
| CBY80 (=FY834) | MATα his3-200 ura3-52 leu2-1 lys2-202 trp1-63 | Winston et al., 1995 |
| CBY740 (=BY4742) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Winzeler et al., 1999 |
| CBY742 (=BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Winzeler et al., 1999 |
| CBY770 | CBY80 with ERV46::HISMX6-PGAL1-3HA | Otte et al., 2001 |
| CBY978 | CBY80 containing pRS424-ERV41 and pRS426-ERV46 | Otte et al., 2001 |
| CBY1086 | CBY740 with gls1Δ::kanR | Winzeler et al., 1999 |
| CBY1168 | CBY740 with erv41Δ::kanR | Winzeler et al., 1999 |
| CBY2732 | CBY740 with pep4Δ::kanR | Winzeler et al., 1999 |
| CBY3306 | CBY740 with erv41Δ::HIS3 pep4Δ::kanR | This study |
| CBY3561 | CBY740 containing pYEX4T-1-GLS1 | This study |
| CBY3612 | CBY740 with erv46Δ::kanR | This study |
| CBY3728 | CBY80 with ERV46::HISMX6-PGAL1-3HA containing pRS425-GLS1 | This study |
| CBY3758 | CBY740 with fpr2Δ::kanR | Winzeler et al., 1999 |
| CBY3785 | CBY80 with ERV46::HISMX6-PGAL1-3HA containing pRS424-ERV41 and pRS425-GLS1 | This study |
| CBY3791 | CBY742 with JEM1::HIS3MX6-TAP | Ghaemmaghami et al., 2003 |
| CBY3793 | CBY742 with FPR2::HIS3MX6-TAP | Ghaemmaghami et al., 2003 |
| CBY3811 | CBY742 with JEM1::HIS3MX6-TAP erv41Δ::kanR | This study |
| CBY3813 | CBY742 with FPR2::HIS3MX6-TAP erv41Δ::kanR | This study |
| CBY3832 | CBY80 containing pRS425-GLS1 | This study |
| CBY3841 | CBY740 with gls1Δ::kanR containing pRS317-HA-GLS1 | This study |
| CBY3854 | CBY740 with pep4Δ::kanR gls1Δ::natR | This study |
| CBY3855 | CBY740 with erv41Δ::HIS3 pep4Δ::kanR gls1Δ::natR | This study |
| CBY3864 | CBY740 with pep4Δ::kanR gls1Δ::natR containing pRS317-HA-GLS1 | This study |
| CBY3867 | CBY740 with erv41Δ::HIS3 pep4Δ::kanR gls1Δ::natR containing pRS317-HA-GLS1 | This study |
| CBY3897 | CBY742 with VPS62::HIS3MX6-TAP | Ghaemmaghami et al., 2003 |
| CBY3908 | CBY742 with VPS62::HIS3MX6-TAP erv41Δ::kanR | This study |
| CBY4002 | CBY740 with erv46Δ::kanR containing pRS316 | This study |
| CBY4003 | CBY740 with erv46Δ::kanR containing pRS316-ERV46 | This study |
| CBY4004 | CBY740 with erv46Δ::kanR containing pRS316-ERV46(KK/RR) | This study |
Plasmid construction
Plasmids and primers used in this study are listed in Tables S1 and S2. Genomic DNA from CBY740 was used as the template for PCR amplification and cloning. The DNA sequences of GLS1 (YGL027C), ERV46 (YAL042W), and FPR2 (YDR519W) were obtained from the Saccharomyces Genome Database. For overexpression of GST-Gls1 in yeast, the GLS1 sequence encoding amino acids 33–833 was amplified by PCR using primers GLS1-EcoRI and GLS1–SalI-1. The PCR product was ligated into the EcoRI and SalI restriction sites of the pYEX4T-1-ERV46 plasmid (obtained from Research Genetics; Macreadie et al., 1991; Ward et al., 1994) to generate plasmid pYEX4T-1–GLS1. To construct pRS425-GLS1 and pRS317-GLS1, GLS1 and ∼300 bp of its flanking upstream and downstream sequences were amplified using primers GLS1-NotI and GLS1–SalI-2 and ligated into the NotI and SalI restriction sites of pRS425 (Christianson et al., 1992) and pRS317 (Eriksson et al., 2004). The correct GLS1 sequences were verified by DNA sequencing using primers GLS1-SalI-1, GLS1-NotI, GLS1-SalI-2, GLS1-XhoI, GLS1-IntF, and GLS1-IntR. The plasmid pRS317-HA-GLS1 was constructed as previously described (Jiang et al., 1996). The triple HA epitope tag was inserted into the unique EcoRI site located between codons 24 and 25 of GLS1 in plasmid pRS317-GLS1.
