Arp2 phosphorylation by the kinase NIK increases the actin-nucleating activity of the Arp2/3 complex and is necessary for plasma membrane protrusion in response to EGF.
Abstract
The nucleating activity of the Arp2/3 complex promotes the assembly of branched actin filaments that drive plasma membrane protrusion in migrating cells. Arp2/3 complex binding to nucleation-promoting factors of the WASP and WAVE families was previously thought to be sufficient to increase nucleating activity. However, phosphorylation of the Arp2 subunit was recently shown to be necessary for Arp2/3 complex activity. We show in mammary carcinoma cells that mutant Arp2 lacking phosphorylation assembled with endogenous subunits and dominantly suppressed actin filament assembly and membrane protrusion. We also report that Nck-interacting kinase (NIK), a MAP4K4, binds and directly phosphorylates the Arp2 subunit, which increases the nucleating activity of the Arp2/3 complex. In cells, NIK kinase activity was necessary for increased Arp2 phosphorylation and plasma membrane protrusion in response to epidermal growth factor. NIK is the first kinase shown to phosphorylate and increase the activity of the Arp2/3 complex, and our findings suggest that it integrates growth factor regulation of actin filament dynamics.
Introduction
The seven subunit actin-related protein (Arp) 2/3 complex nucleates actin filaments that drive diverse cell processes such as vesicle trafficking, plasma membrane protrusion during cell migration, and motility of some pathogens in host cells (Goley and Welch, 2006). Hence, resolving how activity of the Arp2/3 complex is regulated is important for a basic understanding of diverse cell processes and for targeting aberrant actin-dependent cell behaviors in diseases. Arp2/3 complex binding to nucleation promoting factors (NPFs) such as N-WASP was previously thought to be sufficient to increase nucleating activity (Welch and Mullins, 2002). However, we recently showed that the Arp2 subunit must be phosphorylated for the complex to be activated by NPFs (LeClaire et al., 2008; Narayanan et al., 2011; Choi et al., 2013). The necessary role for Arp2 phosphorylation for nucleation activity suggests that the Arp2/3 complex is a coincidence detector requiring both binding to NPFs and phosphorylation of Arp2 for increased activity. Coincidence detection is emerging as a common regulatory mechanism for many actin-binding proteins, including NPFs (Rohatgi et al., 2001; Rivera et al., 2009) and cofilin (Frantz et al., 2008; Magalhaes et al., 2011). In support of coincidence regulation of the Arp2/3 complex, we show that expression of a phosphorylation-defective Arp2 mutant complexes with endogenous Arp2/3 complex subunits and dominantly suppresses increased actin filament assembly and membrane protrusion in response to EGF.
Despite the confirmed importance of phosphorylated Arp2 in the nucleation activity of the Arp2/3 complex, kinases phosphorylating Arp2 have not been identified. We report that the Nck-interacting kinase (NIK), a Ste20/MAP4K4 serine/threonine kinase, phosphorylates Arp2 and primes the Arp2/3 complex for activation by NPFs. NIK and its orthologues have a conserved role in regulating actin cytoskeleton-dependent cell processes. NIK activity is necessary for mesoderm migration (Xue et al., 2001) and for epithelial cell membrane protrusion (Baumgartner et al., 2006) and invasion (Wright et al., 2003). The NIK orthologue misshapen in Drosophila melanogaster functions in determining epithelial polarity, neuronal targeting, and cell invasion (Su et al., 1997; Cobreros-Reguera et al., 2010), and the orthologue MIG-15 in Caenorhabditis elegans controls axonal navigation (Poinat et al., 2002) and neuroblast migration (Chapman et al., 2008). However, a substrate for NIK or an orthologue that directly regulates actin filament assembly has not been identified. We show that NIK activity is necessary for increased Arp2 phosphorylation and membrane protrusion in response to EGF and also that NIK directly binds the Arp2/3 complex. These findings identify a new regulator for increasing Arp2/3 complex activity and suggest that NIK phosphorylation of Arp2 is a mechanism linking growth factor signaling to actin filament assembly.
Results and discussion
Arp2-T237/238A-Y202A dominantly suppresses EGF-increased actin filament assembly and membrane protrusion
Previous studies to reveal cell processes regulated by Arp2/3 complex have mostly used RNA interference or genetic depletion to eliminate expression of individual subunits (Di Nardo et al., 2005; Nicholson-Dykstra and Higgs, 2008; Suraneni et al., 2012; Wu et al., 2012). However, a limitation of these approaches is that expression of other subunits is abolished because subunit stability depends on an assembled complex. Small molecule inhibitors of Arp2/3 complex activity (Nolen et al., 2009) retain an intact complex but disrupt cortical localization of the complex (Yang et al., 2012). We asked whether these limitations could be resolved by heterologous expression of a mutant nonphosphorylatable Arp2 to dominantly inhibit endogenous Arp2/3 complex activity.
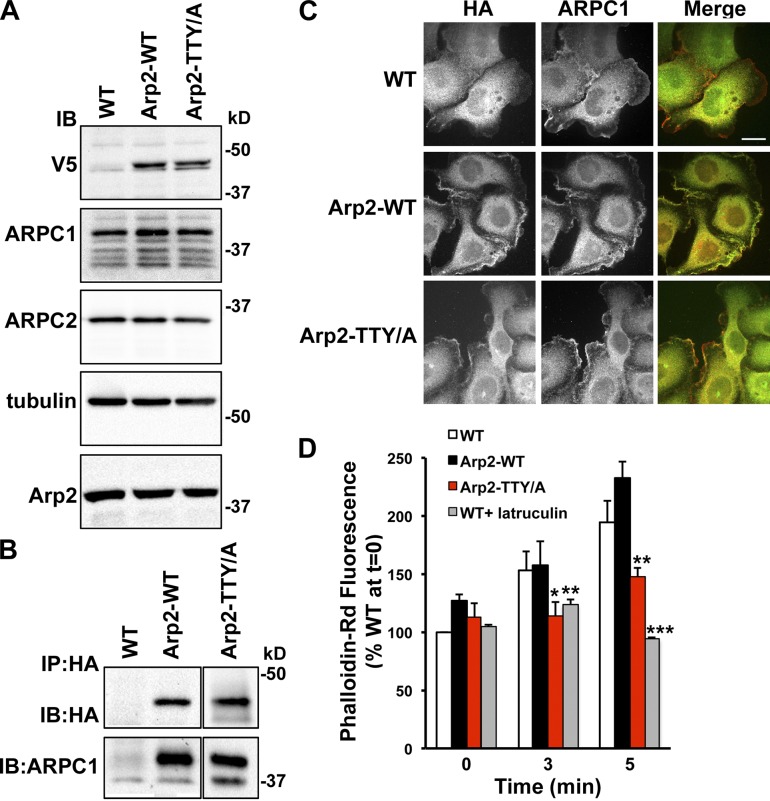
To test this prediction, we stably expressed Arp2 wild type (WT) and a T237/238A-Y202A mutant (TTY/A), tagged at the C terminus with tandem HA and V5 epitopes, in MTLn3 rat carcinoma cells. We used the triple phospho-site Arp2 mutant because we previously showed that phosphorylation of either Thr237/238 or Tyr202 functions as an OR gate for NPF-stimulated Arp2/3 complex activity (LeClaire et al., 2008; Choi et al., 2013). Immunoblotting cell lysates with antibodies to V5 showed similar expression levels of recombinant Arp2-WT and -TTY/A, and neither construct changed the abundance of endogenous ARPC1, ARPC2, or Arp2 (Fig. 1 A). ARPC1 coprecipitated with HA-immune complexes of Arp2-WT and -TTY/A (Fig. 1 B), which suggests that recombinant Arp2 assembles with endogenous Arp2/3 complex subunits.
