Salt bridges at the dynein–microtubule interface couple microtubule binding to ATPase activation and thereby control the directional movement of dynein
Abstract
Dynein is a motor protein that moves on microtubules (MTs) using the energy of adenosine triphosphate (ATP) hydrolysis. To understand its motility mechanism, it is crucial to know how the signal of MT binding is transmitted to the ATPase domain to enhance ATP hydrolysis. However, the molecular basis of signal transmission at the dynein–MT interface remains unclear. Scanning mutagenesis of tubulin identified two residues in α-tubulin, R403 and E416, that are critical for ATPase activation and directional movement of dynein. Electron cryomicroscopy and biochemical analyses revealed that these residues form salt bridges with the residues in the dynein MT-binding domain (MTBD) that work in concert to induce registry change in the stalk coiled coil and activate the ATPase. The R403-E3390 salt bridge functions as a switch for this mechanism because of its reversed charge relative to other residues at the interface. This study unveils the structural basis for coupling between MT binding and ATPase activation and implicates the MTBD in the control of directional movement.
Introduction
Dynein is a large protein complex that moves along microtubules (MTs) toward their minus ends by using the energy of ATP hydrolysis (Roberts et al., 2013). The motility of dynein is essential for various cellular functions including axonal transport, chromosome segregation, and beating of cilia and flagella (Gibbons, 1981; Höök and Vallee, 2006). The cytoplasmic form of dynein (hereafter referred as dynein) is composed of two ∼500-kD heavy chains and can move processively along MTs through multiple cycles of ATP hydrolysis. Compared with other processive motors such as kinesin-1 and myosin V, little is known regarding the molecular mechanism of dynein motility.
Dynein is a unique motor protein because its main site of ATP hydrolysis is located in the AAA1 module in the head, whereas its MT-binding domain (MTBD) is located at the tip of the stalk coiled coil, more than 20 nm from the AAA1 ATPase site (Fig. 1 A). For dynein to move along an MT, the cycles of nucleotide hydrolysis in the AAA1 module should be coupled with the MTBD binding to and releasing from the MT, in spite of this large separation.
Figure 1.
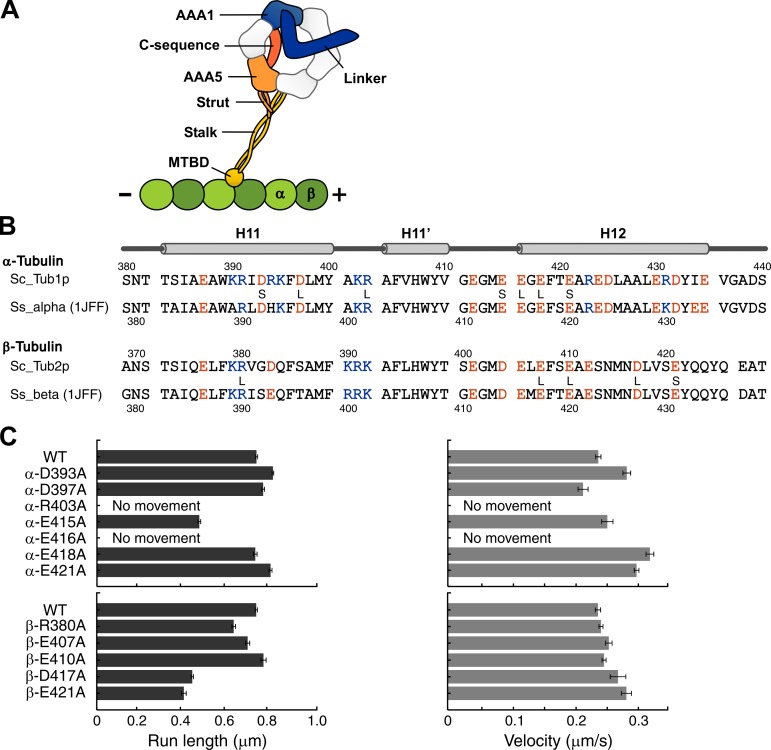
Tubulin mutants and the results of the single-molecule motility assay. (A) Schematic representation of the dynein–MT complex representing the structural elements likely to be involved in allosteric communication between the MT and the ATPase site in dynein. (B) Positions of charged residues targeted for alanine mutagenesis in the sequences of α- and β-tubulin from yeast S. cerevisiae (Sc), aligned with the corresponding sequences in pig S. scrofa (Ss) tubulin (PDB ID: 1JFF; Löwe et al., 2001). Each of the positively (blue) or negatively (red) charged residues was substituted with alanine. The mutations that resulted in haploid lethality and slow growth in yeast cells are marked by the letters L and S, respectively (Uchimura et al., 2010). (C) Results of the single-molecule motility assay using the two-headed cytoplasmic dynein GST380. Error bars represent the standard error of the mean.
As a structural basis for this long-distance communication, biochemical studies have implied that changes in registration between the two helices of the stalk coiled coil may mediate the coupling between ATPase and MT-binding activities (Gibbons et al., 2005; Kon et al., 2009a). Moreover, x-ray crystallographic analysis has revealed that junctional structures of the dynein molecule called the strut (or a buttress) and C-sequence are essential for the coupling mechanism (Carter et al., 2011; Kon et al., 2012). Thus, the structural communication between the MTBD and AAA1 module is thought to have one end starting at the MTBD; connect to the stalk coiled coil, strut, and C-sequence; and then reach the other end of AAA1 (Fig. 1 A).
For a complete understanding of the coupling mechanism, however, it is necessary to determine the molecular basis for signal transmission at the MTBD–MT interface. Which residues of tubulin provide a signal to the MTBD critical for ATPase activation? How does it induce a registry change in the stalk coiled coil? Earlier work using charged-to-alanine mutational analyses of dynein implied that 15 amino acid residues located in α helices H1, H3, and H6 of the MTBD are involved in the interaction with the MT (Koonce and Tikhonenko, 2000), but, little is known regarding the binding partners of these residues within the MT. Recently, a structural model for the MTBD–MT complex in the strong binding state was presented by using electron cryomicroscopy (cryoEM) and image analyses (Redwine et al., 2012), and the model depicted an MT-dependent conformational change of the MTBD. However, these questions remained unresolved. They predicted several salt bridges possibly involved in the complex formation by molecular dymanics simulation based on a cryoEM structure, but without a functional analysis of tubulin it was impossible to identify the tubulin residues responsible for ATPase activation.
We addressed these issues by applying mutational analysis of tubulin combined with a single-molecule motility assay and biochemical analyses. The preparation of recombinant tubulin has long been hampered by the technical difficulties of tubulin expression, but the recently developed yeast Saccharomyces cerevisiae expression system has opened the way for mutational analysis of tubulin in vitro (Davis et al., 1993; Gupta et al., 2003; Uchimura et al., 2006). Using this system, we found that two highly conserved residues of α-tubulin, R403 and E416, are critical for dynein motility and ATPase activation. The structure of the MTBD–MT complex obtained by cryoEM image analysis revealed that E3390 in α helix H1 of MTBD was the binding partner for α-R403 and that R3469 and/or K3472 in H6 of MTBD were the binding partners for α-E416 in the strong binding state. These two salt bridges are located at the points connecting the MTBD and the stalk coiled coil and thus may function in concert to induce the registry change of the coiled-coil helix, leading to ATPase activation. Notably, a comparison of this result with the results of a previous mutagenesis analysis of the kinesin–MT interface revealed that dynein and kinesin use the same loop of α-tubulin (415EEGE) to activate their ATPase (Uchimura et al., 2010), indicating that a specific part of the MT plays an essential role in their motility mechanism.
Note: In this paper, we adopted numbering of amino acid residues for S. cerevisiae yeast tubulin, which is slightly different from that of the Sus scrofa pig tubulin used in the PDB file 1JFF (Fig. 1 B).