For overexpression of Fpr2 fused to the C terminus of GST (GST-Fpr2) in Escherichia coli, the FPR2 sequence encoding amino acids 18–135 was amplified by PCR using primers FPR2-BamHI and FPR2-EcoRI. The PCR product was ligated into the BamHI and EcoRI restriction site of the pGEX2T plasmid (GE Healthcare) to generate plasmid pGEX2T-FPR2.
To convert the Erv46 lysine residues in positions 412 and 413 to arginine residues, ERV46 and ∼300 bp of upstream sequence were amplified using pRS316-ERV46 as a template for primers YAL042w-NotI and ERV46(KK/RR)-R followed by gel purification (QIAGEN). Next, ERV46 and ∼300 bp of the downstream sequence were amplified using pRS316-ERV46 and the primers YAL042w-BamHI and ERV46(KK/RR)-F followed by gel purification. Both gel-purified PCR products were then mixed and used as the template to amplify ERV46 containing the point mutations using primers YAL042w-NotI and YAL042w-BamHI. This PCR product was ligated into the NotI and BamHI restriction site of pRS316 (Sikorski and Hieter, 1989) to obtain plasmid pRS316-ERV46(KK/RR). The correct ERV46 sequences were verified by DNA sequencing using primers YAL042w-NotI, YAL042w-BamHI, ERV46-IntF, and ERV46-IntR.
Strain construction
To generate the erv41Δ pep4Δ double knockout strain (CBY3306), ERV41 in the pep4Δ strain (CBY2732) was targeted for gene disruption with the HIS3 gene using plasmid pFA6a-His3MX6 and primers Erv41-KO-F and Erv41-KO-R (Longtine et al., 1998). To generate the erv46Δ strain (CBY3612), ERV46 in the WT strain (CBY740) was targeted for gene disruption with the kanamycin-resistance (kanR) gene using plasmid pFA6a-kanMX6 and primers Erv46-KO-F and Erv46-KO-R (Longtine et al., 1998). For CBY3854 and CBY3855, GLS1 in the pep4Δ strain (CBY2732) and the erv41Δ pep4Δ strain (CBY3306) was deleted with the nourseothricin-resistance (natR) gene using plasmid pFA6a-natMX6 and primers Gls1-KO-F and Gls1-KO-R (Hentges et al., 2005). All strains with C-terminally TAP-tagged ORFs were purchased from Thermo Fisher Scientific, and ERV41 in these strains was deleted with the kanR gene using plasmid pFA6a-kanMX6 and primers Erv41-KO-F and Erv41-KO-R (Longtine et al., 1998). All yeast transformations were performed using the lithium acetate technique (Elble, 1992).
Antibodies and immunoblotting
Rabbit polyclonal antiserum against Erv41 and Erv46 (Otte et al., 2001), CPY (Rothblatt et al., 1989), Kar2 (Brodsky and Schekman, 1993), ALP (Haas et al., 1995), Erv26 (Bue et al., 2006), and Yet3 (Wilson and Barlowe, 2010) have been described previously. Anti-Erv26p and anti-Yet3 rabbit antibodies were raised against GST fusion proteins and used for detection of GST where indicated. Polyclonal antiserum against Fpr2 was raised in rabbits using purified GST-Fpr2 as the antigen (Thermo Fisher Scientific). GST-Fpr2 was expressed in DH5α cells, lysed, and purified by glutathione-agarose chromatography as described by the manufacturer (GE Healthcare). Polyclonal antiserum for Ssp120 was raised in rabbits using purified 6×His-tagged Ssp120 expressed from the pET15b plasmid (EMD Millipore) as an antigen. Sheep anti–mouse and donkey anti–rabbit secondary horseradish peroxidase–linked antibodies were purchased from GE Healthcare, and mouse monoclonal antibodies against HA (HA.11) were obtained from Covance. All primary antibodies were used at a 1:1,000 dilution except for CPY (1:1,500 dilution), and secondary antibodies were at a 1:20,000 dilution. Chemiluminescence from immunoblots was enhanced with SuperSignal West Pico Chemiluminescent substrate (Thermo Fisher Scientific) or Luminata Crescendo Western HRP substrate (EMD Millipore) and detected using a G:BOX Chemi XR5 (Syngene). For densitometric analysis, captured images of immunoblots were quantified with GeneTools image analysis software (Syngene).