Figure 1.
Arp2-T237/238A-Y202A coprecipitates with endogenous Arp2/3 complex subunits and dominantly suppresses EGF-increased actin filament assembly. (A) Immunoblot of lysates from WT MTLn3 cells (WT) and MTLn3 cells stably expressing recombinant Arp2 WT (Arp2-WT) and mutant T237/238A-Y202A (Arp2-TTY/A) tagged with VA and HA epitopes show that expression of recombinant Arp2 does not change the abundance of endogenous ARPC1, ARPC2, or Arp2. (B) Immunoblot of proteins in anti-HA immune complexes shows coprecipitation of endogenous ARPC1 with recombinant Arp2 WT and mutant TTY/A. (C) Immunolabeling with anti-HA and anti-ARPC1 antibodies indicates colocalization at the distal margin of plasma membrane protrusions. Bar, 5 µm. (D) Total F-actin in MTLn3 cells in the absence and presence of EGF for the indicated times shows a similar time-dependent increase in WT cells and cells expressing Arp2-WT that is significantly attenuated in cells expressing Arp2-TTY/A or WT cells treated with latrunculin. Data are means ± SEM (error bars) of triplicate determinations in four separate cell preparations. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with WT at the same time point (Student’s t test).
In MTLn3 cells maintained in growth medium, HA immunolabeling was seen at the distal margin of cells expressing Arp2-WT and -TTY/A but was not in vector control cells, although all cells showed a nonspecific cytoplasmic signal (Fig. 1 C). Co-labeling for endogenous ARPC1 showed localization at the cell distal margin and overlap with HA labeling of both Arp2-WT and -TTY/A in merged images (Fig. 1 C), which indicates that localization of the Arp2/3 complex is not disrupted by expression of Arp2-TTY/A.
Expression of Arp2-TTY/A dominantly suppressed increased actin filament assembly with EGF. In MTLn3 cells, EGF induces a marked increase in F-actin (Chan et al., 1998; Yang et al., 2012), which we confirmed by measuring fluorescence of fixed cells labeled with rhodamine phalloidin. In WT cells, a time-dependent increase in F-actin with EGF was maximal at 5 min (Fig. 1 D). At all time points with EGF, F-actin abundance was not different with expression of Arp2-WT compared with WT cells but was significantly less with expression of Arp2-TTY/A and with latrunculin B (1 µM), which binds and sequesters G-actin (Fig. 1 D). In the absence of EGF, the mean F-actin abundance was not different with Arp2-TTY/A or latrunculin compared with WT cells.
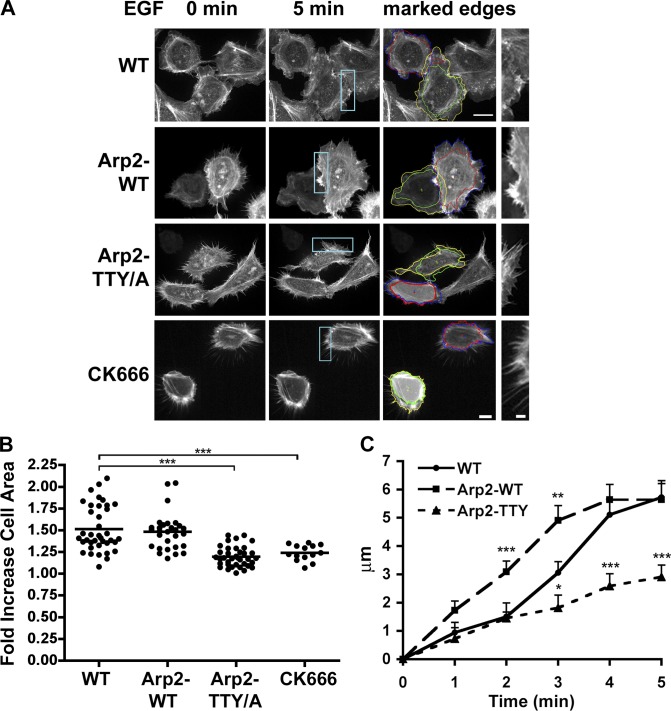
Compared with WT cells and Arp2-WT, expression of Arp2-TTY/A decreased the extent and rate of membrane protrusions with EGF. In growth medium, there were no obvious morphological differences between WT cells and cells expressing Arp2-WT or Arp2-TTY/A, as indicated in Fig. 1 C. However, marked differences were seen with Arp2-TTY/A in quiescent cells, as indicated in cells expressing the actin filament reporter Lifeact-GFP (Riedl et al., 2008). Images captured by time-lapse microscopy showed that quiescent WT and Arp2-WT cells had marked cortical actin bundles and a ruffled plasma membrane. With EGF, the cortical actin bundle remained mostly unchanged but lamellipodia rapidly extended (Fig. 2 A). Quiescent cells expressing Arp2-TTY/A mostly lacked a distinct cortical actin bundle and had fewer and smaller ruffles but an increase in filopodia containing actin filaments in linear arrays (Fig. 2 A). With EGF, extension of lamellipodia was markedly reduced but there was little change in the number or length of filopodia. We also saw this stellate phenotype with filopodia in MTLn3 cells treated with CK666 (Fig. 2 A), and, as we previously reported, in Drosophila S2 cells with RNAi knockdown of Arp2 and expression of Arp2-T237/238A-Y202A (LeClaire et al., 2008). Additionally, the stellate phenotype is seen in fibroblasts lacking ARPC2 (Wu et al., 2012) or ARPC3 (Suraneni et al., 2012) subunits. Hence, inhibiting the Arp2/3 complex may unmask actin assemblies regulated by other nucleators such as formins, which can generate contractile actin filaments. Consistent with this prediction, cells with Arp2-TTY/A and CK666 were smaller and appeared more contracted. Additional analysis showed that an increase in the area of WT and Arp2-WT cells with EGF was significantly less with Arp2-TTY/A and CK666 (Fig. 2 B). Compared with WT cells, the rate of membrane protrusion was significantly faster, with Arp2-WT at 2 and 3 min and significantly slower with Arp2-TTY/A at 3, 4, and 5 min (Fig. 2 C). It is unclear why the phenotype of cells expressing Arp2-TTY/A is different with quiescence compared with being maintained in growth medium but may reflect an unresolved role for Arp2 phosphorylation. Although cells expressing Arp2-TTY/A had a slower proliferation rate than WT and Arp2-WT cells (Fig. S1), they were viable, which is in contrast to the lethality of Dictyostelium discoideum cells lacking this subunit (Zaki et al., 2007). The ability of Arp2-TTY/A to assemble with endogenous Arp2/3 complex subunits and suppress EGF-induced actin filament assembly and membrane protrusion without disrupting localization of the complex suggests a new strategy to determine functions of the Arp2/3 complex in diverse cell processes.
