During mitosis in human cells, separate mechanisms involving Aurora B and CENP-T promote anchoring of the microtubule- and checkpoint-receptor complex KMN at kinetochores.
Abstract
During mitosis, the spindle checkpoint senses kinetochores not properly attached to spindle microtubules and prevents precocious sister-chromatid separation and aneuploidy. The constitutive centromere-associated network (CCAN) at inner kinetochores anchors the KMN network consisting of Knl1, the Mis12 complex (Mis12C), and the Ndc80 complex (Ndc80C) at outer kinetochores. KMN is a critical kinetochore receptor for both microtubules and checkpoint proteins. Here, we show that nearly complete inactivation of KMN in human cells through multiple strategies produced strong checkpoint defects even when all kinetochores lacked microtubule attachment. These KMN-inactivating strategies reveal multiple KMN assembly mechanisms at human mitotic kinetochores. In one mechanism, the centromeric kinase Aurora B phosphorylates Mis12C and strengthens its binding to the CCAN subunit CENP-C. In another, CENP-T contributes to KMN attachment in a CENP-H-I-K–dependent manner. Our study provides insights into the mechanisms of mitosis-specific assembly of the checkpoint platform KMN at human kinetochores.
Introduction
The kinetochore is a multilayered protein assembly on centromeric chromatin and acts as a platform on sister chromatids for the attachment of spindle microtubules during mitosis (Cleveland et al., 2003). The constitutive centromere-associated network (CCAN) of proteins binds to centromeric chromatin throughout the cell cycle and forms the inner kinetochore (Foltz et al., 2006; Okada et al., 2006; Black and Cleveland, 2011; Takeuchi and Fukagawa, 2012). It provides the foundation for the mitosis-specific assembly of the outer kinetochore. Among the outer kinetochore proteins, the KMN network, which consists of the Knl1 complex (Knl1C, which consists of Knl1 and Zwint in humans), the Mis12 complex (Mis12C, which comprises Dsn1, Nsl1, Mis12, and Nnf1), and the Ndc80 complex (Ndc80C, which comprises Ndc80, Nuf2, Spc25, and Spc24), acts as a receptor for spindle microtubules (Cheeseman et al., 2006; Cheeseman and Desai, 2008). In this network, Mis12C directly binds to both Knl1 and Ndc80C, thus bridging an interaction between the two (Gascoigne and Cheeseman, 2013; Petrovic et al., 2014).
Accurate chromosome segregation relies on proper kinetochore–microtubule attachment during mitosis, which entails the capturing of a pair of sister kinetochores by microtubules originating from the two opposite spindle poles (a state termed bi-orientation; Cheeseman and Desai, 2008). Unattached or improperly attached kinetochores activate the spindle checkpoint to delay anaphase onset (Lara-Gonzalez et al., 2012; Foley and Kapoor, 2013; Jia et al., 2013). Furthermore, the centromeric kinase Aurora B severs improper kinetochore–microtubule attachments through phosphorylating multiple KMN components (Tanaka et al., 2002; Ruchaud et al., 2007; Welburn et al., 2010; Lampson and Cheeseman, 2011), thus promoting sister-chromatid bi-orientation. After all pairs of sister kinetochores reach bi-orientation, the spindle checkpoint is inactivated to allow synchronous dissolution of sister-chromatid cohesion and equal partition of the separated sister chromatids into the two daughter cells.
In addition to microtubule binding, KMN recruits spindle checkpoint proteins to outer kinetochores during mitosis. Knl1 is the kinetochore receptor for the Bub1–Bub3 and BubR1–Bub3 checkpoint complexes (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Primorac et al., 2013; Vleugel et al., 2013; Krenn et al., 2014). Ndc80C is required for the optimal kinetochore targeting of Mps1 and Mad1–Mad2 (Martin-Lluesma et al., 2002; Stucke et al., 2004). Furthermore, antibody-mediated Ndc80C inactivation in Xenopus laevis or temperature-sensitive mutations of Ndc80C components in the budding yeast compromises the spindle checkpoint (McCleland et al., 2003). These findings suggest a role for KMN components in communicating microtubule attachment status to the spindle checkpoint.
Despite its importance, our understanding of how KMN is installed at human mitotic kinetochores is incomplete. During mitosis, CCAN connects centromeric chromatin and the outer kinetochore through simultaneously engaging both the centromere-defining CENP-A nucleosome and outer kinetochore components, including KMN. CENP-T and CENP-C directly interact with Ndc80C and Mis12C, respectively (Gascoigne et al., 2011; Screpanti et al., 2011; Schleiffer et al., 2012). These findings are seemingly consistent with a simple model in which KMN is anchored to CCAN through the bipartite CENP-C–Mis12C and CENP-T–Ndc80C interactions.
CENP-T binds to Ndc80C through a conserved phospho-motif in its N-terminal region (residues 85–99 in human CENP-T; Schleiffer et al., 2012), termed the Ndc80-binding motif (NBM) hereafter. Recent studies have shown, however, that CENP-T NBM and Mis12C bind competitively to overlapping sites on Ndc80C (Bock et al., 2012; Malvezzi et al., 2013; Nishino et al., 2013). Ndc80C bound to CENP-T NBM cannot simultaneously engage Mis12C. Thus, there are at least two kinetochore complexes containing Ndc80C: Ndc80C bound to CENP-T NBM and intact KMN bound to CENP-C. The CENP-C–Mis12C interaction remains the only well-characterized connection between CCAN and the intact KMN.
The mitosis-specific installation of Ndc80C bound to CENP-T NBM at human kinetochores is mediated by the cytoplasm-to-nucleus translocation of Ndc80C shortly before mitotic entry and by Cdk1-dependent phosphorylation of CENP-T NBM (Gascoigne and Cheeseman, 2013). The mitosis-specific targeting of CENP-C–bound KMN to kinetochores is not understood, however. In particular, Mis12C resides in the nucleus throughout interphase, but does not localize to kinetochores until mitosis (Gascoigne and Cheeseman, 2013), which suggests that the CCAN–Mis12C interaction is cell cycle regulated. The regulatory mechanism has not been established.