Polyclonal antiserum against Gls1 was raised in rabbits using purified GST-Gls1 as an antigen (Covance). To prepare GST-Gls1, yeast cells containing pYEX4T-1–GLS1 (CBY3561) were precultured in YMD containing complete supplement mixture–URA and back diluted into 7.5 liters of YPD. Cells were grown to ∼0.5 OD600, and copper sulfate was added to a final concentration of 0.5 mM. After induction for 3 h, cells were harvested and washed with TBS (50 mM Tris/Cl, pH 7.4, and 150 mM NaCl). Cell pellets were resuspended to 1.8 g/ml in TBS and frozen into cell beads by dripping into liquid nitrogen. Cells were lysed in liquid nitrogen using a Waring blender to mix the beads for 2 min to obtain a fine cell powder. The cell powder was resuspended in 175 ml of TBS containing 1 mM PMSF and 1 mM DTT and centrifuged for 5 min at 4°C in rotor (SS-34; Sorvall) at 3,500 rpm. The resulting supernatant fraction was then centrifuged for 30 min at 4°C in SS-34 rotor at 15,000 rpm. This supernatant was then applied to a Q Sepharose (GE Healthcare) column (2.5 × 10 cm; Bio-Rad Laboratories) that contained a 70-ml packed bed volume using a fast protein liquid chromatography system (AKTA; GE Healthcare). After washing with TBS, the GST-Gls1 protein was eluted with a 250-ml continuous buffer gradient from 150 mM to 0.5 M NaCl. The fractions containing GST-Gls1 were monitored by Western blotting using GST-Erv26 antiserum for detection of GST-tagged proteins, and peak fractions were combined to obtain ∼50 ml of semipurified GST-Gls1. This fraction was mixed with 5 ml of glutathione-agarose (Sigma-Aldrich) for 1 h with rotation at 4°C and then applied into a 1.5 × 10–cm column followed by wash with TBS and elution with TBS containing 10 mM glutathione. The fractions containing highly purified GST-Gls1 protein were determined by Coomassie blue–stained polyacrylamide gels and pooled to yield 3 ml of 0.8 mg/ml protein.
Membrane preparations and subcellular fractionation
Yeast semi-intact cells were prepared as previously described (Baker et al., 1988) with minor modification as follows. Cells were grown to the mid–log phase, harvested by centrifugation at 5,000 rpm for 5 min at room temperature in a rotor (GS-3; Sorvall), and resuspended in Tris/DTT buffer (100 mM Tris-HCl, pH 9.4, and 10 mM DTT). After incubation at room temperature for 10 min, cells were harvested by centrifugation, resuspended in lyticase buffer (20 mM Tris-HCl, pH 7.5, 0.7 M sorbitol, 0.5% glucose, 2 mM DTT, and lyticase), and incubated for 15 min at room temperature until the OD600 decreased to <20% of the initial value. Spheroplasted cells were harvested by centrifugation at 5,000 rpm for 10 min at 4°C in an SS-34 rotor and resuspended at 75 OD280 U/ml in semi-intact cells lysis buffer (20 mM Hepes, pH 7.0, 0.4 M sorbitol, 150 mM KOAc, and 2 mM MgOAc) at 4°C. 100-µl aliquots of this spheroplast suspension were transferred to tubes, frozen in liquid N2, and stored at −80°C.
Yeast ER microsomes were prepared as previously described (Wuestehube and Schekman, 1992) with modifications as follows. Spheroplasted cells as prepared in the previous paragraph were resuspended in JR lysis buffer (20 mM Hepes, pH 7.4, 0.1 M sorbitol, 50 mM KOAc, 2 mM EDTA, 1 mM PMSF, and 1 mM DTT) and homogenized with 10 strokes of a chilled-glass Potter–Elvehjem homogenizer. Homogenates were centrifuged for 5 min at 4°C in an SS-34 rotor at 3,500 rpm. This supernatant fraction was then centrifuged for 10 min at 4°C in an SS-34 rotor at 15,000 rpm. The resulting membrane pellets were resuspended in JR lysis buffer and loaded on top of a 1.2 M/1.5 M sucrose step gradient. Gradients were spun at 40,000 rpm for 1 h in a rotor (SW-60; Beckman Coulter) at 4°C. The microsome band that sedimented to the 1.2 M/1.5 M sucrose interface was collected and washed with B88 buffer (20 mM Hepes, pH 6.8, 250 mM sorbitol, 150 mM KOAc, and 5 mM MgOAc) by centrifugation at 15,000 rpm for 10 min at 4°C in an SS-34 rotor. The final microsome pellet was resuspended at 16 OD280 U/ml in B88 buffer at 4°C. 100-µl aliquots were transferred to plastic tubes, frozen in liquid N2, and stored at −80°C.
For subcellular fraction of membranes, whole cell lysates were resolved on sucrose density gradients as described previously (Powers and Barlowe, 1998) with minor modifications as follows. Spheroplasted cells were resuspended in lysis buffer (10 mM Hepes, pH 7.4, 12.5% sucrose, 1 mM EDTA, and 0.5 mM PMSF) and homogenized with 10 strokes of a chilled-glass Potter–Elvehjem homogenizer. The homogenates were centrifuged for 5 min at 4°C in an SS-34 rotor at 3,500 rpm. The supernatant fraction was loaded onto the top of a sucrose step gradient. For preparing sucrose gradients, 1 ml of 60% sucrose was placed at the bottom of an ultracentrifuge tube (SW-40; Beckman Coulter) and overlaid with 0.75-ml steps of sucrose from 54% to 18% (54, 50, 46, 42, 38, 34, 30, 26, 22, and 18%) in Hepes buffer (10 mM Hepes, pH 7.4, and 1 mM MgCl2). Gradients were centrifuged at 4°C for 3 h in an SW-40 rotor at 36,000 rpm, and 0.75-ml fractions were collected from the top of the gradient. Fractions were diluted 1:1 in SDS-PAGE sample buffer, heated at 75°C for 5 min, resolved on 10.5% polyacrylamide gels, and immunoblotted for Gls1, CPY, Ssp120, and Yet3.