Figure 2.
Expression of recombinant Arp2-TTY/A inhibits membrane protrusion with EGF. (A) Time-lapse images of MTLn3 cells expressing Lifeact-GFP in the absence (0 min) and presence (5 min) of EGF show differences in morphology and changes in plasma membrane protrusions for WT, Arp2-WT, Arp2-TTY/A, and WT cells treated with the Arp2/3 inhibitor CK666. Cells expressing Arp2-TTY/A or treated with CK666 had prominent filopodia in the absence of EGF that did not increase in size or abundance with EGF. Also shown are overlapped tracings of the cell periphery in the absence (red and green outlines) and presence of EGF (yellow and blue outlines) that were used to quantify cell area, as well as high-magnification images of the cell periphery at 5 min with EGF (shown on the right, taken from the indicated cyan boxes). Bars: (left) 5 µm; (right) 2 µm. (B) WT and Arp2-WT cells had increased areas after 5 min with EGF that was significantly less with Arp2-TTY/A and WT cells treated with the Arp2/3 complex inhibitor CK666. Data are from four separate cell preparations, with each filled circle representing a single cell (for WT, n = 39; for Arp2-WT, n = 34; for Arp2-TTY, n = 36; for CK666, n = 14). (C) The rate of membrane protrusion was significantly faster with Arp2-WT at 2 and 3 min and significantly slower with Arp2-TTY/A at 3, 4, and 5 min compared with WT cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with WT (Student’s unpaired t test).
The Arp2/3 complex is an NIK substrate
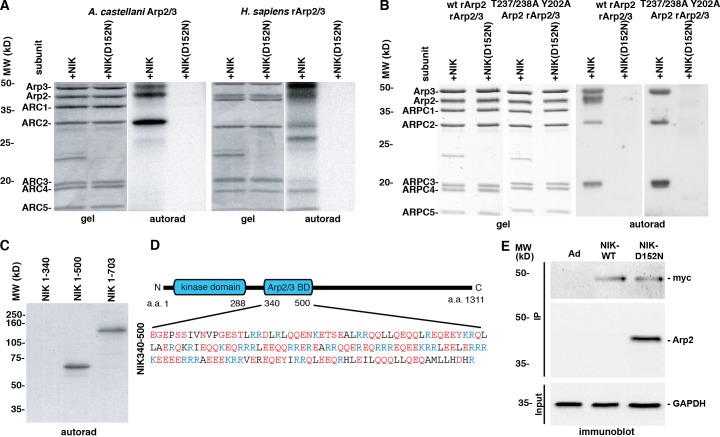
To identify kinases that phosphorylate Arp2, we used in vitro kinase assays with Arp2/3 complex purified from Acanthamoeba castellanii and dephosphorylated with Antarctic phosphatase (AP). We tested a subset of kinases previously shown to associate with the Arp2/3 complex or regulate actin or membrane dynamics, including p21-activated kinase (PAK), the Src tyrosine kinase (Src), focal adhesion kinase (FAK), and NIK. Neither PAK1, a Ste20 family member like NIK, nor FAK phosphorylated any Arp2/3 complex subunit, although they did phosphorylate the generic substrate myelin basic protein (unpublished data). Src phosphorylated Arp3 and Arp2 subunits (Fig. S2 A), but as described below it had no effect on nucleating activity. In contrast, NIK phosphorylated Arp3, Arp2, and ARPC2 subunits of the A. castellanii Arp2/3 complex and recombinant human Arp2/3 complex generated by baculovirus expression (Fig. 3 A). NIK also phosphorylated the ARPC5 subunit of human but not A. castellanii Arp2/3 complex. Phosphorylation of subunits was not detected in the absence of NIK or with a mutant kinase-inactive NIK (NIK-D152N) that lacks ATP binding but retains substrate binding (Baumgartner et al., 2006; Fig. 3 A).
Figure 3.
NIK phosphorylates and binds the Arp2/3 complex. (A) In vitro, WT NIK but not kinase-inactive NIK-D152N phosphorylates Arp3, Arp2, and ARPC1 subunits of the Arp2/3 complex purified from A. castellanii and from baculovirus expression of human recombinant Arp2/3 complex (rArp2/3). (B) NIK phosphorylation of Arp2 but not Arp3 and ARPC3 subunits is not seen with rArp2/3 complex containing a mutant Arp2-T237/238A-Y202A. (C) Arp2/3 complex coupled to CH-Sepharose and incubated with recombinant NIK 1–340, NIK 1–500, and NIK 1–703 shows that the NIK region of amino acids 340–500 binds the Arp2/3 complex. (D) Schematic of NIK showing the kinase domain and proposed Arp2/3 complex binding domain, which contains abundant acidic (red) and basic (blue) residues. (E) Arp2 coprecipitates in Myc immune complexes of MTLn3 cells transiently expressing Myc-tagged NIK-D152N by not with Myc-tagged WT NIK or empty adenovirus (Ad).