In this study, we investigated the functions of KMN in the spindle checkpoint in human cells. To avoid complication from checkpoint weakening by partial microtubule occupancy at kinetochores, we treated cells with high concentrations of the microtubule-depolymerizing drug nocodazole. We found that intact KMN is critical for sustaining nocodazole-triggered mitotic arrest. We developed multiple strategies to inactivate KMN below the threshold required for sustaining an active spindle checkpoint. These KMN-inactivation strategies reveal multiple mechanisms of KMN assembly at kinetochores. In one mechanism, Aurora B actively promotes the CCAN–KMN interaction through phosphorylating the Mis12C subunit Dsn1 and strengthening Mis12C binding to CENP-C in mitosis. Strikingly, coexpression of a phospho-mimicking Dsn1 mutant and Ndc80 fused to an NLS is sufficient to install KMN at interphase kinetochores. In another mechanism, CENP-T provides an additional, indirect interaction with Ndc80C, possibly through the CENP-H-I-K subcomplex. This function of CENP-T does not appear to require the NBM. Thus, our results provide insights into the molecular mechanisms of the mitosis-specific attachment of the key checkpoint platform KMN to human kinetochores.
Results
Simultaneous depletion of multiple KMN components causes strong spindle checkpoint defects
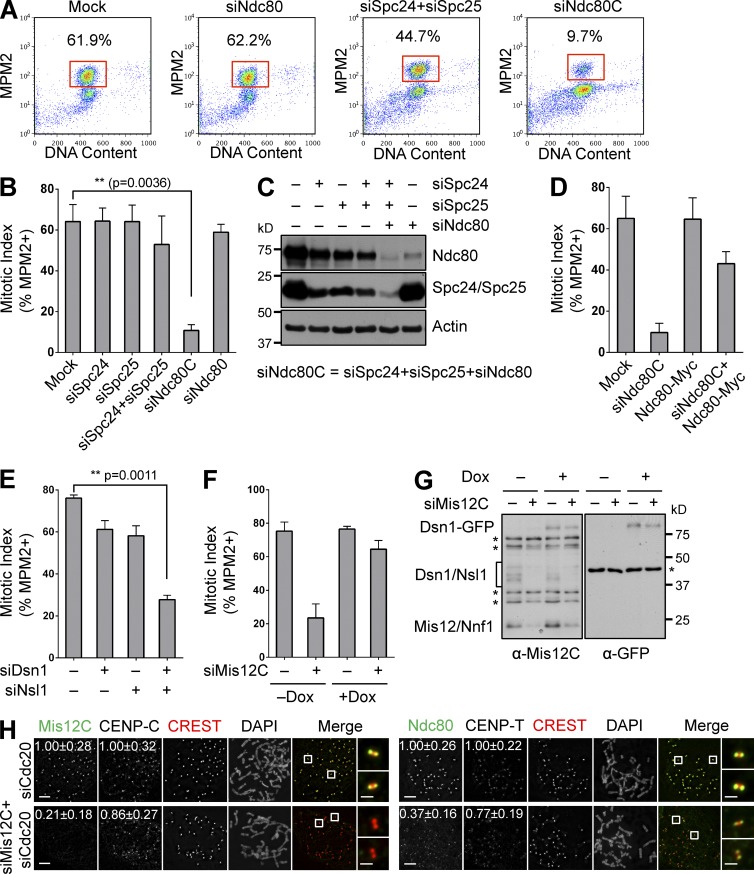
We first revisited the role of Ndc80C (which consisted of four subunits: Ndc80, Nuf2, Spc25, and Spc24) in the spindle checkpoint in human cells. Depletion of a single Ndc80C subunit in human cells caused kinetochore–microtubule attachment defects and checkpoint-dependent mitotic arrest (Martin-Lluesma et al., 2002; Bharadwaj et al., 2004). Consistently, these cells also underwent prolonged mitotic arrest in the presence of high concentrations of nocodazole (Fig. 1, A and B). Thus, depletion of a single subunit of Ndc80C reduces the levels of intact Ndc80C below the threshold needed for proper kinetochore–microtubule attachment. The small amount of residual Ndc80C is still above the threshold needed for spindle checkpoint signaling when all kinetochores are unattached.
Figure 1.
KMN is required for the spindle checkpoint in human cells. (A) Flow cytometry (FACS) analysis of HeLa cells transfected with the indicated siRNAs and treated with nocodazole. The mitotic indices (defined as the percentage of cells with 4C DNA content and high MPM2 staining) are indicated. (B) Quantification of mitotic indices of cells in A, with means and SD (error bars) shown (n = 3). (C) Lysates of cells in A and B were blotted with the indicated antibodies. (D and E) Quantification of mitotic indices of HeLa cells transfected with indicated vectors and siRNAs and treated with nocodazole (mean ± SD [error bars], n = 3). (F) Quantification of mitotic indices of HeLa cells stably expressing Dsn1-WT-GFP that were mock transfected or transfected with siMis12C in the absence (–) or presence (+) of doxycycline (Dox) and treated with nocodazole (mean ± SD [error bars], n = 3). (G) Lysates of cells in F were blotted with the indicated antibodies. Asterisks indicate nonspecific bands. (H) Mitotic HeLa cells transfected with the indicated siRNAs were stained with the indicated antibodies and DAPI. Selected channels (as labeled by the corresponding colors) were included in merged images. Boxed regions were magnified and shown in the rightmost column. Normalized intensities (mean ± SD [error bars], n = 400) of kinetochore staining are shown. Bars, 5 µm (1 µm in magnified images on the right).
We reasoned that simultaneous depletion of multiple subunits of a given protein complex might more efficiently reduce the concentration of the intact complex, as compared with the depletion of each subunit alone. Indeed, HeLa cells transfected with a mixture of siNdc80, siSpc25, and siSpc24 (termed siNdc80C) failed to arrest in mitosis in the presence of nocodazole, whereas cells transfected with siSpc25 and siSpc24 still did (Fig. 1, A and B). As expected, we observed the greatest decrease of protein levels of Ndc80C subunits when cells were transfected with siNdc80C (Fig. 1 C). Importantly, transient transfection of RNAi-resistant Ndc80-Myc partially restored the mitotic arrest in cells transfected with siNdc80C (Fig. 1 D). Therefore, Ndc80C is required for sustaining spindle checkpoint signaling, even when the microtubule is depolymerized and all kinetochores are unattached. Because the spindle checkpoint can be activated by a few unattached kinetochores within the cell (Rieder et al., 1995; Collin et al., 2013; Dick and Gerlich, 2013; Heinrich et al., 2013; Subramanian and Kapoor, 2013), the kinetochore proteins that initiate checkpoint signaling have to be reduced to exceedingly low levels to reveal their requirement for mitotic arrest in the absence of the mitotic spindle.