Results
Tubulin residues critical for dynein motility
CryoEM studies have shown that dynein binds in a groove at the interface between α- and β-tubulin, anchored by the ridge of a protofilament (pf) composed of H11-H12 of both α- and β-tubulin (Mizuno et al., 2004; Carter et al., 2008; Redwine et al., 2012). Thus, to identify the critical residues, we used a set of charged-to-alanine point mutations in the tubulin sequence coding for H11-H12 in α- and β-tubulin in yeast S. cerevisiae, which were generated in previous analyses of kinesin–MT interaction (Fig. 1 B; Uchimura et al., 2010). The sequence in this region was highly conserved among species. Among the 36 mutants expressed in yeast cells, eight mutants appeared to be haploid lethal and four were slow in growth. Earlier studies reported that dynein gene disruption resulted in an abnormal distribution of dividing nuclei between mother and daughter cells, but not lethality or growth inhibition (Eshel et al., 1993; Li et al., 1993). Thus, to screen the 36 tubulin mutants and identify the sites critical for dynein function, we divided them into two groups (those showing normal growth and those with a lethal or slow-growth phenotype) and examined each group separately, as follows.
For the first group, showing normal growth (24 mutants), we analyzed nuclear segregation during cell division. We found that segregation was normal in all strains (Fig. S1), indicating that none of them affected dynein function. In the second group, showing lethality or growth inhibition, their phenotypes might have arisen from the perturbation of MT interaction with kinesin, MAPs, or other MT-interacting proteins (Pellman et al., 1995; Saunders et al., 1995; Wang and Huffaker, 1997). Because nuclear segregation cannot be used for screening mutants belonging to this group, we purified tubulin from each mutant, polymerized it into MTs, and tested its ability to support dynein motility in a single-molecule motility assay using two-headed, cytoplasmic dynein GST380, derived from cellular slime mold Dictyostelium discoideum (Numata et al., 2011). The result showed that dynein moved along most of these mutant MTs at velocities comparable to that of the wild type, except for two α-tubulin mutants, α-R403A and α-E416A (Fig. 1 C). In these mutants, even brief binding of the dynein molecule to an MT was scarcely observed. In other mutants, namely, α-E415A, β-D417A, and β-E421A, dynein moved along the MTs, but the run lengths were reduced to approximately half of that of the wild type.
In the MT structure, α-R403 and α-E416 are located in the α-tubulin H11-H11′ loop and H12, respectively, and are likely to form a salt bridge that stabilizes the C-terminal hairpin structure of α-tubulin (Löwe et al., 2001). Therefore, the impaired motility in the α-R403A and α-E416A mutants may mean either that these residues directly mediate the interaction of dynein with the MTs or that the intramolecular salt bridge within α-tubulin is critical for structural integrity of MT. To examine which of the aforementioned interpretations is correct, we tested the motility in an α-R403E/α-E416R double mutant. In this mutant, the salt bridge between the pair should be maintained, and thus the structure of the MT is likely to be intact. The result showed that dynein could not move on this double mutant MT, indicating that the impaired motility by either α-R403A or α-E416A was a result of the loss of a dynein-interacting residue in the MT.
In summary, the screening of 36 mutants led to the identification of two residues in α-tubulin critical for dynein motility and three additional residues in α- and β-tubulin that moderately affect motility.
α-R403 and α-E416 are critical for coupling MT binding and ATPase activation
The lack of motility of single dynein molecules along α-R403A and α-E416A MTs does not mean that these mutations inhibit dynein–MT interactions under all conditions. In the case of kinesin, charged-to-alanine mutations in tubulin resulted in impaired motility of single kinesin but did not impair the gliding movement of MT on multiple kinesin molecules (Uchimura et al., 2010).
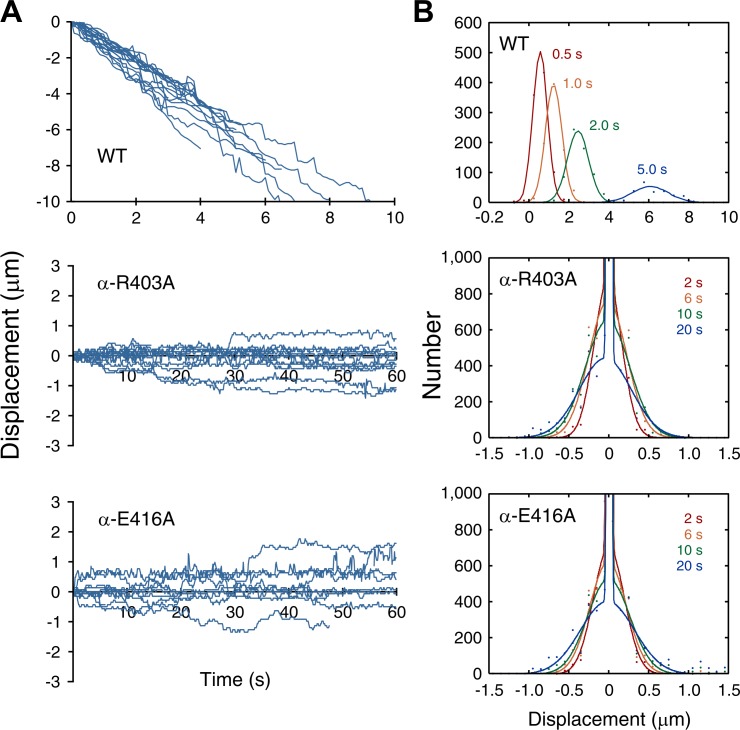
To test if α-R403A and α-E416A MTs can move on multiple dynein molecules, the surface of the chamber for the motility assay was coated with monomeric dynein MD380 and interactions with either α-R403A or α-E416A MTs were examined in the presence of ATP. Whereas wild-type MTs showed minus end–directed movement at a velocity of 1.21 ± 0.02 µm/s, the mutant MTs showed stochastic back-and-forth movements along the long axes of the MTs (Fig. 2 A and Video 1). The distributions of MT displacement were fitted well by Gaussian functions whose variance increased linearly with time, indicating that the movement was caused by Brownian motion (Fig. 2 B and Fig. S2). The mean displacement was virtually zero, indicating that the Brownian motion was not biased in any direction.
Figure 2.
Effect of α-R403A and α-E416A mutations analyzed in an MT-gliding assay. (A) Time course of the positions of single MTs in a direction parallel to the long axes of the MTs measured in the presence of 1 mM ATP. Negative displacement corresponds to motion toward the minus end of the MT. (B) Distributions of displacements at the indicated time intervals. The diffusion coefficient, calculated from the variance of the distribution, was 0.0021 and 0.0022 µm2/s for α-R403A and α-E416A, respectively (Fig. S2).
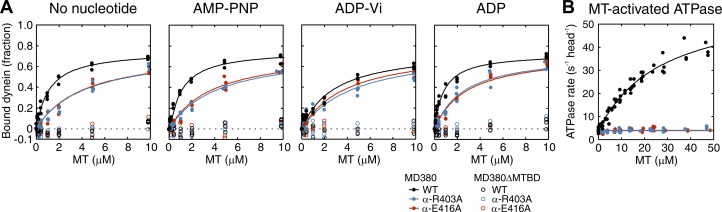
This observation indicates that dynein can bind to α-R403 and α-E416 MTs at least in some states in the ATPase cycle, but it cannot enter a state critical for directional movement. Therefore, to examine which state of the ATPase cycle is affected by the mutation, binding of MD380 to either α-R403A or α-E416A MTs was measured by a cosedimentation assay in various nucleotide conditions (Fig. 3 A and Table 1). In all nucleotide conditions (no nucleotide, AMP-PNP, ADP-Vi, and ADP), MT affinity was reduced by the mutations. In both mutants, the reduction was most significant in the strong binding state, where the dissociation constants increased by three- to fourfold compared with the wild type.
Figure 3.
Effect of tubulin mutation on dynein binding and MT-activated ATPase. (A) Binding of MD380 and MD380ΔMTBD to wild-type, α-R403A, and α-E416A MTs. See Materials and methods for the composition of the solution used. Each curve shows a hyperbolic fitting to all the data obtained from three independent experiments, with the dissociation constant and maximum binding (Bmax) given in Table 1. (B) Effect of tubulin mutations on MT-activated ATPase activity of MD380. A curve shows a fitting of all the data obtained from seven independent experiments to the Michaelis-Menten equation, with the kcat and K0.5MT values given in Table 2.
Table 1.
Dissociation constant of MD380
| Dissociation constant | ||||
| Construct | No nucleotide | AMP-PNP | ADP-Vi | ADP |
| μM | μM | μM | μM | |
| WT | 1.1 ± 0.1 | 1.0 ± 0.1 | 2.8 ± 0.6 | 0.8 ± 0.1 |
| α-R403A | 4.2 ± 0.4 | 4.2 ± 0.2 | 4.7 ± 0.5 | 2.5 ± 0.2 |
| α-E416A | 4.3 ± 0.3 | 3.8 ± 0.3 | 4.0 ± 0.3 | 2.8 ± 0.2 |
Errors indicate the errors in curve fitting. In wild-type MTs, the maximum binding Bmax was calculated to be 0.76, 0.76, 0.77, and 0.74 for no nucleotide, AMP-PNP, ADP-Vi, and ADP conditions, respectively. These values were used as the Bmax for mutants measured in the same nucleotide conditions.