To determine whether Gls1 is an integral membrane protein, fractionation and solubilization procedures were performed as previously described (Otte et al., 2001) using microsomes from the WT strain (CBY740). Microsomes were suspended in buffer pH 7.0 (20 mM Hepes, pH 7, 150 mM KOAc, and 2 mM EDTA), in buffer pH 7.0 with 1% Triton X-100, or in buffer pH 11.0 (0.1 M sodium carbonate, pH 11, and 2 mM EDTA) and incubated on ice for 10 min. After centrifugation at 60,000 rpm in a rotor (TLA100.3; Beckman Coulter), equivalent amounts of the total, supernatant, and pellet fractions were resolved on 10.5% polyacrylamide gels and immunoblotted for Gls1, Erv41, Erv46, and Yet3.
In vitro assays
In vitro COPII budding assays were performed by incubation of ER microsomes in the presence or absence of purified COPII proteins (Sar1, Sec23-24, and Sec13-31) at 25°C for 20 min as previously described (Barlowe et al., 1994). After centrifugation of these reactions at 14,000 rpm in a rotor (FA45-30-11; Eppendorf) to pellet ER membranes, budded vesicles in the supernatant fraction were collected by centrifugation in a TLA100.3 at 55,000 rpm for 12 min. The resulting total and vesicle pellet fractions were heated at 75°C for 5 min in 35 µl of SDS-PAGE buffer, resolved on 10.5% polyacrylamide gels, and immunoblotted for Gls1, Erv41, Erv46, and Yet3.
For GST pull-down assays, fast protein liquid chromatography–purified GST-Gls1 and bacterially expressed GST from plasmid pGEX-2T (GE Healthcare) were immobilized on glutathione-agarose beads. First, 100 µl of glutathione-agarose beads (50% vol/vol; Sigma-Aldrich) were washed twice with 1 ml of TBS by centrifugation at 4°C for 1 min in a rotor (FA45-30-11) at 14,000 rpm. The washed glutathione-agarose beads were incubated with 1 ml of purified GST proteins (∼0.1 mg/ml) and incubated at 4°C for 2 h with rotation. After incubation, the beads were washed twice with 1 ml of TBS and washed once with 1 ml of PD buffer (pH 5.5, 6.0, and 6.5; 50 mM MES and 150 mM NaCl, pH 7.0 and 7.5; and 50 mM Tris-HCl and 150 mM NaCl). Microsomes (100 µl of 16 OD280U/ml) were solubilized with 1% Triton X-100 in 1 ml of each PD buffer containing 1 mM PMSF on ice for 10 min and centrifuged for 5 min at 14,000 rpm in a FA45-30-11 rotor at 4°C. The supernatants were incubated with GST-bound glutathione-agarose beads at 4°C overnight with rotation. Agarose beads were centrifuged at 4°C for 1 min in a FA45-30-11 rotor at 14,000 rpm, washed twice with 1 ml of each PD buffer containing 1% Triton X-100, eluted with 50 µl of SDS-PAGE buffer, and resolved on 10.5% polyacrylamide gels. To compare the GST protein levels bound to the agarose, eluted samples were immunoblotted on a single blot using GST-Yet3 antiserum for detection of GST and GST-Gls1 proteins.
IP assay
Protein A Dynabeads (Novex) were washed with 1 ml of IP buffer (pH 5.5, 6.0, and 6.5; 50 mM MES and 150 mM NaCl, pH 7.0 and 7.5; and 50 mM Tris-HCl and 150 mM NaCl). Dynabeads (30 µl of 30 mg/ml) were incubated with 1 µl of anti-Gls1 antiserum for 20 min at room temperature with rotation followed by washing with 1 ml of IP buffer. To prepare membrane extracts, 0.1 ml of semi-intact cells (75 OD280U/ml) were solubilized with 1% Triton X-100 in 1 ml of IP buffer containing 1 mM PMSF for 10 min on ice followed by a centrifugation for 5 min at 14,000 rpm in a FA45-30-11 rotor at 4°C. The supernatants were mixed with antibody-bound Dynabeads and incubated for 5 h at 4°C with rotation. The beads were washed five times with cold IP buffer, eluted by heating at 75°C for 5 min in 30 µl of SDS-PAGE sample buffer, and resolved on 10.5% polyacrylamide gels. For mixing-IP assays, 0.1-ml semi-intact cells were solubilized with 1% Triton X-100 in 0.5 ml of IP buffer containing 1 mM PMSF. The supernatants from solubilized semi-intact cells were combined just before mixing with the antibody-bound Dynabeads.