We next tested whether Mis12C was critical for spindle checkpoint signaling. We depleted Mis12C, which consisted of Dsn1, Nsl1, Mis12, and Nnf1, from HeLa cells, and investigated the ability of these cells to undergo mitotic arrest in the presence of nocodazole. Depletion of both Dsn1 and Nsl1 with a mixture of siDsn1 and siNsl1 (termed siMis12C) caused a major defect in mitotic arrest, whereas depletion of either protein alone did not (Fig. 1 E). The mitotic-arrest deficiency produced by siMis12C was rescued by the inducible, stable expression of RNAi-resistant Dsn1-GFP (Fig. 1, F and G). These results indicate that Mis12C is indispensable for the spindle checkpoint, and suggest that the CENP-C–bound intact KMN is required for the spindle checkpoint.
CENP-T–bound Ndc80C is insufficient to sustain checkpoint signaling without Mis12C
To examine the effect of Mis12C depletion on the localization of Ndc80C and vice versa, we transfected cells with siMis12C or siNdc80C together with siCdc20 to prevent mitotic exit, and visualized Mis12C and Ndc80 by immunofluorescence (Fig. 1 H). Transfection of siMis12C reduced the kinetochore signal of Mis12C to background levels. The kinetochore signal of Ndc80 was reduced about twofold. In contrast, the signals of CENP-C and CENP-T at kinetochores were not greatly reduced. These results are consistent with earlier studies, and suggest that there are multiple pools of Ndc80C at kinetochores. The remaining signal of Ndc80 at kinetochores in siMis12C cells was further reduced by CENP-T depletion (Fig. S1 A), which confirms that this pool of Ndc80C is likely bound to CENP-T. Because siMis12C cells fail to undergo mitotic arrest in nocodazole, this pool of Ndc80C is insufficient to maintain the spindle checkpoint in the absence of Mis12C.
The kinetochore signal of Ndc80 was undetectable in cells treated with siNdc80C (Fig. S1 B). While depletion of Ndc80C did not alter the kinetochore localization of CENP-C or CENP-T, the kinetochore signal of Mis12C was reduced by twofold (Fig. S1 C). This finding was surprising, as CENP-C is the major kinetochore receptor of Mis12C, and Ndc80C binding to Mis12C does not affect the binding of Mis12C to CENP-C in vitro (Screpanti et al., 2011). The fact that Ndc80C contributes the kinetochore localization of Mis12C thus suggests that, aside from the CENP-C–Mis12C interaction, attachment of intact KMN to kinetochores involves an additional Ndc80C- dependent mechanism.
Aurora B regulates the kinetochore targeting of KMN during mitosis
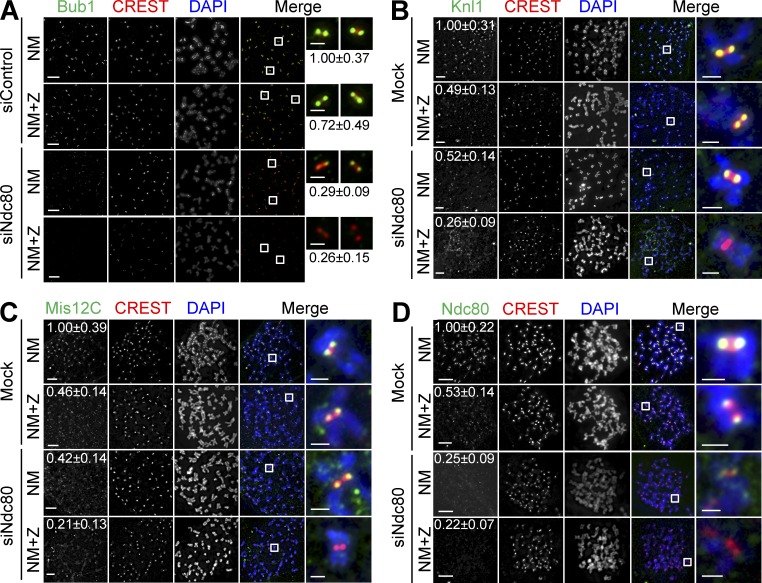
Aurora B inhibition in Ndc80 RNAi cells caused them to escape from nocodazole-induced mitotic arrest (Santaguida et al., 2011; Saurin et al., 2011). We decided to investigate the mechanism underlying the synergy between Aurora B inhibition and Ndc80 depletion in abolishing the spindle checkpoint. As expected, we found that several checkpoint proteins, including Bub1, BubR1, Mad1, and Mad2, were absent from kinetochores in siNdc80/ZM447439 (ZM)-treated cells that were maintained in mitosis by the proteasome inhibitor MG132 (Fig. 2 A and not depicted). Because KMN is required for the kinetochore localization of these checkpoint proteins and because Aurora B contributes to outer kinetochore assembly in various organisms (Emanuele et al., 2008), we examined the localization of KMN in these cells. Aurora B inhibition or siNdc80 alone diminished the kinetochore staining of Knl1 and Mis12C about twofold (Fig. 2, B and C). Aurora B inhibition did not alter CENP-C and -T localization (Fig. S1, D and E). To our surprise, Aurora B inhibition in Ndc80 RNAi cells abolished the kinetochore localization of Knl1 and Mis12C (Fig. 2, B and C). These results indicate that, when Ndc80C is compromised, the kinetochore targeting of Knl1 and Mis12C requires Aurora B. Thus, a critical microtubule-independent function of Aurora B in the spindle checkpoint is to cooperate with Ndc80C to target Knl1, Mis12C, and downstream checkpoint proteins to kinetochores.
Figure 2.
Aurora B is critical for KMN kinetochore targeting in cells with compromised Ndc80C. (A–D) HeLa cells were transfected with the indicated siRNAs and treated with nocodazole (N). Mitotic cells were collected and divided into two wells: one well was incubated with the proteasome inhibitor MG132 (NM), whereas the other was treated with both MG132 and ZM (NM+Z). Cells were stained with the indicated antibodies and DAPI. Selected channels (as labeled by the corresponding colors) were included in the merged images. Boxed regions are magnified and shown in the rightmost columns. The relative kinetochore intensities of KMN (mean ± SD, n = 400) were shown. The CREST and DAPI channels in D are shown again in Fig. S2 E alongside labeling for an additional protein. Bars, 5 µm (1 µm for the magnified images).