Despite the partial binding of MD380, the mutant MTs utterly lacked the ability to activate dynein ATPase (Fig. 3 B). Whereas wild-type MT increased the basal ATPase activity (3.7 ± 0.6 s−1) by >10-fold to generate kcat and K0.5MT values of 60.6 ± 4.0 s−1 and 27.2 ± 4.0 µM, respectively, activation by α-R403A or α-E416A MTs was so limited that we could not estimate the kcat and K0.5MT. This result contrasts with those obtained for the α-E415A, β-D417A, and β-E421A mutants showing a reduced run length of dynein in the single-molecule motility assay (Fig. 1 C). These mutant MTs activated ATPase, though their affinity for MTs (K0.5MT) was reduced two- to fourfold relative to the wild type (Table 2). In summary, our results demonstrate that the residues α-R403 and α-E416 are critical for coupling MT binding and ATPase activation.
Table 2.
MT-activated ATPase of MD380
| Construct | kcat | K0.5MT | |
| Basal | MT activated | ||
| s−1 head−1 | s−1 head−1 | μM | |
| WT | 3.7 ± 0.6 | 60.6 ± 4.0 | 27.2 ± 4.0 |
| α-R403A | 4.0 ± 0.1a | ND | ND |
| α-E416A | 3.9 ± 0.1a | ND | ND |
| α-E415A | 2.8 ± 0.6 | 59.1 ± 12.2 | 54.7 ± 18.7 |
| β-D417A | 2.6 ± 0.5 | 55.4 ± 12.3 | 67.2 ± 24.3 |
| β-E421A | 4.0 ± 0.3 | 64.0 ± 17.3 | 104.7 ± 41.0 |
Errors indicate the errors in curve fitting.
Mean ± SEM of all data points.
Binding partners of α-R403 and α-E416
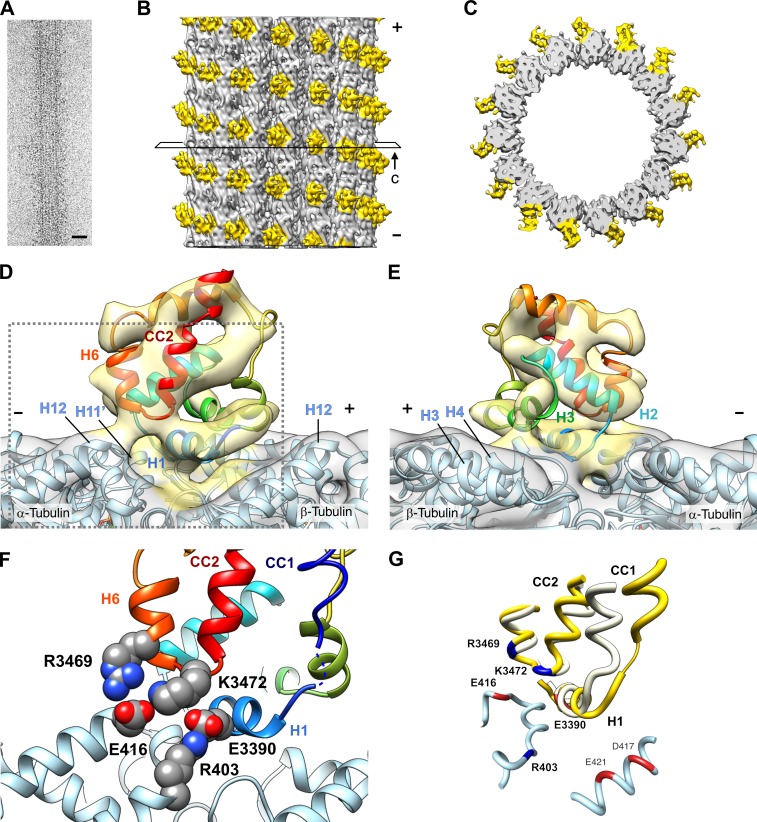
To elucidate how the MTBD–MT interactions mediated by α-R403 and α-E416 lead to ATPase activation, we solved the structure of the MTBD–MT complex in the strong binding state by cryoEM image analysis. The distal part of slime mold dynein stalk (V3350-K3514) was fused to an artificial coiled coil with its registry fixed to a so-called α registry, high-affinity configuration for MTs (Fig. S3 A; Gibbons et al., 2005; Kon et al., 2009a). To obtain a high-resolution structure, the purified MTBD was bound to an MT with 15 pf polymerized from Caenorhabditis elegans nematode tubulin (MEC-12 and MEC-7; Chalfie, 1982). The 15-pf MT is ideal for image reconstruction because of its perfect helical symmetry (Chrétien and Wade, 1991). It was confirmed that the residue substitutions α-R403A and α-E416A reduced the binding affinity of the MTBD for an MT (Fig. S3, A and B) and that the substitution of the corresponding residue in 15-pf nematode MTs disrupted the motility of single dynein molecules (Fig. S3, C and D). The 3D image of the MTBD–MT complex was reconstructed by helical image analysis at a resolution of 8.2 Å (Fig. 4, A–C; and Fig. S4).
Figure 4.
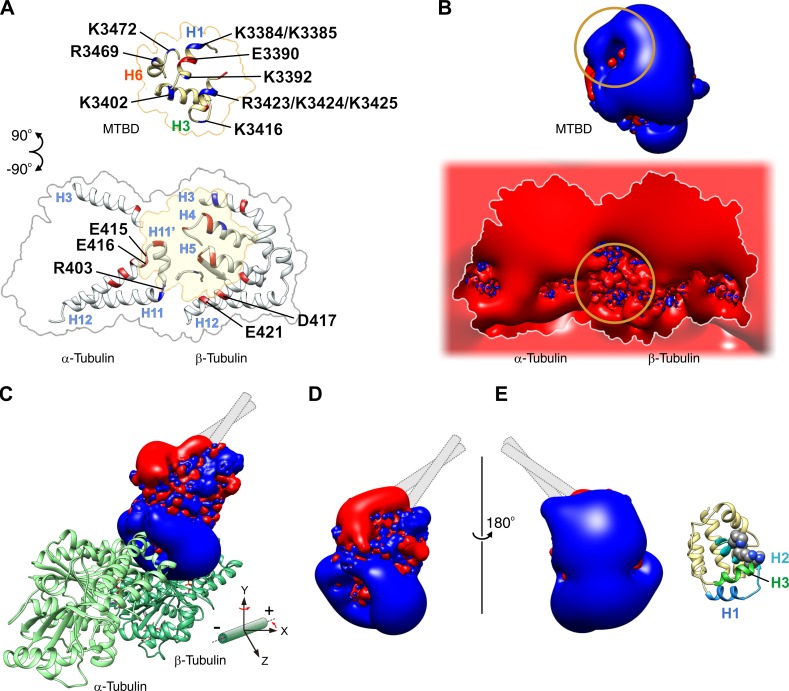
Structure of the MTBD–MT complex. (A) CryoEM image of a 15-pf MT decorated with the high-affinity MTBD. Bar, 25 nm. (B) Three-dimensional image reconstruction of the MTBD–MT complex. The open square surrounding the MTBD-decorated MT density represents the plane of a slice through the volume, shown in C. (C) The top half of the density is shown in B, cut out at the plane and viewed from the minus end of the MT. (D and E) Fitting of the x-ray crystal structure of MTBD (PDB ID: 3VKH; Kon et al., 2012) into the map, viewed from the side of the MT with its minus end on the left (D) and right (E) sides. See Fig. S5 for the details of the modeling procedure. (F) Magnified view of the boxed area in D. Positively and negatively charged residues are shown in CPK representation with nitrogen atoms in blue and oxygen atoms in red. The putative position of CC1, inferred from the model of the MTBD of mouse cytoplasmic dynein (PDB ID: 3J1T; Redwine et al., 2012), is indicated by a thin helix. The color coding scheme for the MTBD helices used in D–F is that used by Carter et al. (2008). (G) Superposition of the crystal structure of the MTBD (PDB ID: 3VKH; white) and the model of the MTBD in the MTBD–MT complex (yellow). The structures of the MTBD spanning H2-CC2 (T3399-E3489) were superposed using the MatchMaker tool of UCSF Chimera (Meng et al., 2006).