To test whether NIK, a Ser/Thr kinase, phosphorylates Arp2-Thr237/238, we used as a substrate recombinant Arp2/3 (rArp2/3) complex containing Arp2-WT or -TTY/A, generated by baculovirus expression as we described previously (Narayanan et al., 2011). Recombinant Arp2-TTY/A had no effect on the stoichiometric assembly of rArp2/3 complex subunits (Fig. 3 B, gel). Active NIK but not NIK-D152N phosphorylated Arp3, Arp2, and ARPC2 subunits of WT rArp2/3 complex (Fig. 3 B). With mutant Arp2-T237/238A-Y202A, NIK phosphorylation of Arp2 was abolished but phosphorylation of Arp3 and ARPC2 was unchanged compared with WT. Kinetic analysis revealed a Km of 37.7 ± 8 µM for total phosphorylation of the Arp2/3 complex by NIK (Fig. S2 C). These data confirm Thr237 and Thr238 as the Arp2 residues phosphorylated by NIK in vitro. The significance of NIK-phosphorylated residues in Arp3 and ARPC2 is unknown; however, as described below, NIK-dependent phosphorylation of Arp2 but not Arp3 or ARPC2 was seen in cells (see Fig. 5).
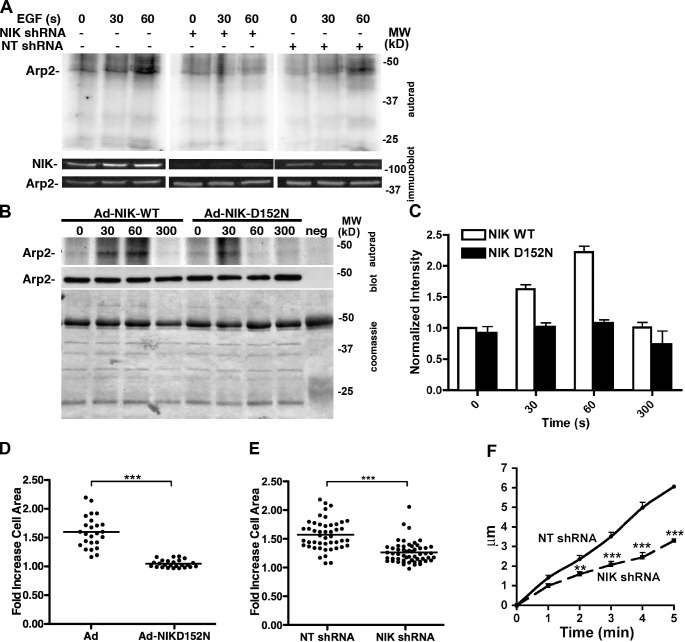
Figure 5.
NIK activity is necessary for increased Arp2 phosphorylation with EGF in MTLn3 cells. (A) The Arp2/3 complex was immunoprecipitated from 32P-labeled MTLn3 cells stimulated with EGF. Increased Arp2 phosphorylation with EGF for 60 s in ARPC1 immune complexes from WT MTLn3 cells and with nontargeting shRNA (NT shRNA) was not seen in cells expressing NIK shRNA. Top, autorad indicating 32P incorporation; bottom, immunoblot for Arp2. (B) Phosphorylation of Arp2, determined as in A, increased in cells infected with control Ad-NIK WT but not with Ad-NIK-D152N-GFP. (C) Arp2 phosphorylation after EGF stimulation increased approximately twofold more in cells expressing NIK WT compared with NIK D152N (error bars indicate SD). (D and E) In response to EGF, the fold increase in cell area with Ad-GFP and NT-shRNA was significantly less with Ad-NIK-D152N-GFP and NIK shRNA. (F) Rates of membrane protrusion with EGF were significantly less with NIK shRNA compared with NT shRNA. Data represent means of values from 25–50 cells in three to four separate cell preparations. **, P < 0.01; ***, P < 0.001 (Student’s t test).
The C-terminal regulatory domain of NIK directly binds substrates, including the mitogen-activated protein kinase kinase MEKK1, within residues 906–1,233 (Su et al., 1997), the ERM proteins ezrin, radixin, and moesin within 288–321 (Baumgartner et al., 2006), and the Na-H exchanger NHE1 within 407–520 (Yan et al., 2001). Arp2/3 complex immobilized on CH-Sepharose bound in vitro–translated NIK 1–500 and NIK 1–703 but not NIK 1–340 (Fig. 3 C), which suggests that there is an Arp2/3 complex binding region within amino acids 340–500 that overlaps with the NHE1-binding region. A. castellanii Arp2/3 complex coprecipitated with GST-NIK 340–500 with an apparent Kd ∼0.213 µM (Fig. S2 D), which is similar to high-affinity binding by other Arp2/3-binding proteins, including Scar (Pan et al., 2004; LeClaire et al., 2008) and N-WASP (Kelly et al., 2006; LeClaire et al., 2008). The Arp2/3 complex binding region in NIK includes multiple polybasic regions with the motif KRRX (Fig. 3 D). These regions are similar to those found in ActA, WASP, N-WASP, and Scar that are necessary to bind the complex (Marchand et al., 2001). When transiently expressed in MTLn3 cells, Myc-tagged NIK-D152N but not WT NIK coprecipitated endogenous Arp2 (Fig. 3 E), which suggests binding in cells and also that with phosphorylation by WT NIK, the Arp2/3 complex may undergo conformational changes that disrupt binding in cells.