We noticed that there were detectable Ndc80 kinetochore signals in siNdc80-treated cells (Fig. 2 D). This residual Ndc80 signal was abolished by Aurora B inhibition. Because this residual Ndc80 signal was barely above background, quantification of the intensity of the Ndc80 signal in siNdc80 cells did not show significant differences with or without Aurora B inhibition (Fig. 2 D). These results again suggest that the spindle checkpoint requires a very small amount of Ndc80C (presumably as a part of KMN) at kinetochores.
Aurora B contributes to KMN kinetochore targeting through Dsn1 phosphorylation
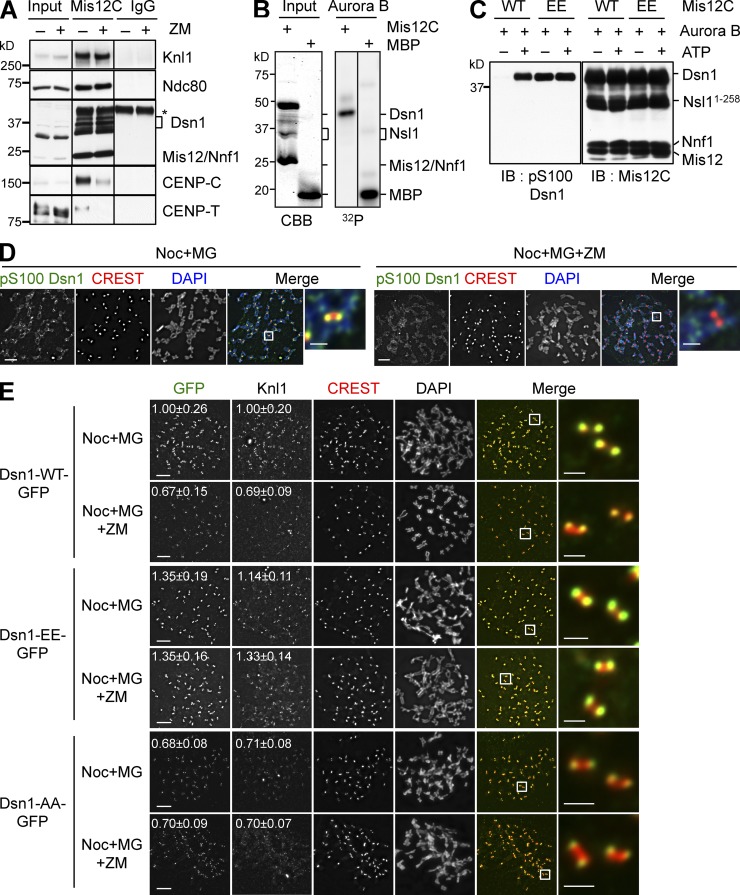
Consistent with the fact that Aurora B inhibition reduced the kinetochore localization of KMN without affecting CENP-C localization, Aurora B inhibition weakened the binding between KMN and CENP-C in coimmunoprecipitation (IP) assays (Fig. 3 A). We next sought to identify the relevant Aurora B substrates in this process. Aurora B phosphorylated Dsn1 of recombinant Mis12C in vitro (Fig. 3 B). Consistent with previous studies (Yang et al., 2008; Welburn et al., 2010), our mass spectrometry analysis identified S100 and S109 in Dsn1 as the major Aurora B phosphorylation sites (unpublished data). The functions of these phosphorylation events were controversial, however (Yang et al., 2008; Welburn et al., 2010). We made a phospho-specific antibody against phospho-S100 Dsn1 (Fig. 3 C), and confirmed that phospho-S100 Dsn1 localized to kinetochores, and that this phosphorylation required Aurora B activity (Fig. 3 D).
Figure 3.
Aurora B phosphorylates Dsn1 and strengthens the CCAN–KMN interaction. (A) Lysates (Input), α-Mis12C IP (Mis12C), and IgG IP of mitotic HeLa cells treated with nocodazole and MG132 in the presence or absence of ZM447439 were blotted with the indicated antibodies. The asterisk indicates the IgG heavy chain in the IP samples. Dsn1 migrated as multiple bands due to phosphorylation. Mis12 and Nnf1 co-migrated. (B) Mis12C was incubated with Aurora B–INCENP and γ-[32P]ATP, separated by SDS-PAGE, and analyzed by Coomassie brilliant blue (CBB) staining (left) or with a phosphorimager (right). Mis12 and Nnf1 co-migrated. Nsl1 underwent proteolysis. Myelin basic protein (MBP) was used as a positive control. (C) Mis12C containing Dsn1-WT or Dsn1-EE was incubated with Aurora B–INCENP with (+) or without (–) cold ATP. Samples were separated by SDS-PAGE and blotted with the indicated antibodies. Dsn1-EE was recognized by α-pS100 Dsn1. Black lines indicate that intervening lanes have been spliced out. (D) HeLa cells were arrested in prometaphase with nocodazole and incubated with MG132 alone or with MG132 and ZM447439, and then stained with DAPI (blue in the merge), CREST (red), and α-pS100 Dsn1 (green). The boxed regions were magnified and shown in the rightmost column. (E) Cells stably expressing Dsn1-WT, S100E/S109E (EE), or S100A/S109A (AA)-GFP were transfected with siDsn1, treated as in D, and stained with the indicated antibodies. Boxed regions in merged images were magnified and shown in the rightmost column. The normalized kinetochore intensities (mean ± SD, n = 400) of Dsn1-GFP and Knl1of cells were quantified and shown. Bars, 5 µm (1 µm for magnified images).
We constructed HeLa cell lines that stably expressed Dsn1-WT-GFP, the phospho-deficient Dsn1-S100A/S109A (AA)-GFP, and the phospho-mimicking Dsn1-S100E/S109E (EE)-GFP in a doxycycline-inducible manner (Fig. S2 A), and monitored the kinetochore localization of these Dsn1-GFP proteins (Fig. 3 E). Similar to that of the endogenous Mis12C, the kinetochore signal of Dsn1-WT-GFP was reduced by ZM (Fig. 3 E). The kinetochore signal of the phospho-mimicking Dsn1-EE-GFP was slightly stronger than that of Dsn1-WT-GFP, and was not reduced by ZM. The kinetochore signal of Dsn1-AA-GFP was similar to that of Dsn1-WT-GFP in ZM-treated cells, and was not further reduced by ZM. The intensities of the kinetochore signals of Knl1 were similarly affected in these samples (Fig. 3 E). These results suggest that phosphorylation of Dsn1 by Aurora B contributes to KMN kinetochore localization in mitosis.