The cryoEM density map of the complex showed that the density corresponding to the MTBD was wedged at the intra-dimer interface of α- and β-tubulin, with a spacing of 8 nm along the MT axis. The stalk coiled coil was not visible in our map, probably because the coiled-coil structure proximal to the MTBD is intrinsically unstable in slime mold dynein (Kon et al., 2009a, 2012; Nishikawa et al., 2014).
To build a structural model for the MTBD–MT complex, the MTBD crystal structure derived from slime mold dynein in the ADP bound state (PDB ID: 3VKH; Kon et al., 2012) was initially fitted as a rigid body into the map. Although most MTBD α helices can be roughly fitted into the map, a tubular density aligned parallel to α helix H12 of β-tubulin remained unfilled by the structure (Fig. S5). The map was best fit when H1 and its N-terminal chain in MTBD were shifted to fill the empty space of the tubular density by flexible fitting (see Fig. S5 and Video 2 for details of model building; Topf et al., 2008; Wang and Schröder, 2012). In the final model, H1, H3, and H6 of the MTBD appeared to be the major structural elements at the interface for the MT, which is consistent with the previous results of the cryoEM and mutational analyses (Koonce and Tikhonenko, 2000; Carter et al., 2008; Redwine et al., 2012).
From a side view (Fig. 4 D), H1 and the chain spanning H6 and coiled coil 2 (CC2) of the MTBD are located in the vicinity of H12 of β-tubulin and H11′ and H12 of α-tubulin, respectively. On the opposite side (Fig. 4 E), H3 of the MTBD is in the vicinity of H3, H4, and H5 of β-tubulin. The tubulin residue α-E416 is in close proximity to R3469 in H6 and K3472 in the H6-CC2 loop of the MTBD, and the tubulin residue α-R403 is close to E3390 in H1 of the MTBD (Fig. 4 F). Considering the results of mutational analysis (Koonce and Tikhonenko, 2000; this study), it is highly likely that these pairs of charged residues form salt bridges across the MTBD–MT interface. Particularly, the interaction between α-R403 and E3390 is convincing because E3390 is the only negatively charged residue in the MTBD identified to be involved in the dynein–MT interaction. This contrasts with a previous model of the MTBD–MT complex (Redwine et al., 2012). In that model, neither R403 nor E3390 was identified as the residue at the MTBD–MT interface, probably because it was led by molecular dynamics simulation based on a cryoEM structure of limited resolution, counting only the result of the mutational analysis of MTBD.
Both E3390 and R3469 are highly conserved among species (Koonce and Tikhonenko, 2000; Carter et al., 2008). R3469 and K3472 are in the loop connecting H6 and CC2, and E3390 is in H1 immediately adjacent to CC1 (Fig. 4 F). Both are located in ideal positions to transmit the signal of MT binding to the dynein stalk coiled coil. Therefore, the present structure strongly indicates that the two pairs of charged residues, R403-E3390 and E416-R3469/K3472, across the MTBD–MT interface are critical for coupling MT binding and ATPase activation.
Discussion
α-Tubulin R403 and E416 function as a structural signal for ATPase activation
Using alanine-scanning mutagenesis, we identified R403 and E416 in α-tubulin to be critical for dynein motility and ATPase activation. Alanine substitution in either of these residues resulted in complete loss of single-molecule motility and MT-activated ATPase activity (Figs. 1 C and 3 B). This is the first time that coupling between binding at specific tubulin residues and ATPase activation has been demonstrated.
In these mutants, despite the complete loss of MT-activated ATPase, dynein was able to interact with MTs in a cosedimentation assay (Fig. 3 A). In the MT-gliding assay, the mutant MTs showed stochastic back-and-forth movements on a dynein-coated glass surface, whereas wild-type MTs showed directional movement (Fig. 2). These results collectively indicate that α-R403 and α-E416 are critical for dynein to switch from a weak to a strong binding state, which is coupled to the acceleration of the rate-limiting step in the ATPase cycle. Because the state transition was blocked in the α-R403A and α-E416A mutants, dynein runs off from the normal mechanochemical cycle, remaining in the weak binding state. Consequently, the mutant MTs exhibited unbiased Brownian motion with the cycle of ATP hydrolysis uncoupled from MT binding.
In the MT structure, α-R403 and α-E416 are located in the C-terminal region of α-tubulin and are likely to form an intramolecular salt bridge that stabilizes the hairpin structure of the C terminus (Löwe et al., 2001). Interestingly, a cluster of negatively charged residues including α-E416 (415EEGE) is also critical for ATPase activation of the other motor protein, kinesin (Uchimura et al., 2010). Charged-to-alanine mutations in α-E416 and α-E415 in this loop drastically reduce kcat for dynein and kinesin ATPase, respectively. Considering that dynein and kinesin belong to different enzyme classes and show distinct enzyme kinetics (Mallik and Gross, 2004), it is surprising that these two motors use the same short loop/helix of α-tubulin composed of only four peptides to accelerate their ATPase. The two motors may have independently evolved to use this site (Mizuno et al., 2004), probably because the loop plays an essential role in the motility mechanism.
H1 serves as an MT sensor to transmit the activation signal
Earlier studies have shown that long-distance communication between the MTBD and the AAA1 ATPase site is mediated by a change in the registration between the two helices of the stalk coiled coil (Gibbons et al., 2005; Kon et al., 2009a). According to those results, the relative position of the two helices is inherently flexible and fluctuates among multiple registries termed −β, α, and +β registries, but MT binding shifts this structural equilibrium toward the α registry, leading to the acceleration of the rate limiting step in the ATPase cycle. Thus, to transmit a signal required for ATPase activation, the MTBD–MT interaction via α-R403 and α-E416 may somehow induce or stabilize the α registry in the helices of the stalk coiled coil.
To clarify the coupling mechanism, we analyzed the structure of the MTBD–MT complex by cryoEM imaging wherein the alignment of helices was artificially fixed in the α registry (Fig. S3). The results revealed that H1 of MTBD was rotated from its original position and orientation in the crystal structure (PDB ID: 3VKH) and became parallel to β-tubulin H12 (Fig. 4, D–F; Fig. S5; and Video 2). As the MTBD crystal structure presumably represents a configuration in the low-affinity, weak binding state (Carter et al., 2008; Kon et al., 2012; Redwine et al., 2012), we interpret the repositioning of H1 to be a consequence of MT binding.
A comparison of the structures before and after MT binding allows us to propose that H1 functions as an MT sensor that detects α-R403 (in tubulin) and transmits the binding signal to CC1 (Fig. 4 G). The close location of the acidic residue E3390 in H1 to the basic residue R403 of α-tubulin in the MTBD–MT complex suggests that the salt bridge between these residues may serve as the pivot point for the H1 rotation, leading to the sliding of CC1 relative to CC2 in the stalk. In contrast, the acidic residue E416 of α-tubulin is located in the vicinity of R3469 and/or K3472 in H6 of the MTBD. Thus, these pairs of charged residues may link the N-terminal end of CC2 to the MT by forming salt bridges across the protein interfaces. Therefore, these two (or three) salt bridges are expected to function in concert to induce the α registry. Whereas the R403-E3390 salt bridge triggers a rotation of H1 and causes sliding of CC1, the E416-R3469/K3472 salt bridge fixes CC2 firmly on the MT.
As implied in a previous study (Redwine et al., 2012) and by our model (Fig. 4 G), the rotated H1 will be further stabilized by ionic interactions between H1 of MTBD and the acidic patch in H12 of β-tubulin (β-D417 and β-E421). Our ATPase measurement showed that the interaction via D417 and E421 contributes to the affinity of dynein to the MT (K0.5MT), but not to the rate of ATP hydrolysis (kcat; Table 2). This is again similar to the case of kinesin, where the interaction via H12 of β-tubulin regulates the affinity of the motor with the MT (K0.5MT; Uchimura et al., 2010).