Indirect immunofluorescence microscopy
For indirect immunofluorescence microscopy as previously described (Powers and Barlowe, 1998), formaldehyde-fixed yeast cells were converted to spheroplasts and adhered to polylysine-coated slides. Cells were washed and incubated at room temperature in blocking buffer (1% BSA and 0.5% Triton X-100 in phosphate-buffered saline) followed by development with primary antibodies. The anti-HA monoclonal antibody (HA.11) was used at 1:200 dilution. Polyclonal sera against Kar2 and ALP were used at 1:400 and 1:200, respectively. Secondary fluorescein-conjugated anti–rabbit IgG and Texas red–conjugated anti–mouse IgG antibodies were obtained from Vector Laboratories and used at 1:200. Images were obtained at room temperature using a DeltaVision Imaging System (Applied Precision), comprising a customized inverted wide-field microscope (IX-71; Olympus), an UPlanS Apochromat 100×/1.40 NA lens (Olympus) with DeltaVision immersion oil (n = 1.516; GE Healthcare), a camera (CoolSNAP HQ2; Photometrics), and an Insight solid-state illumination unit. Images were processed by deconvolution in SoftWoRx (Applied Precision) and prepared with Photoshop (Adobe) and ImageJ (National Institutes of Health).
Mass spectrometry–based quantitative proteomics
To determine protein abundance fold changes, WT (BY4742) and isogenic erv41Δ (CBY1168) and erv46Δ (CBY3612) strains were compared in SILAC experiments following the protocol described in Fröhlich et al. (2013). In brief, cell labeling was conducted in synthetic medium at 30°C. Equal amounts (40 OD600 units) of light-labeled control and heavy-labeled mutant cells (erv41Δ and erv46Δ) were mixed before cell lysis. Cells were lysed in buffer containing 50 mM Tris-HCl, pH 9.0, 5% SDS, and 100 mM DTT for 30 min at 55°C. Lysates were cleared by centrifugation at 17,000 g for 10 min. Supernatants were diluted with buffer UA (8 M urea and 0.1 M Tris-HCl, pH 8.5) to a final concentration of 0.5% SDS. Proteins were digested with the endoproteinase LysC by filter-aided sample preparation. Peptides were recovered in 0.5 M NaCl, acidified by the addition of trifluoroacetic acid, and cleared of precipitates via centrifugation at 17,000 g for 5 min. Peptides (5 µg) were desalted and injected into the high-performance liquid chromatography. Reversed-phase chromatography was performed on a liquid chromatograph system (EASY-nLC 1000; Thermo Fisher Scientific) connected to a mass spectrometer (Q Exactive; Thermo Fisher Scientific) through a nano-electrospray ion source. Eluted peptides from the column were directly electrosprayed into the mass spectrometer. Mass spectra were acquired on the Q Exactive in a data-dependent mode to automatically switch between full scan mass spectrometry and ≤10 data-dependent tandem mass spectrometry scans. The resulting mass spectrometry and tandem mass spectrometry spectra were analyzed using MaxQuant (version 1.3.0.5), using its integrated Andromeda search algorithms. Triplicate liquid chromatography runs coupled to mass spectrometry were performed for independent biological duplicates. Data analysis was performed, and significance values were determined as previously described (Fröhlich et al., 2013).
Fpr2 and Gls1 secretion
Extracellular secretion of proteins was analyzed as previously described (Belden and Barlowe, 2001). In brief, cells were precultured in YPD or complete synthetic media and back diluted in YPD to an OD600 of 0.15, and cultures were grown to an OD600 of ∼0.8. Cultures were then centrifuged at 14,000 rpm, and proteins contained in the supernatant fraction (1.3 ml) were precipitated by addition of TCA to a final concentration of 10%. TCA-precipitated proteins were collected by centrifugation, washed with 96% ethanol, resolved on 15% polyacrylamide gels, and immunoblotted for Gls1, Erv41, Yet3, and Fpr2.
Bafilomycin A1 treatment
WT (CBY740) cells were grown in YPD to ∼0.5 OD600, and 2 mM bafilomycin A1 in DMSO was added to a final concentration of 20 µM. After treatment for 2 h, cells were harvested and separated into intracellular and extracellular fractions. Pellets in the intracellular fraction were lysed, and proteins secreted to the extracellular fraction were precipitated by TCA as described in the previous section.