NIK-phosphorylated Arp2/3 complex has increased nucleating activity
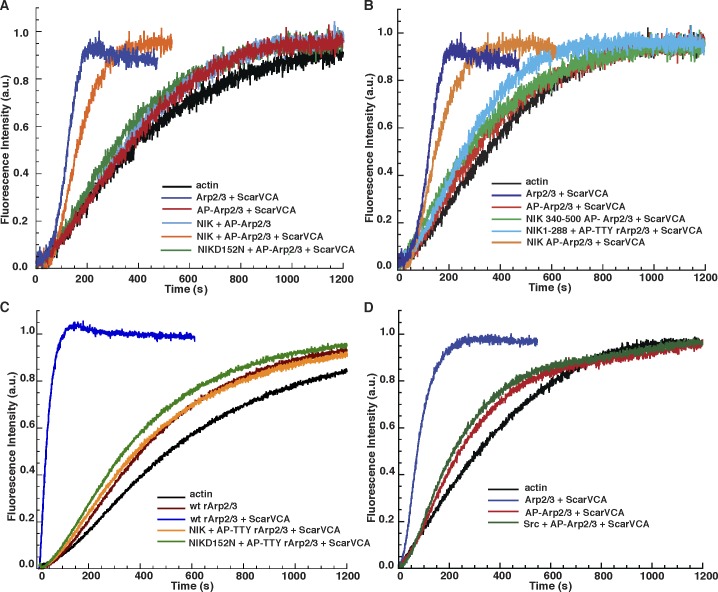
Based on computational modeling and in vitro studies with rArp2/3 complex, we previously proposed a model predicting that phosphorylation of Arp2 Thr236/237 disrupts an electrostatic interaction with ARPC4 Arg104/105 to relieve an autoinhibited Arp2/3 complex and allow activation by NPFs (Narayanan et al., 2011). Consistent with this model, full-length WT NIK but not NIK-D152N restored the actin nucleation activity of Arp2/3 complex dephosphorylated with AP to ∼80% of the Arp2/3 complex rate without AP treatment (Fig. 4 A). The time to half maximal actin polymerization (T1/2) of AP-treated Arp2/3 complex was reduced from 900 s to <400 s with NIK (Fig. S3). The NIK kinase domain (NIK 1–288) lacking the Arp2/3 complex binding site increased Arp2/3 activity to 30% of untreated Arp2/3 complex, which suggests that NIK-increased nucleating activity is in part dependent on binding the Arp2/3 complex (Fig. 4 B). However, NIK 340–500 had no effect on the nucleating activity of untreated or AP-treated Arp2/3 complex (Fig. 4 B), which indicates that the binding domain without the kinase domain is not sufficient to activate the complex. Additionally, rArp2/3 with Arp2-TTY/A was inactive in the presence of Scar-VCA both with and without NIK (Fig. 4 C), further confirming that the Arp2 phosphorylation sites T237/238-Y202 are necessary for NIK-increased Arp2/3 complex nucleating activity. Although Src phosphorylated several Arp2/3 complex subunits (Fig. S1 A), it did not restore the nucleation activity of the AP-Arp2/3 complex (Fig. 4 D). The functional significance of Src-increased phosphorylation and whether Src phosphorylates Arp2-Y202 remain to be determined.
Figure 4.
NIK restores the actin nucleation activity of the Arp2/3 complex. (A) Pyrene actin assays show that WT NIK but not kinase-inactive NIK-D152N restores activity of alkaline phosphatase–treated Arp2/3 complex with but not without the VCA domain of Scar. (B) The NIK kinase domain (NIK 1–288) lacking Arp2/3 complex binding sites activates Arp2/3 complex to ∼30% of the rate of Arp2/3 complex phosphorylated by full-length NIK. The NIK Arp2/3 binding domain (NIK 340–500) lacking the kinase domain has no effect on Arp2/3 complex activity. (C) Actin-nucleation activity of Arp2/3 complex containing T237/238A-Y202A Arp2 is not restored with NIK. (D) Src does not restore activity of dephosphorylated Arp2/3 complex. The data shown for pyrene actin traces are from a single representative experiment out of three repeats.
NIK activity is necessary for EGF-increased Arp2 phosphorylation and membrane protrusion
Arp2 phosphorylation increases in MTLn3 mammary carcinoma cells treated with EGF (LeClaire et al., 2008) and in D. discoideum cells treated with cAMP (Choi et al., 2013); however, a kinase mediating either effect has not been identified. To test a possible role for NIK, we used metabolically labeled MTLn3 cells stably expressing shRNA for NIK or a nontargeting (NT) shRNA, confirming the specificity of decreased NIK expression by immunoblotting (Fig. 5 A). Incorporation of [32P]orthophosphate into Arp2/3 complex subunits was determined in immune complexes obtained using ARPC1 subunit antibodies. Quiescent control and NT cells had a low basal level of phosphorylated Arp2 that was markedly increased with EGF by 60 s (Fig. 5 A). A second phosphorylated polypeptide migrating at ∼45 kD did not correspond to Arp2/3 complex subunits and was not identified. Phosphorylation of Arp3 and ARPC2, subunits that NIK phosphorylated in vitro (Fig. 3 A), was not detected in cells. Cells expressing NIK shRNA had lower basal phosphorylation of Arp2 that did not increase with EGF (Fig. 5 A). Arp2 phosphorylation with EGF was also markedly less in MTLn3 cells infected with adenovirus expressing kinase-inactive NIK-D152N-GFP compared with adenovirus controls (Fig. 5, B and C), which suggests the significance of NIK activity and not just NIK binding.
Consistent with NIK increasing phosphorylation and activity of the Arp2/3 complex in vitro, the EGF-induced increase in MTLn3 cell area seen in controls was significantly attenuated with expression of NIK-D152N-GFP and NIK shRNA (Fig. 5, D and C; and Fig. S3 B). Additionally, the rate of membrane protrusion with EGF was significantly less with NIK shRNA compared with NT shRNA (Fig. 5 F). We previously showed that NIK-dependent membrane protrusion with EGF is in part dependent on NIK phosphorylation of ERM proteins (Baumgartner et al., 2006). The ERM protein ezrin stabilizes membrane blebs by tethering cortical actin filaments to the plasma membrane (Charras et al., 2006). Hence, we propose that in response to EGF, NIK activates the Arp2/3 complex to initiate membrane protrusions, which are stabilized by NIK activation of ERM proteins.
Our findings identify NIK as the first kinase shown to both phosphorylate Arp2/3 complex subunits and increase nucleation activity. Of particular interest is that in addition to NIK binding the Arp2/3 complex, it also binds and phosphorylates ERM proteins (Baumgartner et al., 2006) and NHE1 (Yan et al., 2001), which also regulate actin dynamics at the plasma membrane (Denker and Barber, 2002; Patel and Barber, 2005; Frantz et al., 2008; Neisch and Fehon, 2011). As its acronym indicates, NIK binds the receptor tyrosine kinase adaptor protein Nck, and Nck binds to and activates N-WASP (Rohatgi et al., 2001; Rivera et al., 2009). Together, these findings suggest that NIK coordinates actin filament and membrane dynamics in response to activated receptor tyrosine kinases.