ATPase activation coupled with a transition of the MT-binding state
The involvement of the basic residue α-R403 in dynein motility is unconventional; the tubulin residues thus far implicated in kinesin and dynein motility were all acidic (Uchimura et al., 2010; Redwine et al., 2012). In fact, when we mapped the charged residues possibly involved in the electrostatic interaction across MTBD–MT based on our model, the residues on the MT surface were dominated by negative charges, whereas residues on the MTBD were dominated by positive charges (Fig. 5 A). The basic tubulin residue R403 and the acidic dynein residue E3390 are the very few exceptions. Reflecting such charge distributions, the surface potentials of the two molecular surfaces exhibit unique electrostatic complementarity, with their polarity locally reversed in the vicinity of the salt bridge R403-E3390 (Fig. 5 B, circled).
Figure 5.
Charged residues involved in the electrostatic interaction between the MTBD and MT together with the surface potential of the interfaces. (A) Distribution of charged residues on the surface of the MTBD (top) and tubulin (bottom), possibly involved in salt bridge formation across the interfaces of the MTBD and MT in the strong binding state (Fig. 4, D and E). Because the resolution of the cryoEM map is limited and does not allow visualization of side chains, we searched for pairs of charged residues located within 10 Å of each other to identify candidate residues. Positively and negatively charged residues are colored blue and red, respectively. The residues whose mutation impaired the dynein–MT interactions are labeled (Koonce and Tikhonenko, 2000). (B) 3D isopotential contours for the MTBD (top) and tubulin (bottom) calculated for the interfaces shown in A. The values of the contours are −2.5 kT/e (red) and 2.5 kT/e (blue) for the MTBD and −26 kT/e (red) and 26 kT/e (blue) for tubulin, respectively, where k is the Boltzmann constant, T is the temperature, and e is the magnitude of the electron charge. The circles indicate the areas where the direction of the electrostatic field is inverted. (C) Isopotential contours for the MTBD bound on the MT. The MTBD–MT complex shown in Fig. 4 D is rotated by 40° around the y axis and viewed from the direction of the z axis. Also see Video 3. (D) Isopotential contours for the x-ray crystal structure of the MTBD (PDB ID: 3VKH), presumably representing the weak binding state (Carter et al., 2008; Kon et al., 2012), are viewed in the same orientation as in C. (E) The model in D is turned 180° around the y axis, with the corresponding structural model shown alongside it. In the weak binding state, the MTBD may exhibit diffusion along the MT with its electropositive surface containing H2 and H3 facing toward the MT. Mutagenesis analysis showed that residue substitution in this area of the MTBD (K3402, K3415, and K3416, represented by the spheres in the structural model) resulted in decreased affinity of dynein for the MT (Koonce and Tikhonenko, 2000).
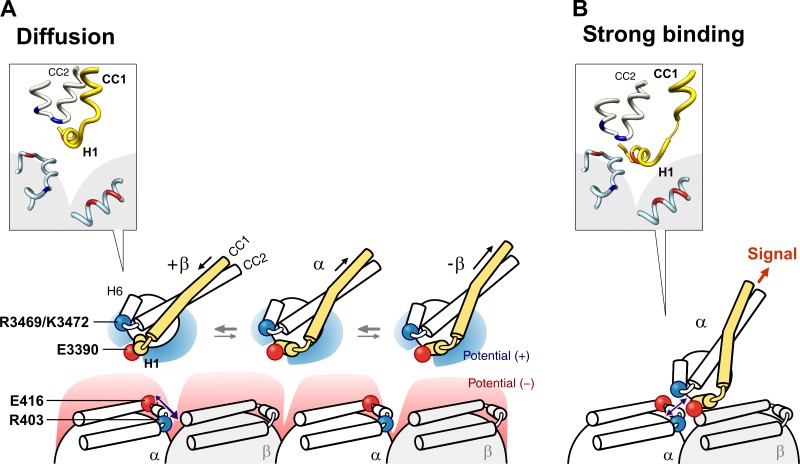
These potential landscapes may underlie the weak-to-strong state transition in the MTBD–MT interaction. In the weak binding state, when dynein exhibits diffusional motion along the negative potential field of MT, the flipped charge on the MT may provide a spatial cue to the MTBD (Fig. 5, B–D). This hypothesis is consistent with the results of a previous mutagenesis analysis showing that E3390 is the only acidic residue among more than a dozen MT-interacting residues on the MTBD (Koonce and Tikhonenko, 2000). Thus, considering the electrostatic properties, we present a model that correlates the weak-to-strong transition of the binding mode with ATPase activation in dynein (Fig. 6).
Figure 6.
A pair of salt bridges mediating coupling between the weak-to-strong state transition and ATPase activation. (A and B) Model of the MTBD–MT interaction in the weak and strong binding states, respectively. In the weak binding state (A), MTBD exhibited diffusional motion constrained in the electronegative potential valley of MT, with its electropositive surface orienting toward MT (Fig. 5 E). In the strong binding state (B), navigated by the flipped ion pair of E3390 and R403, MTBD found its binding site on MT (Fig. 5 C). The molecular structures of the MTBD and α- and β-tubulin at the MTBD–MT interface in the respective states are shown in the insets.
In the weak binding state, the MTBD diffuses along the MT, orienting its electropositive surface containing H2 and H3 toward the electronegative surface of MT (Figs. 5 E and 6 A; Vale et al., 1989; Wang and Sheetz, 2000; Minoura et al., 2010). During this diffusional search, the stalk coiled coil of the dynein molecule undergoes conformational changes among multiple registries, with the +β registry comprising a major fraction (Kon et al., 2009a). On the MT side, residues R403 and E416 of α-tubulin form an intra-tubulin salt bridge.
Navigated by the flipped charge (Fig. 5 B, circled), the MTBD and the MT face each other at a specific position of the MT to form the R403-E3390 salt bridge (Figs. 5 C and 6 B, and Video 3). Because α-R403 is a counterpart of the intra-tubulin R403-E416 salt bridge, formation of the R403-E3390 salt bridge inevitably forces another salt bridge to form between E416 and R3469/K3472, and, thus, the MTBD and the MT are interlocked with each other in a specific orientation through this pair of salt bridges. This cooperative rearrangement of salt bridges simultaneously shifts the structural equilibrium of stalk coiled-coil helices toward the α registry, transmitting a structural signal required for acceleration of the rate-limiting step in the ATPase cycle at the AAA1 ATPase site (Fig. 6 B, inset).
Because the α-R403A and α-E416A MTs could not produce directional movement (Fig. 2), this switching mechanism must be somehow linked to the directional movement of dynein. One possibility is that the stationary binding of the MTBD on MTs triggers the swing of dynein’s linker domain, leading to the displacement of the N terminus of the linker toward the minus end (Burgess et al., 2003; Kon et al., 2005). Alternatively, a molecular switch from diffusional to stationary binding is a strain sensor by itself, and thus a transition to stationary binding depends on the direction of the load. Recent analyses of dynein stepping showed that coordination of dynein’s two heads emerges as the distance between motor domains increases, indicating the possibility that the MTBD binds stationary to MTs when an internal load is imposed against the direction of movement (DeWitt et al., 2012; Qiu et al., 2012). Such asymmetry in MT binding, if it indeed happens, may facilitate or drive the directional movement of dynein.
Whatever mechanism controls the directionality, the R403-E3390 salt bridge plays a central role in our model because of its flipped charge as compared with the surrounding residues (Fig. 6). Indeed, the importance of residue R403 is supported by the in vivo observation that the mutation R402H of the human TUBA3 protein (R403 in yeast α-tubulin) causes the neurodevelopmental disorder lissencephaly (Keays et al., 2007). The symptoms of patients carrying the α-R402H mutation closely resemble those caused by mutations in LIS1, an adaptor protein that controls the binding mode of dynein with the MT (Vallee et al., 2001). The mutation R402H might have prevented dynein from switching from diffusional motion to strong binding, thereby impairing dynein motility and causing brain malformation.
The present study demonstrates the importance of studying the role of the MT in motility. Because coupling of ATP hydrolysis and motor binding/release is achieved through a dynamic interplay between dynein and MTs, it is only possible to clarify the mechanism by studying a complete system composed of both elements and not just motor protein alone. Our finding that the negatively charged short loop of α-tubulin (415EEGE) is critical for ATPase activation of both kinesin and dynein indicates that the MT may play an essential role in the motility mechanism of motor proteins.