Online supplemental material
Fig. S1 shows that the erv46Δ mutation reduces cellular levels of Gls1 similarly to erv41Δ and is consistent with a functional requirement for both subunits of the Erv41–Erv46 complex. Fig. S2 indicates that the absence of Erv41 causes mislocalization of Gls1 to vacuoles by visualizing HA-Gls1 colocalized with the vacuolar marker ALP in an immunofluorescence microscopy experiment. Fig. S3 shows that mutation of a C-terminal COPI binding motif in Erv46 causes Gls1 mislocalization. Table S1 lists the plasmids used in this study. Table S2 lists primers used in this study. Table S3 provides the mass spectrometry peptide data for the erv41Δ and erv46Δ strains from SILAC experiments and proteome changes sorted from smallest to largest log2 (erv41Δ/control) scores and is provided online in an Excel (Microsoft) spreadsheet. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201408024/DC1.
Supplementary Material
Acknowledgments
We thank the Moseley laboratory for assistance with fluorescence microscopy.
This work was supported by National Institutes of Health grants GM052549 (to C. Barlowe) and GM095982/GM097194 (to T.C. Walther).
The authors declare no competing financial interests.
Footnotes
Abbreviation used in this paper:
- ALP
- alkaline phosphatase
- COPI
- coat protein complex I
- COPII
- coat protein complex II
- CPY
- carboxypeptidase Y
- Gls1
- glucosidase I
- IP
- immunoprecipitation
- mCPY
- mature CPY
- SILAC
- stable isotope labeling by amino acids in culture
- TAP
- tandem affinity purification
- WT
- wild type
References
- Ammerer G., Hunter C.P., Rothman J.H., Saari G.C., Valls L.A., and Stevens T.H.. 1986. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 6:2490–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller C., Andersson H., Kappeler F., and Hauri H.-P.. 1999. The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1:330–334 10.1038/14020 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Roche A.-C., Nufer O., and Hauri H.-P.. 2004. pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J. Biol. Chem. 279:12943–12950 10.1074/jbc.M313245200 [DOI] [PubMed] [Google Scholar]
- Baker D., Hicke L., Rexach M., Schleyer M., and Schekman R.. 1988. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 54:335–344 10.1016/0092-8674(88)90196-1 [DOI] [PubMed] [Google Scholar]
- Banta L.M., Robinson J.S., Klionsky D.J., and Emr S.D.. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369–1383 10.1083/jcb.107.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., and Schekman R.. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895–907 10.1016/0092-8674(94)90138-4 [DOI] [PubMed] [Google Scholar]
- Belden W.J., and Barlowe C.. 2001. Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell. 12:957–969 10.1091/mbc.12.4.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biterova E.I., Svärd M., Possner D.D.D., and Guy J.E.. 2013. The crystal structure of the lumenal domain of Erv41p, a protein involved in transport between the endoplasmic reticulum and Golgi apparatus. J. Mol. Biol. 425:2208–2218 10.1016/j.jmb.2013.03.024 [DOI] [PubMed] [Google Scholar]
- Bowman E.J., Siebers A., and Altendorf K.. 1988. Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA. 85:7972–7976 10.1073/pnas.85.21.7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F., and Barlowe C.. 2013. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14:382–392 10.1038/nrm3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuza L., Halbeisen R., Jenö P., Otte S., Barlowe C., Hong W., and Hauri H.-P.. 2004. Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279:47242–47253 10.1074/jbc.M406644200 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., and Schekman R.. 1993. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123:1355–1363 10.1083/jcb.123.6.1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bue C.A., Bentivoglio C.M., and Barlowe C.. 2006. Erv26p directs pro-alkaline phosphatase into endoplasmic reticulum-derived coat protein complex II transport vesicles. Mol. Biol. Cell. 17:4780–4789 10.1091/mbc.E06-05-0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., and Hieter P.. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene. 110:119–122 10.1016/0378-1119(92)90454-W [DOI] [PubMed] [Google Scholar]
- Cosson P., and Letourneur F.. 1994. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 263:1629–1631 10.1126/science.8128252 [DOI] [PubMed] [Google Scholar]
- Dancourt J., and Barlowe C.. 2010. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79:777–802 10.1146/annurev-biochem-061608-091319 [DOI] [PubMed] [Google Scholar]
- Dhanawansa R., Faridmoayer A., van der Merwe G., Li Y.X., and Scaman C.H.. 2002. Overexpression, purification, and partial characterization of Saccharomyces cerevisiae processing α glucosidase I. Glycobiology. 12:229–234 10.1093/glycob/12.3.229 [DOI] [PubMed] [Google Scholar]
- Elble R.1992. A simple and efficient procedure for transformation of yeasts. Biotechniques. 13:18–20. [PubMed] [Google Scholar]