We then transfected cells with siMis12C and measured the mitotic index in the presence of nocodazole, with or without inducing the expression of Dsn1-WT-GFP or Dsn1-AA-GFP (Fig. S2, B and C). Although Dsn1-WT was able to rescue the mitotic escape caused by Mis12C depletion, Dsn1-AA was partially defective in rescuing the phenotype. Thus, Aurora B–dependent phosphorylation contributes to, but is not required for, the spindle checkpoint.
Phospho-mimicking Dsn1 mutation and forced nuclear localization of Ndc80 suffice to install KMN at interphase kinetochores
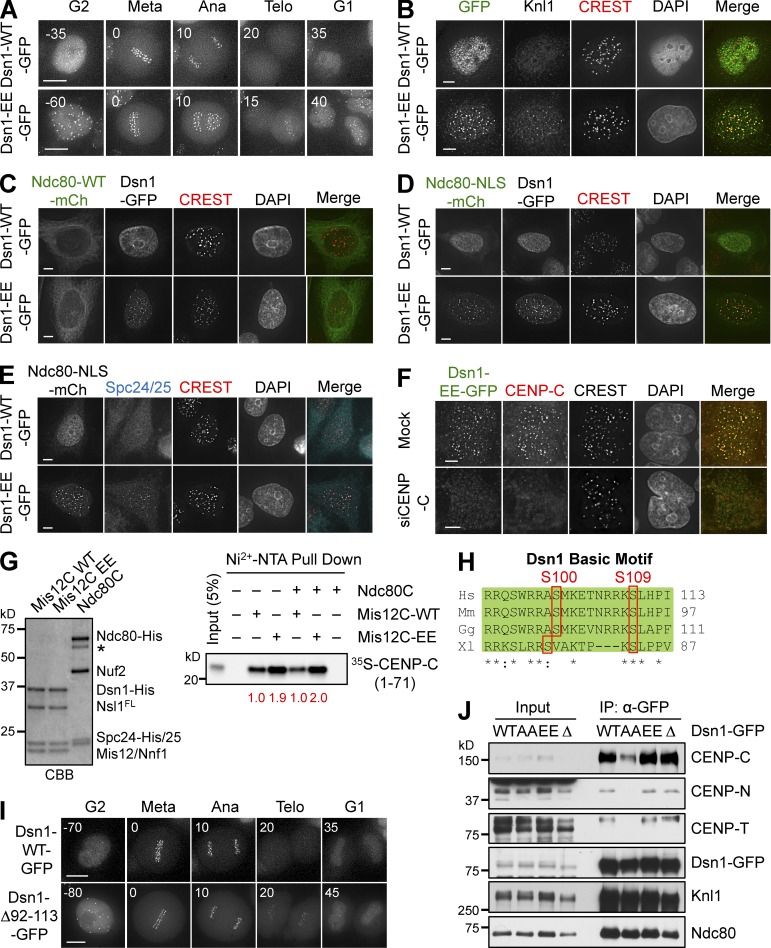
We monitored the subcellular localization of Dsn1-WT-GFP and Dsn1-EE-GFP using live-cell imaging. Dsn1-WT-GFP localized to kinetochores during mitosis, but not in interphase (Fig. 4 A). In contrast, Dsn1-EE-GFP constitutively localized to kinetochores throughout the cell cycle (Figs. 4 A and S3, A and B). Dsn1-EE-GFP was sufficient to target Knl1 to kinetochores during interphase (Fig. 4 B), but was insufficient to recruit Ndc80 or Mad1, which remained localized at the cytosol and nuclear pores, respectively (Fig. S3 C). Therefore, although Dsn1 phosphorylation by Aurora B is not strictly required for Mis12C kinetochore localization in mitosis, untimely phosphorylation is likely sufficient to target the Knl1–Mis12C complex to kinetochores in interphase.
Figure 4.
Phospho-mimicking Dsn1 mutation enhances Mis12C binding to CENP-C. (A) HeLa cells expressing Dsn1-WT/EE-GFP were analyzed by time-lapse microscopy. GFP images of representative cells at the indicated times (in minutes) were shown. Metaphase was used as the reference point (time 0). (B) Interphase cells expressing Dsn1-WT/EE-GFP were stained with the indicated antibodies and DAPI. (C–E) HeLa cells expressing Dsn1-WT/EE-GFP were transfected with Ndc80-WT/NLS-mCherry plasmids, and stained with the indicated antibodies and DAPI. (F) HeLa cells expressing Dsn1-EE-GFP transfected with siCENP-C and treated with thymidine were stained with the indicated antibodies and DAPI. (G) In vitro binding assays between recombinant Dsn1-WT/EE-containing Mis12C with or without Ndc80C and 35S-labeled CENP-C (residues 1–71; right). The relative CENP-C binding intensity is indicated at the bottom. Mis12C and Ndc80C were stained with CBB (left). The asterisk indicates a degradation band of Ndc80. (H) Sequence alignment of the basic motif encompassing the two Aurora B phosphorylation sites in Dsn1 proteins from different species (Hs, Homo sapiens; Mm, Mus musculus; Gg, Gallus gallus; Xl, Xenopus laevis). (I) HeLa cells expressing Dsn1-WT/Δ92-113-GFP were analyzed by time-lapse microscopy. (J) Lysates and α-GFP IP of mitotic HeLa cells expressing Dsn1-WT, -AA, -EE, or -Δ92–113-GFP were blotted with the indicated antibodies. Bars: (A and I) 10 µm; (B–F) 5 µm.
An elegant, previous study showed that coexpression of a phospho-mimicking mutant of CENP-T and an Ndc80 protein fused to the NLS was sufficient to install Ndc80C, but not Mis12C, at interphase kinetochores (Gascoigne and Cheeseman, 2013). That study clearly demonstrated that the mitosis-specific kinetochore targeting of Ndc80C bound to CENP-T NBM relied on two processes: Cdk1-dependent phosphorylation of CENP-T and the nuclear translocation of Ndc80C. We tested whether forced nuclear targeting of Ndc80 enabled the kinetochore targeting of Ndc80C in Dsn1-EE cells in interphase. Similar to endogenous Ndc80, ectopically expressed Ndc80-mCherry remained in the cytosol in Dsn1-EE cells (Fig. 4 C). Ndc80-NLS-mCherry was enriched in the nucleus in Dsn1-WT cells (Fig. 4 D). Strikingly, Ndc80-NLS-mCherry localized to interphase kinetochores in Dsn1-EE cells. Other components of Ndc80C, Spc24/25, were also found at interphase kinetochores in cells expressing both Dsn1-EE and Ndc80-NLS (Fig. 4 E). These results suggest that Aurora B–dependent phosphorylation of Dsn1 and nuclear translocation of Ndc80C are key regulatory events during mitosis-specific assembly of KMN at kinetochores.