Materials and methods
Preparation of MTs
Mutant MTs were expressed and purified using a yeast S. cerevisiae expression system. To prepare MTs from tubulin with lethal mutations, harmless tubulin with numerous glutamates at the C terminus were coexpressed and biochemically separated from each other by Mono Q anion-exchange chromatography (Uchimura et al., 2006, 2010). For polarity-marked MTs used in the MT-gliding assay, at first, tetramethylrhodamine-labeled MT seeds were generated (Hyman et al., 1991). In brief, 100 µM of yeast MTs in TL buffer (100 mM Hepes-KOH, 1 mM MgCl2, 1 mM EGTA, and 40% glycerol; pH 8.6) containing 1 mM GTP and 200 µM taxol was labeled by 5-(and-6)-carboxytetramethylrhodamine succinimidyl ester at a dye/tubulin dimer ratio of 20:1 at room temperature for 15 min, and free dyes were separated from the MTs by two wash cycles with BRB-2Mg buffer (80 mM Pipes-KOH, 2 mM MgCl2, and 1 mM EGTA; pH 6.8) and centrifugation. In the second cycle, the MTs were suspended without taxol to induce partial depolymerization, thereby generating the fluorescently labeled MT seeds ∼1 µm in length. To make polarity-marked MTs, the MT seeds were mixed with nonlabeled tubulin (10 µM) at a molar ratio of 1:10 and incubated at 30°C for 20 min. To avoid end-to-end annealing (Rothwell et al., 1986), the resultant block copolymers were diluted to 1 µM with BRB-2Mg buffer containing 20 µM taxol and kept on ice until use. The polarity-marked MTs were used within a day of the preparation. In the MT-gliding assay using kinesin, ∼80% of the MTs had a longer extension of the nonlabeled segment at the trailing end (i.e., plus end).
For MTs used in the cryoEM analysis, recombinant tubulin was expressed from the mec-12 α-tubulin and mec-7 β-tubulin genes of C. elegans (Chalfie, 1982; Chalfie and Au, 1989) using a baculovirus-insect cell expression system (Minoura et al., 2013). Synthetic DNA fragments encoding mec-12 α-tubulin with its C terminus fused to the His8 tag and mec-7 β-tubulin with its C terminus fused to the FLAG tag (DYKDDDDK) were designed, each with a joint sequence encoding a Factor Xa cleavage site (IEGR) and linker (GGSGG) inserted between tubulin and the tag. The tubulin dimer expressed in HighFive cells was purified by successive use of a DEAE anion exchange column, His8 tag affinity column, and FLAG tag affinity column chromatography, followed by a cycle of polymerization and depolymerization. The second round of polymerization was conducted in BRB-5Mg buffer (80 mM Pipes-KOH, 5 mM MgCl2, and 1 mM EGTA; pH 6.8) containing 5% DMSO. The polymerized MTs were centrifuged and resuspended in PK buffer (20 mM Pipes-KOH, 10 mM potassium acetate, 4 mM MgSO4, and 1 mM EGTA; pH 7.0) containing 1 mM GTP and 20 µM taxol. For proteolytic digestion of tags, Factor Xa was added to a 20-µM MT solution at a final concentration of 0.04 mg/ml and reacted for 16 h at room temperature. The protease and the digested tag fragments were removed by centrifugation, and MTs were resuspended in PK buffer containing 1 mM GTP and 20 µM taxol at a final concentration of 20 µM. Approximately 60% of the MTs prepared by this method were composed of 15 pfs.
Preparation of dynein
For the monomeric dynein construct MD380, the C-terminal 380-kD motor domain of cytoplasmic dynein from the cellular slime mold D. discoideum was fused to a His6 tag, a FLAG tag, and a biotin tag in tandem at its N terminus, with an additional SNAP tag inserted at the AAA2 domain (Numata et al., 2011). For dimeric dynein GST380, GST was further inserted between the biotin tag and motor domain. To create MTBD-removed monomeric construct MD380ΔMTBD, the MTBD coding region A3372-K3495 was replaced by linker sequence TG (Kon et al., 2012; also see Fig. S3 A). These dynein constructs were expressed in D. discoideum and purified by His6 and FLAG tag affinity column chromatography (Kon et al., 2009b). For fluorescent labeling, SNAP-Surface Alexa Fluor 546 (New England BioLabs, Inc.) was covalently attached to the dynein SNAP tag. For the dynein MTBD construct (Fig. S3 A), synthetic DNA was optimized for expression in Escherichia coli. Purification was performed using a His6 tag affinity column and gel filtration chromatography.
Observation of nuclear segregation in yeast cells
Nuclear segregation was examined in synchronous culture of yeast cells, as previously reported (Eshel et al., 1993; Li et al., 1993). The cells grown to mid-logarithmic phase at 30°C were fixed by 3.7% formaldehyde, their nuclei were stained with DAPI, and the stained cells were observed under a fluorescence microscope (IX71; Olympus). As a positive control, a yeast dynein disruptant, Δdyn1, was generated by homologous recombination.
Motility assays
The single-molecule motility assay of two-headed dynein GST380 was conducted under a total internal reflection fluorescence microscope (IX70; Olympus) equipped with a 100×/1.40 NA oil-immersion lens (Plan Apochromat; Olympus) and an electron multiplying charge coupled device camera (ImagEM C9100-13; Hamamatsu Photonics) at 25°C. The flow chamber was prepared according to a method previously described for kinesin (Uchimura et al., 2010), except that the ×2 MA buffer was replaced by PK buffer; it was successively flushed with anti–α-tubulin antibody (Sigma-Aldrich), MTs, casein (Nacalai Tesque), and dynein with oxygen scavengers (Harada et al., 1990). The final concentration of fluorescently labeled dynein in the chamber was 0.37 nM. For MT-gliding assay, a flow chamber composed of a coverslip and a glass slide was sequentially coated with biotinamidocaproyl BSA (Sigma-Aldrich), streptavidin (Life Technologies), α-casein (Sigma-Aldrich), and biotinylated dynein monomers MD380 (surface dynein density = ∼190 µm−2; Shima et al., 2006). The behavior of the polarity-marked MTs was observed under the dark-field microscope (BX50; Olympus) at 25°C (Video 1).
Analyses of motility
In the single-molecule motility assay, the movement of individual dynein molecules was analyzed as previously described (Uchimura et al., 2010). The mean run length was determined by nonlinear least squares fitting of the cumulative probability distribution of the data (Thorn et al., 2000) and the mean velocity was calculated by fitting the Gaussian distribution. For each wild-type and mutant MT, the movements of 68–234 dynein molecules on 5–13 MTs were analyzed. In the analyses of the MT-gliding assay, only MTs with approximate lengths of 4 µm were subjected to the analysis. For each wild-type and mutant MTs, the motions of multiple MTs in three independent experiments were recorded at a frame rate of 30 frames/s, resulting in measurements totaling 18, 14, and 11 MTs for wild type, α-R403A, and α-E416A, respectively. The movement of individual MTs was converted to a kymograph using ImageJ (National Institutes of Health), and the displacement of MTs in a direction parallel to their long axis was measured using R (The R Foundation) and software that we developed in house. Diffusion coefficients were calculated based on a total of 7,216 and 5,271 measurements of net displacement (for α-R403A and α-E416A mutant, respectively) taken at 2-s intervals.
Biochemical measurements
The equilibrium dissociation constant of monomeric dynein MD380 was measured by cosedimentation with the MTs (Ma and Taylor, 1995) in PK buffer under four nucleotide conditions: 1 U/ml apyrase (no nucleotide); 1 mM AMP-PNP and 1 U/ml apyrase (AMP-PNP); 1 mM ATP and 1 mM vanadate (ADP-Vi); and 1 mM ADP, 1 U/ml hexokinase, and 0.01% glucose (ADP). Fluorescently labeled dynein (50 nM) was incubated with various concentrations of MTs at 25°C for 10 min, and the sample was filtered through Ultrafree-MC (EMD Millipore). The filtrate was analyzed by fluorometry (FP-6500; JASCO). The amount of the steady-state MT-activated ATPase of MD380 was measured by using the malachite green method (Kodama et al., 1986), in which the time course of phosphate release was colorimetrically quantified with an absorbance at 650 nm. Reactions were performed in PK buffer containing 10 nM dynein, 0–50 µM MTs, 50 µM taxol, 1 mg/ml BSA, and 2 mM ATP at 25°C.