- Eriksson P., Thomas L.R., Thorburn A., and Stillman D.J.. 2004. pRS yeast vectors with a LYS2 marker. Biotechniques. 36:212–213. [DOI] [PubMed] [Google Scholar]
- Faridmoayer A., and Scaman C.H.. 2004. An improved purification procedure for soluble processing α-glucosidase I from Saccharomyces cerevisiae overexpressing CWH41. Protein Expr. Purif. 33:11–18 10.1016/j.pep.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Forgac M.2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Fröhlich F., Christiano R., and Walther T.C.. 2013. Native SILAC: metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol. Cell. Proteomics. 12:1995–2005 10.1074/mcp.M112.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.-K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., and Weissman J.S.. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Haas A., Scheglmann D., Lazar T., Gallwitz D., and Wickner W.. 1995. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 14:5258–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmings B.A., Zubenko G.S., Hasilik A., and Jones E.W.. 1981. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 78:435–439 10.1073/pnas.78.1.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., and Carr A.M.. 2005. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 22:1013–1019 10.1002/yea.1291 [DOI] [PubMed] [Google Scholar]
- Hitt R., and Wolf D.H.. 2004. DER7, encoding α-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 4:815–820 10.1016/j.femsyr.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Huh W.-K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jackson L.P., Lewis M., Kent H.M., Edeling M.A., Evans P.R., Duden R., and Owen D.J.. 2012. Molecular basis for recognition of dilysine trafficking motifs by COPI. Dev. Cell. 23:1255–1262 10.1016/j.devcel.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B., Sheraton J., Ram A.F., Dijkgraaf G.J., Klis F.M., and Bussey H.. 1996. CWH41 encodes a novel endoplasmic reticulum membrane N-glycoprotein involved in β 1,6-glucan assembly. J. Bacteriol. 178:1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., and Schuldiner M.. 2009. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 323:1693–1697 10.1126/science.1167983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J., Gilstring C.F., and Ljungdahl P.O.. 2007. Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD. J. Cell Biol. 176:617–628 10.1083/jcb.200612100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M.J., Herrmann J.M., and Schekman R.. 1998. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 391:187–190 10.1038/34438 [DOI] [PubMed] [Google Scholar]
- Le Tallec B., Barrault M.-B., Courbeyrette R., Guérois R., Marsolier-Kergoat M.-C., and Peyroche A.. 2007. 20S proteasome assembly is orchestrated by two distinct pairs of chaperones in yeast and in mammals. Mol. Cell. 27:660–674 10.1016/j.molcel.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Lewis M.J., and Pelham H.R.B.. 1990. A human homologue of the yeast HDEL receptor. Nature. 348:162–163 10.1038/348162a0 [DOI] [PubMed] [Google Scholar]
- Llopis J., McCaffery J.M., Miyawaki A., Farquhar M.G., and Tsien R.Y.. 1998. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 95:6803–6808 10.1073/pnas.95.12.6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Losev E., Reinke C.A., Jellen J., Strongin D.E., Bevis B.J., and Glick B.S.. 2006. Golgi maturation visualized in living yeast. Nature. 441:1002–1006 10.1038/nature04717 [DOI] [PubMed] [Google Scholar]
- Macreadie I.G., Horaitis O., Verkuylen A.J., and Savin K.W.. 1991. Improved shuttle vectors for cloning and high-level Cu(2+)-mediated expression of foreign genes in yeast. Gene. 104:107–111 10.1016/0378-1119(91)90474-P [DOI] [PubMed] [Google Scholar]
- Moremen K.W., Trimble R.B., and Herscovics A.. 1994. Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology. 4:113–125 10.1093/glycob/4.2.113 [DOI] [PubMed] [Google Scholar]
- Mossessova E., Bickford L.C., and Goldberg J.. 2003. SNARE selectivity of the COPII coat. Cell. 114:483–495 10.1016/S0092-8674(03)00608-1 [DOI] [PubMed] [Google Scholar]
- Munro S., and Pelham H.R.B.. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 48:899–907 10.1016/0092-8674(87)90086-9 [DOI] [PubMed] [Google Scholar]
- Nielsen J.B., Foor F., Siekierka J.J., Hsu M.J., Ramadan N., Morin N., Shafiee A., Dahl A.M., Brizuela L., Chrebet G., et al. 1992. Yeast FKBP-13 is a membrane-associated FK506-binding protein encoded by the nonessential gene FKB2. Proc. Natl. Acad. Sci. USA. 89:7471–7475 10.1073/pnas.89.16.7471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., and von Heijne G.. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 10.1093/protein/10.1.1 [DOI] [PubMed] [Google Scholar]