Dsn1 phosphorylation by Aurora B strengthens the CENP-C–Mis12C interaction
The simplest model to explain the interphase kinetochore targeting of Dsn1-EE is that Dsn1 phospho-mimicking mutations strengthened the Mis12C–CCAN interaction. Among CCAN components, CENP-C is a direct binding partner of Mis12C (Screpanti et al., 2011). The interphase kinetochore localization of Dsn1-EE was abolished in CENP-C RNAi cells (Fig. 4 F), but was still detectable in cells transfected with siCENP-T, -I, -L, -N, or -U (Fig. S3 D). Dsn1-EE-GFP, but not Dsn1-WT-GFP, coimmunoprecipitated with CENP-C in G1/S cells (Fig. S3 E). Therefore, CENP-C is still the major CCAN receptor for Dsn1-EE and, by inference, the phosphorylated form of Dsn1, in interphase cells.
We next compared the CENP-C–binding affinities of recombinant Mis12C-WT and -EE. His6-tagged Mis12C containing Dsn1-EE (Mis12C-EE) pulled down more 35S-labeled CENP-C1–71 (Fig. 4 G). The interaction between CENP-C and Mis12C-EE was not further enhanced by Ndc80C. We then quantitatively measured the binding affinities of Mis12C-WT and -EE toward a fluorescently labeled CENP-C peptide (residues 1–28) with microscale thermophoresis. The dissociation constant (Kd) of the Mis12C-WT–CENP-C1–28 interaction was 2.1 ± 0.4 µM, whereas the Kd between Mis12C-EE and CENP-C1–28 was 0.74 ± 0.15 µM (Fig. S3 F). These results suggest that Dsn1-EE and, quite possibly, phosphorylated Dsn1 bind to CENP-C more tightly than Dsn1-WT, although we do not know whether the threefold higher affinity suffices to explain the interphase targeting of Mis12-EE.
The two Aurora B phosphorylation sites of Dsn1—S100 and S109—reside in a basic motif conserved in vertebrate Dsn1 proteins (Fig. 4 H). To test whether this motif of Dsn1 mediated CENP-C binding, we created a Dsn1 mutant with this motif deleted (Δ91–113). Surprisingly, Dsn1-Δ91-113-GFP behaved like Dsn1-EE-GFP. It localized to kinetochores properly during mitosis (Fig. S3 G), and showed premature kinetochore localization during interphase (Figs. 4 I and S3, A and B). Similar amounts of CCAN components, including CENP-C, -N, and -T, were associated with Dsn1-WT, -EE, or Δ91–113 in mitotic cell lysates, whereas the CCAN interaction with Dsn1-AA was weaker (Fig. 4 J). Therefore, phospho-S100/S109 residues of Dsn1 do not create a direct CCAN-binding motif. We speculate that this basic motif of Dsn1 might mask the CENP-C–binding site of Mis12C through an autoinhibitory mechanism. Phosphorylation of this motif, as well as phospho-mimicking or deletion mutations, might release this autoinhibition and expose the binding site for CENP-C, thus enhancing the Mis12C–CCAN interaction. This Aurora B–dependent mechanism for strengthening the Mis12C–CCAN interaction is likely conserved in budding yeast (Akiyoshi et al., 2013).
Collectively, our results so far indicate that Aurora B contributes to the kinetochore targeting of KMN through phosphorylating Dsn1 and strengthening the CENP-C–Mis12C interaction. During normal mitosis, Aurora B is not strictly required for the kinetochore localization of KMN. Aurora B becomes critical, however, when the Ndc80C function is compromised. Thus, multiple mechanisms install KMN at kinetochores during mitosis: an Aurora B–dependent mechanism involving the CENP-C–Mis12C interaction, and an Aurora B–independent, Ndc80C-dependent mechanism.
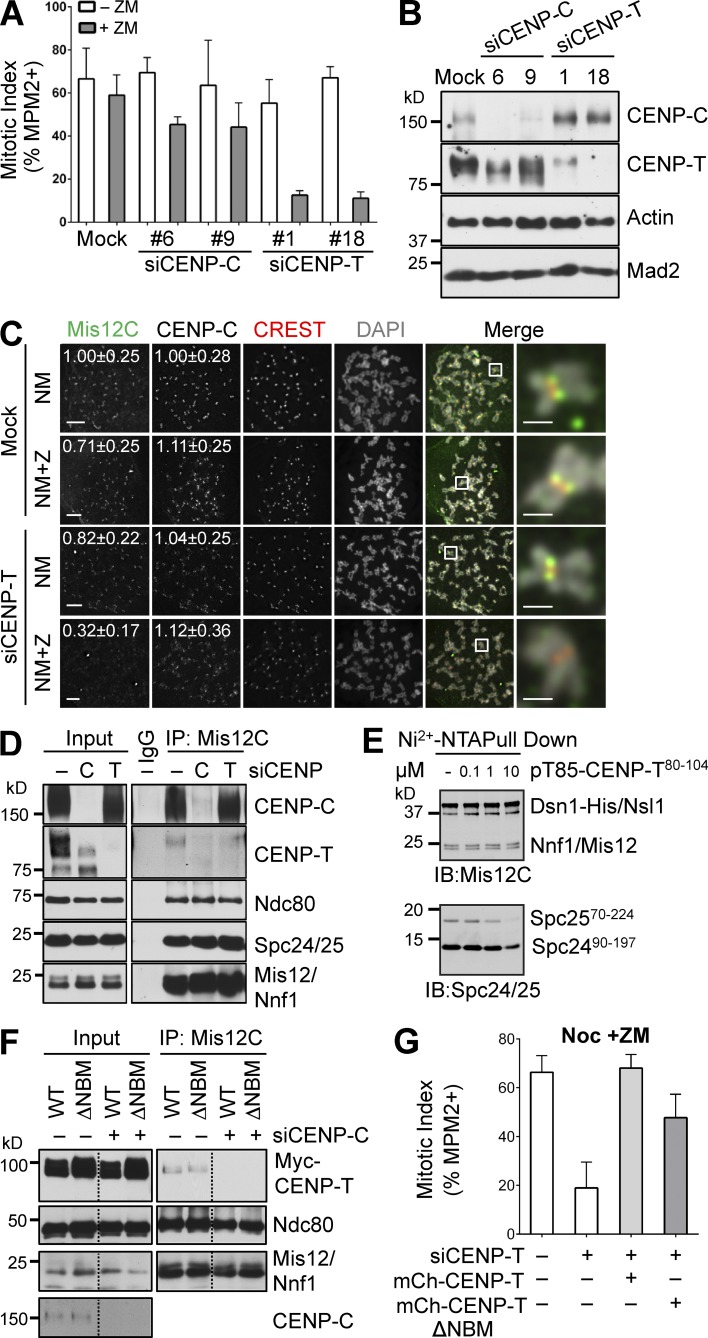
CENP-T contributes to KMN kinetochore targeting independently of its NBM
We next examined whether this Aurora B–independent, Ndc80C-dependent mechanism involved CENP-T. Similar to Aurora B inhibition in siNdc80 cells, Aurora B inhibition in siCENP-T cells caused mitotic arrest deficiency in the presence of nocodazole, whereas Aurora B inhibition had marginal effects on siCENP-C cells (Fig. 5, A and B). Moreover, Aurora B inhibition and siCENP-T reduced Mis12C and Ndc80 kinetochore signals to background levels, without affecting CENP-C signals (Figs. 5 C and S4 A). These results suggest that CENP-T is involved in the Aurora B–independent, Ndc80C-dependent mechanism of KMN attachment to kinetochores.