CryoEM grid preparation and imaging
Sample grids for cryoEM were prepared as follows. First, a 3-µl aliquot of 0.2 µM MTs in PK buffer containing 20 µM taxol was applied onto a glow-discharged holey carbon molybdenum grid (R1.2/1.3; Quantifoil). To enhance the attachment of MTs to the grid surface, the sample solution was manually blotted using a wet filter paper from the opposite side. Subsequently, a 3-µl aliquot of 1 µM MTBD construct, diluted with MilliQ water containing 20 µM taxol (Sindelar and Downing, 2007), was added. After incubation for 1 min, the solution was blotted by touching the edge of the grid with a piece of wet filter paper, and this adding and blotting cycle was repeated twice. Finally, 1.5 µl of the 2-µM MTBD construct was applied to the grid, and the grid was quickly blotted and plunged into liquid nitrogen–cooled liquid ethane using an automated device (Vitrobot; FEI). CryoEM images were recorded with a 4,096 × 4,096-pixel charge coupled device camera (TemCam-F415MP; TVIPS) on an electron cryomicroscope (JEM3200FSC; JEOL) with an Ω-type energy filter, operated at 200 kV and a magnification of 109,489, a defocus range of 1.0 to 1.5 µm, and an electron dose of ∼20 electrons/Å2.
Image processing for cryoEM
Image analysis was conducted using EMAN, SPIDER, and their custom scripts according to the Iterative Helical Real Space Reconstruction method (Egelman, 2007). Segment images of 512 × 512 pixels were extracted from micrographs with a step shift of 58 pixels. Each image was corrected for the phase and amplitude contrast transfer functions (CTFs) by multiplying the CTF calculated from the defocus and astigmatism obtained by CTFFIND3 (Mindell and Grigorieff, 2003) as previously described (Fujii et al., 2010). We used a ratio of 7% for the amplitude CTF to the phase CTF (Yonekura et al., 2006). The images were then high-pass filtered (285 Å) to remove a density undulation of low spatial frequency, normalized, and cropped to 400 × 400 pixels. Three-dimensional image reconstructions were conducted by back projection with reference images produced by rotating the model volume azimuthally about the filament axis between 0° and 360° at every 1°. The initial reference images were produced using an undecorated 15-pf MT, and helical symmetry was imposed on the first reconstruction. The volume data and symmetry parameters obtained from the first reconstruction were used for the second round. In the next cycle, the latest volume data were used to produce new reference images and calculate symmetry parameters. Segment images showing poor correlation to the references were discarded in each cycle. This cycle was repeated iteratively until the symmetry values converged to a stable solution. The resulting reconstruction was then modified by multiplying the transform of the reconstruction by 1/[∑CTF2 + 1/SNR] (SNR is the signal-to-noise ratio) to compensate for the amplitude distortion by the CTF. The 15-pf specific symmetry derived from two-start helix was imposed on the map. The final map was sharpened using a top-hat filter between 9 and 12 Å. The image processing statistics are listed in Fig. S4 A and the resolution of the map is provided in Fig. S4 B.
The DireX program (Wang and Schröder, 2012) was used for flexible fitting of the crystal structure of the dynein MTBD (PDB ID: 3KVH; Kon et al., 2012) and tubulin dimer (PDB ID: 1JFF; Löwe et al., 2001) into the map. The structural distortion was eliminated using Flex-EM (Topf et al., 2008) and the angle of the side chains was adjusted in accordance with the result of the mutagenesis analysis (Koonce and Tikhonenko, 2000; this study) using Coot (Emsley and Cowtan, 2004). The density map has been deposited in the Electron Microscopy Data Bank under accession number EMD-5931, and the atomic coordinates of the fitted model have been submitted to the Protein Data Bank with RCSB ID code rcsb160319 and PDB ID code 3J6P.
Estimation of salt bridges and calculation of electrostatic potentials
Charged residues falling within 10 Å of the interface of MTBD and MT were identified based on the structural model of the MTBD–MT complex (Fig. 4), using University of California San Francisco (UCSF) Chimera. Electrostatic potentials of MTBD and MT were calculated using Adaptive Poisson-Boltzmann Solver (Unni et al., 2011).
Online supplemental material
Fig. S1 shows nuclear segregation in yeast tubulin mutants. Fig. S2 shows the diffusion coefficients for α-R403A and α-E416A MTs calculated from the distribution of MT displacements in an MT-gliding assay. Fig. S3 shows a characterization of MTBD and 15-pf MT used in cryoEM analysis. Fig. S4 shows image processing statistics and resolution of the cryoEM map. Fig. S5 represents the process of building a model for the MTBD–MT complex. Video 1 shows the behavior of single MT filaments in an MT-gliding assay. Video 2 shows a comparison of the models built by rigid body docking and a flexible fitting method. Video 3 shows the electrostatic isopotential contour around the MTBD. A ZIP file provides an R script to analyze a kymograph of the MT movements. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407039/DC1.
Supplementary Material
Acknowledgments
We thank Yuji Kohara at the National Institute of Genetics for providing the cDNA clone for the mec-12 and mec-7 genes, Takayuki Kato for maintaining the cryoEM facility in the best possible conditions, and Naoko Mizuno and Ken Sekimoto for critical reading of the manuscript.
This work was supported by Grants-in-Aid for Scientific Research (13403626 to T. Fujii, 20051006 to G. Kurisu, 23370073 to T. Kon, and 21227006 and 25000013 to K. Namba) and by Precursory Research for Embryonic Science and Technology from the Japan Science and Technology Agency (to T. Fujii and T. Kon).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CC2
- coiled coil 2
- cryoEM
- electron cryomicroscopy
- CTF
- contrast transfer function
- MT
- microtubule
- MTBD
- MT-binding domain
- pf
- protofilament
References
- Burgess S.A., Walker M.L., Sakakibara H., Knight P.J., and Oiwa K.. 2003. Dynein structure and power stroke. Nature. 421:715–718 10.1038/nature01377 [DOI] [PubMed] [Google Scholar]
- Carter A.P., Garbarino J.E., Wilson-Kubalek E.M., Shipley W.E., Cho C., Milligan R.A., Vale R.D., and Gibbons I.R.. 2008. Structure and functional role of dynein’s microtubule-binding domain. Science. 322:1691–1695 10.1126/science.1164424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Cho C., Jin L., and Vale R.D.. 2011. Crystal structure of the dynein motor domain. Science. 331:1159–1165 10.1126/science.1202393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M.1982. Microtubule structure in Caenorhabditis elegans neurons. Cold Spring Harb. Symp. Quant. Biol. 46:255–261 10.1101/SQB.1982.046.01.028 [DOI] [PubMed] [Google Scholar]
- Chalfie M., and Au M.. 1989. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 243:1027–1033 10.1126/science.2646709 [DOI] [PubMed] [Google Scholar]
- Chrétien D., and Wade R.H.. 1991. New data on the microtubule surface lattice. Biol. Cell. 71:161–174 10.1016/0248-4900(91)90062-R [DOI] [PubMed] [Google Scholar]
- Davis A., Sage C.R., Wilson L., and Farrell K.W.. 1993. Purification and biochemical characterization of tubulin from the budding yeast Saccharomyces cerevisiae. Biochemistry. 32:8823–8835 10.1021/bi00085a013 [DOI] [PubMed] [Google Scholar]
- DeWitt M.A., Chang A.Y., Combs P.A., and Yildiz A.. 2012. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 335:221–225 10.1126/science.1215804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman E.H.2007. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157:83–94 10.1016/j.jsb.2006.05.015 [DOI] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Eshel D., Urrestarazu L.A., Vissers S., Jauniaux J.C., van Vliet-Reedijk J.C., Planta R.J., and Gibbons I.R.. 1993. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA. 90:11172–11176 10.1073/pnas.90.23.11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T., Iwane A.H., Yanagida T., and Namba K.. 2010. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 467:724–728 10.1038/nature09372 [DOI] [PubMed] [Google Scholar]