- Noda Y., Hara T., Ishii M., and Yoda K.. 2014. Distinct adaptor proteins assist exit of Kre2-family proteins from the yeast ER. Biol. Open. 3:209–224 10.1242/bio.20146312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orci L., Ravazzola M., Mack G.J., Barlowe C., and Otte S.. 2003. Mammalian Erv46 localizes to the endoplasmic reticulum-Golgi intermediate compartment and to cis-Golgi cisternae. Proc. Natl. Acad. Sci. USA. 100:4586–4591 10.1073/pnas.0730885100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S., and Barlowe C.. 2002. The Erv41p-Erv46p complex: multiple export signals are required in trans for COPII-dependent transport from the ER. EMBO J. 21:6095–6104 10.1093/emboj/cdf598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte S., Belden W.J., Heidtman M., Liu J., Jensen O.N., and Barlowe C.. 2001. Erv41p and Erv46p: new components of COPII vesicles involved in transport between the ER and Golgi complex. J. Cell Biol. 152:503–518 10.1083/jcb.152.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroutis P., Touret N., and Grinstein S.. 2004. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology (Bethesda). 19:207–215 10.1152/physiol.00005.2004 [DOI] [PubMed] [Google Scholar]
- Partaledis J.A., and Berlin V.. 1993. The FKB2 gene of Saccharomyces cerevisiae, encoding the immunosuppressant-binding protein FKBP-13, is regulated in response to accumulation of unfolded proteins in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 90:5450–5454 10.1073/pnas.90.12.5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H.R., Hardwick K.G., and Lewis M.J.. 1988. Sorting of soluble ER proteins in yeast. EMBO J. 7:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., and Barlowe C.. 1998. Transport of axl2p depends on erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J. Cell Biol. 142:1209–1222 10.1083/jcb.142.5.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers J., and Barlowe C.. 2002. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol. Biol. Cell. 13:880–891 10.1091/mbc.01-10-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raykhel I., Alanen H., Salo K., Jurvansuu J., Nguyen V.D., Latva-Ranta M., and Ruddock L.. 2007. A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179:1193–1204 10.1083/jcb.200705180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P.A., Dijkgraaf G.J.P., Shahinian S., Herscovics A., and Bussey H.. 1997. The yeast CWH41 gene encodes glucosidase I. Glycobiology. 7:997–1004 10.1093/glycob/7.7.997 [DOI] [PubMed] [Google Scholar]
- Rothblatt J.A., Deshaies R.J., Sanders S.L., Daum G., and Schekman R.. 1989. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 109:2641–2652 10.1083/jcb.109.6.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K., and van Deurs B.. 2002. Membrane traffic exploited by protein toxins. Annu. Rev. Cell Dev. Biol. 18:1–24 10.1146/annurev.cellbio.18.011502.142107 [DOI] [PubMed] [Google Scholar]
- Sato K., Sato M., and Nakano A.. 1997. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc. Natl. Acad. Sci. USA. 94:9693–9698 10.1073/pnas.94.18.9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel A.A., and Pelham H.R.B.. 1996. Purification and characterization of the human KDEL receptor. Biochemistry. 35:10203–10209 10.1021/bi960807x [DOI] [PubMed] [Google Scholar]
- Semenza J.C., Hardwick K.G., Dean N., and Pelham H.R.B.. 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 61:1349–1357 10.1016/0092-8674(90)90698-E [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., and Schekman R.. 1982. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 30:439–448 10.1016/0092-8674(82)90241-0 [DOI] [PubMed] [Google Scholar]
- Thor F., Gautschi M., Geiger R., and Helenius A.. 2009. Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic. 10:1819–1830 10.1111/j.1600-0854.2009.00989.x [DOI] [PubMed] [Google Scholar]
- Ward A.C., Castelli L.A., Macreadie I.G., and Azad A.A.. 1994. Vectors for Cu(2+)-inducible production of glutathione S-transferase-fusion proteins for single-step purification from yeast. Yeast. 10:441–449 10.1002/yea.320100403 [DOI] [PubMed] [Google Scholar]
- Welsh L.M., Tong A.H.Y., Boone C., Jensen O.N., and Otte S.. 2006. Genetic and molecular interactions of the Erv41p-Erv46p complex involved in transport between the endoplasmic reticulum and Golgi complex. J. Cell Sci. 119:4730–4740 10.1242/jcs.03250 [DOI] [PubMed] [Google Scholar]
- Wilson D.W., Lewis M.J., and Pelham H.R.. 1993. pH-dependent binding of KDEL to its receptor in vitro. J. Biol. Chem. 268:7465–7468. [PubMed] [Google Scholar]
- Wilson J.D., and Barlowe C.. 2010. Yet1p and Yet3p, the yeast homologs of BAP29 and BAP31, interact with the endoplasmic reticulum translocation apparatus and are required for inositol prototrophy. J. Biol. Chem. 285:18252–18261 10.1074/jbc.M109.080382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Dollard C., and Ricupero-Hovasse S.L.. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 11:53–55 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 285:901–906 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Wuestehube L.J., and Schekman R.W.. 1992. Reconstitution of transport from endoplasmic reticulum to Golgi complex using endoplasmic reticulum-enriched membrane fraction from yeast. Methods Enzymol. 219:124–136 10.1016/0076-6879(92)19015-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.