Figure 5.
CENP-T contributes to KMN kinetochore targeting independently of its NBM. (A) HeLa cells were mock transfected or transfected with the indicated siRNAs, treated with thymidine for 14 h, released into nocodazole-containing medium for 12 h, and treated with ZM for 2 h. The mitotic indices of these cells (mean ± SD [error bars], n = 3) were quantified with FACS. (B) Lysates of cells in A were blotted with the indicated antibodies. (C) HeLa cells were mock transfected or transfected with siCENP-T and arrested in mitosis by nocodazole. Mitotic cells were collected by shake-off. Each sample was divided into two fresh wells. One well was incubated with MG132 (MG) for 2 h (NM) while the other well was treated with both MG and ZM for 2 h (NM+Z). Cells were stained with the indicated antibodies and DAPI. The boxed regions of the merged images of the selected channels were magnified and shown in the rightmost column. The relative kinetochore intensities (mean ± SD, n = 400) of CENP-C and Mis12C were quantified and shown. Bars, 5 µm (1 µm for magnified images). (D) Lysates (Input), α-Mis12C IP (Mis12C), and IgG IP of mitotic HeLa cells transfected with the indicated siRNAs and treated with nocodazole were blotted with the indicated antibodies. (E) In vitro binding assay between recombinant Mis12C and Spc24-25 in the presence of increasing concentrations of a synthetic peptide containing residues 80–104 of human CENP-T with phospho-T85. The reaction mixtures were blotted with anti-Mis12C and anti-Spc24/25 antibodies. (F) Lysates (Input) and α-Mis12C IP (Mis12C) of mitotic HeLa cells transfected with the indicated plasmids and siCENP-C and treated with nocodazole were blotted with the indicated antibodies. Residues 85–99 were deleted in CENP-T ΔNBM. Lines indicate that intervening lanes have been spliced out. (G) Quantification of mitotic indices of HeLa cells expressing indicated proteins that were transfected with siCENP-T and treated with nocodazole and ZM (mean ± SD [error bars], n = 3).
To further dissect the role of CENP-T in KMN recruitment, we examined the CENP-C/T–Mis12C interactions in siCENP-C or siCENP-T cells (Fig. 5 D). A small amount of CENP-T was pulled down in the Mis12C IP. This weak CENP-T–Mis12C interaction was dependent on CENP-C, as CENP-T was no longer detectable in Mis12C IP of siCENP-C cells. In contrast, depletion of CENP-T did not appreciably affect the CENP-C–Mis12C interaction. This result suggests that the CENP-T–Mis12C interaction at the endogenous protein levels is likely indirect, and is bridged through CENP-C and other CCAN components.
Although CENP-T can interact directly with Ndc80C through its conserved NBM, this NBM and Mis12C have been shown to bind competitively to overlapping sites on Spc24/25 (Nishino et al., 2013). We further confirmed this competition. As shown in Fig. 5 E, a phospho-NBM–containing peptide at high concentration reduced Spc24/25 binding to Mis12C in vitro. Thus, Ndc80C bound to NBM cannot simultaneously interact with Mis12C. Consistent with this notion, both Myc-CENP-T WT and ΔNBM bound to Mis12C, and these interactions were completely dependent on CENP-C (Fig. 5 F). Therefore, CENP-T NBM is not required for the CENP-T–Mis12C interaction in human cells. Moreover, stable expression of CENP-T ΔNBM partially rescued the spindle checkpoint defects in cells treated with siCENP-T and ZM (Figs. 5 G and S4 B), and restored the kinetochore localization of Mis12C in these cells (Fig. S4 C). Thus, CENP-T has an NBM-independent role in promoting KMN attachment to kinetochores.