- Gibbons I.R.1981. Cilia and flagella of eukaryotes. J. Cell Biol. 91:107s–124s 10.1083/jcb.91.3.107s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I.R., Garbarino J.E., Tan C.E., Reck-Peterson S.L., Vale R.D., and Carter A.P.. 2005. The affinity of the dynein microtubule-binding domain is modulated by the conformation of its coiled-coil stalk. J. Biol. Chem. 280:23960–23965 10.1074/jbc.M501636200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L. Jr, Bode C.J., Georg G.I., and Himes R.H.. 2003. Understanding tubulin–Taxol interactions: mutations that impart Taxol binding to yeast tubulin. Proc. Natl. Acad. Sci. USA. 100:6394–6397 10.1073/pnas.1131967100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y., Sakurada K., Aoki T., Thomas D.D., and Yanagida T.. 1990. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J. Mol. Biol. 216:49–68 10.1016/S0022-2836(05)80060-9 [DOI] [PubMed] [Google Scholar]
- Höök P., and Vallee R.B.. 2006. The dynein family at a glance. J. Cell Sci. 119:4369–4371 10.1242/jcs.03176 [DOI] [PubMed] [Google Scholar]
- Hyman A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., and Mitchison T.. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Keays D.A., Tian G., Poirier K., Huang G.J., Siebold C., Cleak J., Oliver P.L., Fray M., Harvey R.J., Molnár Z., et al. . 2007. Mutations in α-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 128:45–57 10.1016/j.cell.2006.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T., Fukui K., and Kometani K.. 1986. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J. Biochem. 99:1465–1472. [DOI] [PubMed] [Google Scholar]
- Kon T., Mogami T., Ohkura R., Nishiura M., and Sutoh K.. 2005. ATP hydrolysis cycle–dependent tail motions in cytoplasmic dynein. Nat. Struct. Mol. Biol. 12:513–519 10.1038/nsmb930 [DOI] [PubMed] [Google Scholar]
- Kon T., Imamula K., Roberts A.J., Ohkura R., Knight P.J., Gibbons I.R., Burgess S.A., and Sutoh K.. 2009a. Helix sliding in the stalk coiled coil of dynein couples ATPase and microtubule binding. Nat. Struct. Mol. Biol. 16:325–333 10.1038/nsmb.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T., Shima T., and Sutoh K.. 2009b. Protein engineering approaches to study the dynein mechanism using a Dictyostelium expression system. Methods Cell Biol. 92:65–82 10.1016/S0091-679X(08)92005-7 [DOI] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., and Kurisu G.. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature. 484:345–350 10.1038/nature10955 [DOI] [PubMed] [Google Scholar]
- Koonce M.P., and Tikhonenko I.. 2000. Functional elements within the dynein microtubule-binding domain. Mol. Biol. Cell. 11:523–529 10.1091/mbc.11.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y., Yeh E., Hays T., and Bloom K.. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 90:10096–10100 10.1073/pnas.90.21.10096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K.H., and Nogales E.. 2001. Refined structure of α β-tubulin at 3.5 Å resolution. J. Mol. Biol. 313:1045–1057 10.1006/jmbi.2001.5077 [DOI] [PubMed] [Google Scholar]
- Ma Y.Z., and Taylor E.W.. 1995. Mechanism of microtubule kinesin ATPase. Biochemistry. 34:13242–13251 10.1021/bi00040a040 [DOI] [PubMed] [Google Scholar]
- Mallik R., and Gross S.P.. 2004. Molecular motors: strategies to get along. Curr. Biol. 14:R971–R982 10.1016/j.cub.2004.10.046 [DOI] [PubMed] [Google Scholar]
- Meng E.C., Pettersen E.F., Couch G.S., Huang C.C., and Ferrin T.E.. 2006. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 7:339 10.1186/1471-2105-7-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell J.A., and Grigorieff N.. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347 10.1016/S1047-8477(03)00069-8 [DOI] [PubMed] [Google Scholar]
- Minoura I., Katayama E., Sekimoto K., and Muto E.. 2010. One-dimensional Brownian motion of charged nanoparticles along microtubules: a model system for weak binding interactions. Biophys. J. 98:1589–1597 10.1016/j.bpj.2009.12.4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I., Hachikubo Y., Yamakita Y., Takazaki H., Ayukawa R., Uchimura S., and Muto E.. 2013. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 587:3450–3455 10.1016/j.febslet.2013.08.032 [DOI] [PubMed] [Google Scholar]
- Mizuno N., Toba S., Edamatsu M., Watai-Nishii J., Hirokawa N., Toyoshima Y.Y., and Kikkawa M.. 2004. Dynein and kinesin share an overlapping microtubule-binding site. EMBO J. 23:2459–2467 10.1038/sj.emboj.7600240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa Y., Oyama T., Kamiya N., Kon T., Toyoshima Y.Y., Nakamura H., and Kurisu G.. 2014. Structure of the entire stalk region of the Dynein motor domain. J. Mol. Biol. 426:3232–3245 10.1016/j.jmb.2014.06.023 [DOI] [PubMed] [Google Scholar]
- Numata N., Shima T., Ohkura R., Kon T., and Sutoh K.. 2011. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 585:1185–1190 10.1016/j.febslet.2011.03.036 [DOI] [PubMed] [Google Scholar]
- Pellman D., Bagget M., Tu Y.H., Fink G.R., and Tu H.. 1995. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J. Cell Biol. 130:1373–1385 (published erratum appears in J. Cell Biol. 1995. 131:561) 10.1083/jcb.130.6.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Derr N.D., Goodman B.S., Villa E., Wu D., Shih W., and Reck-Peterson S.L.. 2012. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol. 19:193–200 10.1038/nsmb.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redwine W.B., Hernández-López R., Zou S., Huang J., Reck-Peterson S.L., and Leschziner A.E.. 2012. Structural basis for microtubule binding and release by dynein. Science. 337:1532–1536 10.1126/science.1224151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.J., Kon T., Knight P.J., Sutoh K., and Burgess S.A.. 2013. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14:713–726 10.1038/nrm3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell S.W., Grasser W.A., and Murphy D.B.. 1986. End-to-end annealing of microtubules in vitro. J. Cell Biol. 102:619–627 10.1083/jcb.102.2.619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Koshland D., Eshel D., Gibbons I.R., and Hoyt M.A.. 1995. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J. Cell Biol. 128:617–624 10.1083/jcb.128.4.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima T., Imamula K., Kon T., Ohkura R., and Sutoh K.. 2006. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J. Struct. Biol. 156:182–189 10.1016/j.jsb.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Sindelar C.V., and Downing K.H.. 2007. The beginning of kinesin’s force-generating cycle visualized at 9-Å resolution. J. Cell Biol. 177:377–385 10.1083/jcb.200612090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn K.S., Ubersax J.A., and Vale R.D.. 2000. Engineering the processive run length of the kinesin motor. J. Cell Biol. 151:1093–1100 10.1083/jcb.151.5.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topf M., Lasker K., Webb B., Wolfson H., Chiu W., and Sali A.. 2008. Protein structure fitting and refinement guided by cryo-EM density. Structure. 16:295–307 10.1016/j.str.2007.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S., Oguchi Y., Katsuki M., Usui T., Osada H., Nikawa J., Ishiwata S., and Muto E.. 2006. Identification of a strong binding site for kinesin on the microtubule using mutant analysis of tubulin. EMBO J. 25:5932–5941 10.1038/sj.emboj.7601442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura S., Oguchi Y., Hachikubo Y., Ishiwata S., and Muto E.. 2010. Key residues on microtubule responsible for activation of kinesin ATPase. EMBO J. 29:1167–1175 10.1038/emboj.2010.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni S., Huang Y., Hanson R.M., Tobias M., Krishnan S., Li W.W., Nielsen J.E., and Baker N.A.. 2011. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 32:1488–1491 10.1002/jcc.21720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Soll D.R., and Gibbons I.R.. 1989. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 59:915–925 10.1016/0092-8674(89)90614-4 [DOI] [PubMed] [Google Scholar]
- Vallee R.B., Tai C., and Faulkner N.E.. 2001. LIS1: cellular function of a disease-causing gene. Trends Cell Biol. 11:155–160 10.1016/S0962-8924(01)01956-0 [DOI] [PubMed] [Google Scholar]
- Wang P.J., and Huffaker T.C.. 1997. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139:1271–1280 10.1083/jcb.139.5.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., and Schröder G.F.. 2012. Real-space refinement with DireX: from global fitting to side-chain improvements. Biopolymers. 97:687–697 10.1002/bip.22046 [DOI] [PubMed] [Google Scholar]
- Wang Z., and Sheetz M.P.. 2000. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 78:1955–1964 10.1016/S0006-3495(00)76743-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura K., Braunfeld M.B., Maki-Yonekura S., and Agard D.A.. 2006. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300 kV. J. Struct. Biol. 156:524–536 10.1016/j.jsb.2006.07.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.