Materials and methods
MTLn3 cell culture and infection
MTLn3 rat adenocarcinoma cells (provided by J. Condeelis, Albert Einstein College of Medicine, New York, NY) were maintained in MEMα without nucleosides supplemented with 10% FBS at 37°C and 5% CO2. Cells were made quiescent by decreasing FBS medium from 10% to 0.2% for 4 h. For expression of recombinant Arp2, cells were infected with lentivirus expressing WT Arp2 and Arp2-T237/238A-Y202A tagged at the C terminus with tandem HA and V5 epitopes. Expression was selected by resistance to blasticidin and confirmed by immunoblotting and immunolabeling. NT and NIK shRNA were obtained from SABiosciences with the sequence 5′-CTCGGGAGGTGGAAGATAGATTT-3′ to silence NIK expression. Plasmids were transfected into cells with a 5:2 ratio of Fugene HD (Promega) with a 2 µg shRNA construct. Cells positive for GFP expression were isolated by flow cytometry after 48 h and cultured for an additional 48 h before analysis. The plasmid containing mEGFP-N1-LifeAct was provided by R. Wedlich-Sölder (Max Planck Institute of Biochemistry, Martinsried, Germany). The EGFP-N1 LifeAct sequence was cloned into a pENTR TOPO plasmid (Life Technologies) and recombined into pLenti 6.1 destination vector using Gateway cloning technology (Life Technologies). Lentiviruses were produced and packaged in HEK293TA cells using the ViraPower Lentiviral Packaging kit (Life Technologies) according to the manufacturer’s instructions (Haynes et al., 2011). For lentiviral transduction, MTLn3 cells were infected in growth medium supplemented with 4 µg/ml Polybrene (Sigma-Aldrich). Infected cells were selected with 15 µg/ml blasticidin (Life Technologies). Adenovirus was prepared by cloning WT NIK and D152N NIK into pAD-TRACK-CMV and recombining into pADeasy-1. HEK293Ad cells were transfected with pAd-Track-CMV and the recombined plasmid. Adenovirus was collected from freeze-thawing transfected cells then amplified by infecting monolayers of 293Ad cells in T75 tissue culture flasks (Baumgartner et al., 2006).
Immunoblotting, immunolabeling, and immunoprecipitation
Immunoblotting was performed by lysing cells in ice-cold Nonidet P-40 lysis buffer, collecting postnuclear supernatants, and separating 40 µg of protein by SDS-PAGE. Proteins were transferred onto PVDF membranes, blocked in 5% BSA, and probed with primary antibodies for 60 min. We used antibodies for a mouse monoclonal V5 (1:5,000; Life Technologies), mouse monoclonal HA (1:1,000; ABM), mouse monoclonal tubulin (1:2,000; Sigma-Aldrich), rabbit monoclonal ARPC2 (1:1,000; EMD Millipore), rabbit polyclonal ARPC1 (1:5,000, generated to the C terminus of ARPC1 [CKTLESSIQGLRIM] by OpenBiosystems; Thermo Fisher Scientific), and rabbit polyclonal ZC1-3a for NIK (1:3,000; Baumgartner et al., 2006). After washing, membranes were incubated with HRP-coupled secondary antibodies (1:5,000; Jackson ImmunoResearch Laboratories) for 1 h at room temperature, and immunoreactivity was visualized using enhanced femto chemiluminescence (Thermo Fisher Scientific) with reactions imaged using a Chemidoc XRS (BioRad Laboratories).
For immunolabeling, cells were fixed for 10 min with 4% paraformaldehyde in PBS, washed with PBS, and permeabilized for 10 min with 0.1% Triton X-100. After washing and blocking for 30 min with 10% FCS in PBS, samples were incubated for 1 h with mouse monoclonal anti-HA and rabbit polyclonal-ARPC1 antibodies, washed, and incubated with Alexa Fluor 488– and 548–conjugated secondary antibodies (The Jackson Laboratory). Cells mounted in fluorescence were viewed by using a microscope system (Nikon) with a CSU-10 spinning disk confocal head (Yokogawa Electric Corporation) with a 60× Plan-Apochromat TIRF/1.45 NA oil immersion objective lens at 25°C. Images were collected with a charge-coupled device (CCD) camera (HQ2; Photometrics) and viewed with Elements software (Nikon).
For immunoprecipitation, cells were lysed with modified RIPA buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 10% glycerol, 1% NP-40, 1 mg/ml aprotinin, 1 mM pefabloc, and 1 mg/ml leupeptin) containing phosphatase inhibitors (1 mM EGTA, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM β-glycerophosphate, and 1 mM sodium orthovanadate). For Arp2 phosphorylation, before lysis cells were metabolically labeled by incubating for 4 h in phosphate-free MEMα supplemented with 0.2 mCi/ml [32P]orthophosphate. After preclearing and blocking for 1 h with protein A, lysates were incubated with mouse monoclonal HA or rabbit polyclonal ARPC1 antibodies bound to protein A–conjugated Dynabeads (Life Technologies) for 20 h at 4°C. Beads were washed three times with modified RIPA, resuspended in 1× Laemmli sample buffer, and immune complexes separated by SDS-PAGE followed by either immunoblotting or autoradiography.
Coimmunoprecipitation was performed by infecting MTLn3 cells with adenovirus containing either WT NIK-Myc, kinase inactive NIK (D152N-Myc), or empty vector (Ad) for 48 h. Cell lysates were prepared as described above, precleared with protein G Sepharose, and incubated for 2 h with 3 µg Myc mouse monoclonal antibody (EMD Millipore) at 4°C. Immune complexes were precipitated with protein G for 30 min, separated by SDS-PAGE, transferred to PVDF, and probed with antibodies to Myc and Arp2. Cell lysates were probed with antibodies to GAPDH (EMD Millipore).
Actin polymerization
Cellular F-actin in the absence and presence of EGF was determined in MTLn3 cells plated in 24-well dishes. Latrunculin B (Life Technologies) was added 15 min before EGF treatment. At the indicated times, cells were fixed as described above for immunolabeling and incubated with rhodamine-phalloidin (1:500; Life Technologies) for 30 min. After washing, cells were incubated for 5 min with Hoechst 33342 (1:10,000; Life Technologies) to label nuclei. Labeling was measured with a SpectraMax M5 plate reader (Molecular Devices), with rhodamine fluorescence normalized to Hoerchst signal and data expressed relative to WT cells in the absence of EGF.
In vitro actin polymerization was measured using 4 µM monomeric nonmuscle actin containing 5% pyrene-labeled actin in KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0; LeClaire et. al 2008). Polymerization was determined in the absence or presence of 5 nM rArp2/3 complex, 500 nM of VCA domain of Scar, and recombinant WT or mutant NIK. Pyrene actin was excited in a spectrophotometer (RF-5301PC; Shimadzu) at 365 nm, and fluorescence was measured at 407 nm at 1-s intervals. Purified rArp2/3 complex was dephosphorylated by treatment with 1 U AP (New England Biolabs, Inc.) in Tris-HipH buffer (50 mM Tris, pH 8.0, 1 mM Mg2Cl2, and 0.1 mM ZnCl2) for 1.5 h at 30°C.
Cell proliferation
MTLn3 cells maintained in growth medium were counted 48 h after plating 10 × 105 cells/well in 24-well plates. Doubling time was determined and data are shown for individual wells from four separate cell plating preparations. Significant differences compared with WT MTLn3 cells were determined with an unpaired t test.
Video microscopy
MTLn3 cells plated in MatTek 35-mm glass-bottom dishes (MatTek Corporation) were imaged using the microscope system described above in a climate-regulated chamber at 37°C and 5% CO2. Changes in cell area were measured as described previously (El-Sibai et al., 2008), with the following modifications to quantify changes in cell size after stimulation with growth factors. Images of cells were captured at 20-s intervals, beginning 2 min before adding 5 nM EGF and ending 10 min after adding EGF. Image analysis was performed in NIS Elements software (Nikon), including time-dependent changes in cell size and lamellipodium, designated as the region from the cell body to the distal margin of the plasma membrane. Fold increase in cell (CΔ) area was calculated by the equation CΔ = AEGF/AQ, where AEGF is the area of the lamellipodium measured 5 min after EGF stimulation and AQ is the area of the lamellipodium measured before EGF stimulation. Approximately 30 cells were measured for each condition.
In vitro kinase and binding assays
Kinase reactions were performed in kinase reaction buffer (25 mM Hepes, pH 7.5, 1 mM DTT, 10 mM MgCl2, 2 mM MnCl2, 50 mM nonradioactive ATP, and 5 µCi [32P]γ-ATP) for 20 min at 30°C. Reactions contained 200 ng NIK generated by baculovirus/insect cell expression, as described previously (Baumgartner et al., 2006). In brief, the NIK gene (NM_001252200) was ligated into pFASTBac and recombined into a bacmid genome in DH10BAC cells (Life Technologies). Bacmid encoding NIK was isolated and transfected into sf9 cells, and supernatants containing baculovirus were collected. Viral stocks were used to infect sf9 cells in suspension. Recombinant NIK was purified by nickel affinity chromatography. Native Arp2/3 complex purified from A. castellanii was purified (provided by D. Mullins, University of California, San Francisco, San Francisco, CA), WT rArp2/3 complex was generated by baculovirus/insect cell expression as described previously (Gournier et al., 2001), or rArp2/3 containing mutant Arp2-T237/238A-Y202A was generated as described previously (Narayanan et al., 2011). In brief, Sf21 cells were infected with baculoviruses containing cDNA encoding Arp2/3 complex subunits. Infected cells were grown in suspension for 48°C for 48 h. Recombinant Arp2/3 complexes were purified on Talon affinity resin (Takara Bio Inc.). Purified Arp2/3 complex was passed over a Superdex 200 FPLC column (GE Healthcare) to isolate fully assembled complexes. A range of Arp2/3 concentrations from 1 nM to 500 nM per reaction were performed to determine kinase kinetics. Reactions were stopped by the addition of Laemmli buffer, proteins were separated by SDS/PAGE, and gels were dried and exposed to x-ray film.
Binding assays were performed in binding buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT, 100 µM ATP, 1% BSA, and 5% glycerol) containing rArp2/3 complex coupled to CH-Sepharose (GE Healthcare) and the indicated NIK truncations. Truncated NIK proteins were generated by in vitro expression using a cell-free expression kit (Life Technologies) with Express-Tag EXPRESS Protein Labeling Mix (PerkinElmer) containing [35S]methionine. Arp2/3-coupled Sepharose was incubated with the labeled protein for 30 min at 25°C. Sepharose was pelleted by centrifugation, washed with binding buffer, and analyzed by SDS-PAGE. Gels were dried and visualized to be detected by autoradiography.
The dissociation constant (Kd) of NIK 340–500 for Arp2/3 complex was determined by incubating glutathione-bound GST-NIK 340–500 with progressive dilutions of Arp2/3 complex in Arp2/3 binding buffer (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT) from 2,000 ng/ml to 25 ng/ml for 30 min at 37°C. Sepharose-bound protein complexes were centrifuged at 3,000 g, resuspended in Laemmli buffer, and separated by SDS-PAGE. Gels were stained in Coomassie brilliant blue, imaged on an Odyssey Li-Cor scanner, and quantified using ImageJ. Protein binding curves were generated using GraphPad Prism.
Online supplemental material
Fig. S1 shows that the expression of Arp2/3-TTY slows the grow rate of MTLn3 cells. Fig. S2 shows Arp2/3 complex phosphorylation and binding. Fig. S3 shows that NIK regulates the Arp2/3 actin nucleation rate and membrane protrusion. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404095/DC1.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM47413 (D.L. Barber), and postdoctoral fellowship awards from the Sandler Foundation (L.L. LeClaire) and the Swiss National Science Foundation (M. Baumgartner).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AP
- Antarctic phosphatase
- NIK
- Nck-interacting kinase
- NPF
- nucleation promoting factor
- NT
- nontargeting
- rArp
- recombinant Arp
- WT
- wild type
References
- Baumgartner M., Sillman A.L., Blackwood E.M., Srivastava J., Madson N., Schilling J.W., Wright J.H., and Barber D.L.. 2006. The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc. Natl. Acad. Sci. USA. 103:13391–13396 10.1073/pnas.0605950103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.Y., Raft S., Bailly M., Wyckoff J.B., Segall J.E., and Condeelis J.S.. 1998. EGF stimulates an increase in actin nucleation and filament number at the leading edge of the lamellipod in mammary adenocarcinoma cells. J. Cell Sci. 111:199–211. [DOI] [PubMed] [Google Scholar]
- Chapman J.O., Li H., and Lundquist E.A.. 2008. The MIG-15 NIK kinase acts cell-autonomously in neuroblast polarization and migration in C. elegans. Dev. Biol. 324:245–257 10.1016/j.ydbio.2008.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G.T., Hu C.K., Coughlin M., and Mitchison T.J.. 2006. Reassembly of contractile actin cortex in cell blebs. J. Cell Biol. 175:477–490 10.1083/jcb.200602085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.H., Thomason P.A., Zaki M., Insall R.H., and Barber D.L.. 2013. Phosphorylation of actin-related protein 2 (Arp2) is required for normal development and cAMP chemotaxis in Dictyostelium. J. Biol. Chem. 288:2464–2474 10.1074/jbc.M112.435313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobreros-Reguera L., Fernández-Miñán A., Fernández-Espartero C.H., López-Schier H., González-Reyes A., and Martín-Bermudo M.D.. 2010. The Ste20 kinase misshapen is essential for the invasive behaviour of ovarian epithelial cells in Drosophila. EMBO Rep. 11:943–949 10.1038/embor.2010.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker S.P., and Barber D.L.. 2002. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159:1087–1096 10.1083/jcb.200208050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A., Cicchetti G., Falet H., Hartwig J.H., Stossel T.P., and Kwiatkowski D.J.. 2005. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc. Natl. Acad. Sci. USA. 102:16263–16268 10.1073/pnas.0508228102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai M., Pertz O., Pang H., Yip S.C., Lorenz M., Symons M., Condeelis J.S., Hahn K.M., and Backer J.M.. 2008. RhoA/ROCK-mediated switching between Cdc42- and Rac1-dependent protrusion in MTLn3 carcinoma cells. Exp. Cell Res. 314:1540–1552 10.1016/j.yexcr.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M.J., Jacobson M.P., and Barber D.L.. 2008. Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183:865–879 10.1083/jcb.200804161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley E.D., and Welch M.D.. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7:713–726 10.1038/nrm2026 [DOI] [PubMed] [Google Scholar]
- Gournier H., Goley E.D., Niederstrasser H., Trinh T., and Welch M.D.. 2001. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol. Cell. 8:1041–1052 10.1016/S1097-2765(01)00393-8 [DOI] [PubMed] [Google Scholar]
- Haynes J., Srivastava J., Madson N., Wittmann T., and Barber D.L.. 2011. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol. Biol. Cell. 22:4750–4764 10.1091/mbc.E11-02-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Kranitz H., Dötsch V., and Mullins R.D.. 2006. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 281:10589–10597 10.1074/jbc.M507470200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClaire L.L. III, Baumgartner M., Iwasa J.H., Mullins R.D., and Barber D.L.. 2008. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J. Cell Biol. 182:647–654 10.1083/jcb.200802145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes M.A., Larson D.R., Mader C.C., Bravo-Cordero J.J., Gil-Henn H., Oser M., Chen X., Koleske A.J., and Condeelis J.. 2011. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195:903–920 10.1083/jcb.201103045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand J.B., Kaiser D.A., Pollard T.D., and Higgs H.N.. 2001. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3:76–82 10.1038/35050590 [DOI] [PubMed] [Google Scholar]
- Narayanan A., LeClaire L.L. III, Barber D.L., and Jacobson M.P.. 2011. Phosphorylation of the Arp2 subunit relieves auto-inhibitory interactions for Arp2/3 complex activation. PLOS Comput. Biol. 7:e1002226 10.1371/journal.pcbi.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch A.L., and Fehon R.G.. 2011. Ezrin, Radixin and Moesin: key regulators of membrane-cortex interactions and signaling. Curr. Opin. Cell Biol. 23:377–382 10.1016/j.ceb.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra S.M., and Higgs H.N.. 2008. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil. Cytoskeleton. 65:904–922 10.1002/cm.20312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Egile C., Lipkin T., and Li R.. 2004. ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J. Biol. Chem. 279:54629–54636 10.1074/jbc.M402357200 [DOI] [PubMed] [Google Scholar]
- Patel H., and Barber D.L.. 2005. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J. Cell Biol. 169:321–329 10.1083/jcb.200412145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinat P., De Arcangelis A., Sookhareea S., Zhu X., Hedgecock E.M., Labouesse M., and Georges-Labouesse E.. 2002. A conserved interaction between β1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr. Biol. 12:622–631 10.1016/S0960-9822(02)00764-9 [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. . 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera G.M., Vasilescu D., Papayannopoulos V., Lim W.A., and Mayer B.J.. 2009. A reciprocal interdependence between Nck and PI(4,5)P2 promotes localized N-WASp-mediated actin polymerization in living cells. Mol. Cell. 36:525–535 10.1016/j.molcel.2009.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Nollau P., Ho H.Y., Kirschner M.W., and Mayer B.J.. 2001. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 276:26448–26452 10.1074/jbc.M103856200 [DOI] [PubMed] [Google Scholar]
- Su Y.C., Han J., Xu S., Cobb M., and Skolnik E.Y.. 1997. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 16:1279–1290 10.1093/emboj/16.6.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P., Rubinstein B., Unruh J.R., Durnin M., Hanein D., and Li R.. 2012. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197:239–251 10.1083/jcb.201112113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M.D., and Mullins R.D.. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288 10.1146/annurev.cellbio.18.040202.112133 [DOI] [PubMed] [Google Scholar]
- Wright J.H., Wang X., Manning G., LaMere B.J., Le P., Zhu S., Khatry D., Flanagan P.M., Buckley S.D., Whyte D.B., et al. . 2003. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol. Cell. Biol. 23:2068–2082 10.1128/MCB.23.6.2068-2082.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Asokan S.B., Berginski M.E., Haynes E.M., Sharpless N.E., Griffith J.D., Gomez S.M., and Bear J.E.. 2012. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 148:973–987 10.1016/j.cell.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wang X., Li Z., Gotoh N., Chapman D., and Skolnik E.Y.. 2001. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK). Development. 128:1559–1572. [DOI] [PubMed] [Google Scholar]
- Yan W., Nehrke K., Choi J., and Barber D.L.. 2001. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 276:31349–31356 10.1074/jbc.M102679200 [DOI] [PubMed] [Google Scholar]
- Yang Q., Zhang X.F., Pollard T.D., and Forscher P.. 2012. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 197:939–956 10.1083/jcb.201111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M., King J., Fütterer K., and Insall R.H.. 2007. Replacement of the essential Dictyostelium Arp2 gene by its Entamoeba homologue using parasexual genetics. BMC Genet. 8:28 10.1186/1471-2156-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.