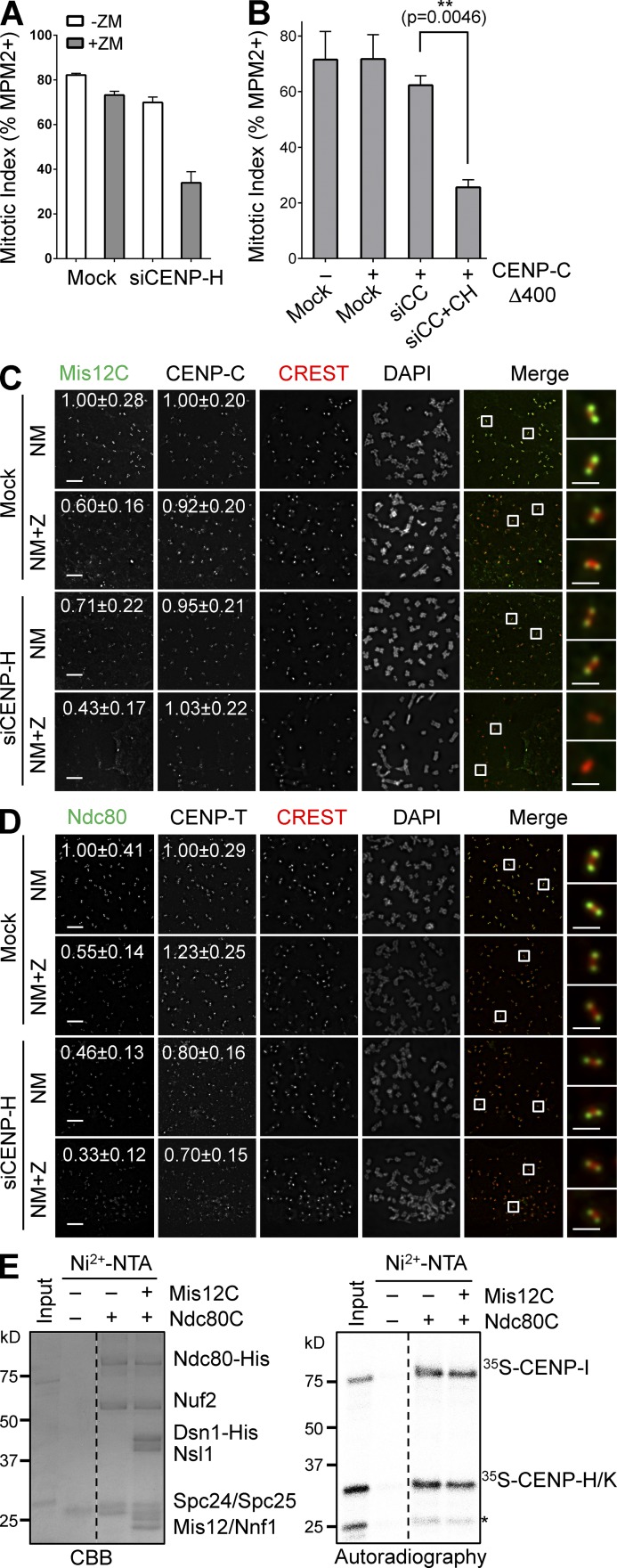
CENP-T contributes to KMN kinetochore attachment through CENP-H-I-K
Because CENP-T is required for the proper localization of other CCAN components including the CENP-H-I-K subcomplex (Hori et al., 2008), we tested whether CENP-T contributed to KMN kinetochore targeting indirectly by maintaining the integrity of CCAN. We first systematically depleted various CCAN components and tested whether their depletion synergized with Aurora B inhibition to cause mitotic arrest deficiency in the presence of nocodazole. Although depletion of CENP-N, CENP-U, or CENP-Q did not have significant effects (Fig. S4, D–F), depletion of CENP-H synergized with Aurora B inhibition to produce checkpoint defects (Figs. 6 A and S4 G), which is consistent with previous studies (Matson et al., 2012; Matson and Stukenberg, 2014). We next tested whether depletion of both CENP-C and CENP-H could produce strong checkpoint defects. A more complete CENP-C depletion caused an interphase delay in HeLa cells, complicating the analysis. Expression of the siRNA-resistant CENP-C Δ400, which was deficient in Mis12C binding but retained kinetochore binding (Fig. S5, A–C), rescued this interphase phenotype (not depicted). Co-depletion of CENP-C and CENP-H from CENP-C Δ400-expressing cells indeed caused strong checkpoint defects (Figs. 6 B and S5 A). Moreover, CENP-H depletion and Aurora B inhibition greatly reduced Mis12C and Ndc80C kinetochore localization, without affecting CENP-C localization (Fig. 6, C and D). These results are consistent with an involvement of CENP-H-I-K in Aurora B–independent KMN attachment to kinetochores.
Figure 6.
CENP-T promotes KMN kinetochore targeting through CENP-H-I-K. (A) HeLa Tet-On cells were mock transfected or transfected with siCENP-H, treated with thymidine for 14 h, released into nocodazole-containing medium for 12 h, and treated with or without ZM for 2 h. Their mitotic index was determined by flow cytometry. Means and SDs (error bars) are shown (n = 3 independent experiments). (B) HeLa cells were transfected with the indicated plasmids and siRNAs (siCC, siCENP-C; siCC+CH, siCENP-C+siCENP-H), and treated with nocodazole. Their mitotic index (mean ± SD [error bars], n = 3) was quantified by flow cytometry. (C and D) Nocodazole-treated mitotic HeLa cells transfected with siCENP-H were further incubated with MG132 (NM) or with both MG132 and ZM (NM+Z), and stained with the indicated antibodies and DAPI. Boxed regions of merged images were magnified and shown in the rightmost column. The relative kinetochore intensities (mean ± SD, n = 400) in certain channels were quantified and shown. Bars, 5 µm (1 µm for magnified images). (E) Recombinant Ndc80C was preincubated with or without recombinant Mis12C, immobilized on beads, and incubated with 35S-labeled CENP-H-I-K. Bound proteins and input were separated by SDS-PAGE, stained with CBB (left), and analyzed with a phosphorimager (right). CENP-H and -K co-migrate. The asterisk indicates a CENP-K fragment. Broken lines indicate that intervening lanes have been spliced out.
We then examined the potential interdependence between CENP-T and CENP-H in their kinetochore localization. Consistent with a previous report (Hori et al., 2008), depletion of CENP-H only slightly reduced CENP-T kinetochore signals (Fig. 6 D). In the same experiment, the Ndc80 kinetochore signal was reduced about twofold by CENP-H depletion, and was further reduced when Aurora B was inhibited (Fig. 6 D). Despite being incomplete, depletion of CENP-T reduced the kinetochore signals of CENP-H twofold (Fig. S5 D). Importantly, expression of either CENP-T or CENP-T ΔNBM restored the CENP-H kinetochore localization.
We next tested whether CENP-H-I-K physically interacted with Ndc80C. Purified recombinant His6-Ndc80C efficiently pulled down in vitro translated 35S-labeled CENP-H-I-K (Fig. 6 E). This Ndc80C–CENP-H-I-K interaction was not blocked by Mis12C, which suggests that Ndc80C can bind simultaneously to Mis12C and CENP-H-I-K. In contrast to a previous report that implicated CENP-H in Ndc80 binding (Mikami et al., 2005), we found that CENP-I on its own bound efficiently to Ndc80C, and it could also bridge an interaction between Ndc80C and CENP-H-K only when both CENP-H and -K were present (Fig. S5 E). Very recently, CENP-T-W has been shown to directly bind to the CENP-H-I-K-M complex (Basilico et al., 2014). Collectively, these results are consistent with a model in which CENP-T recruits CENP-H-I-K to kinetochores (Fig. 7 A). CENP-H-I-K then contributes to Aurora B–independent KMN attachment to kinetochores through physically interacting with Ndc80C.
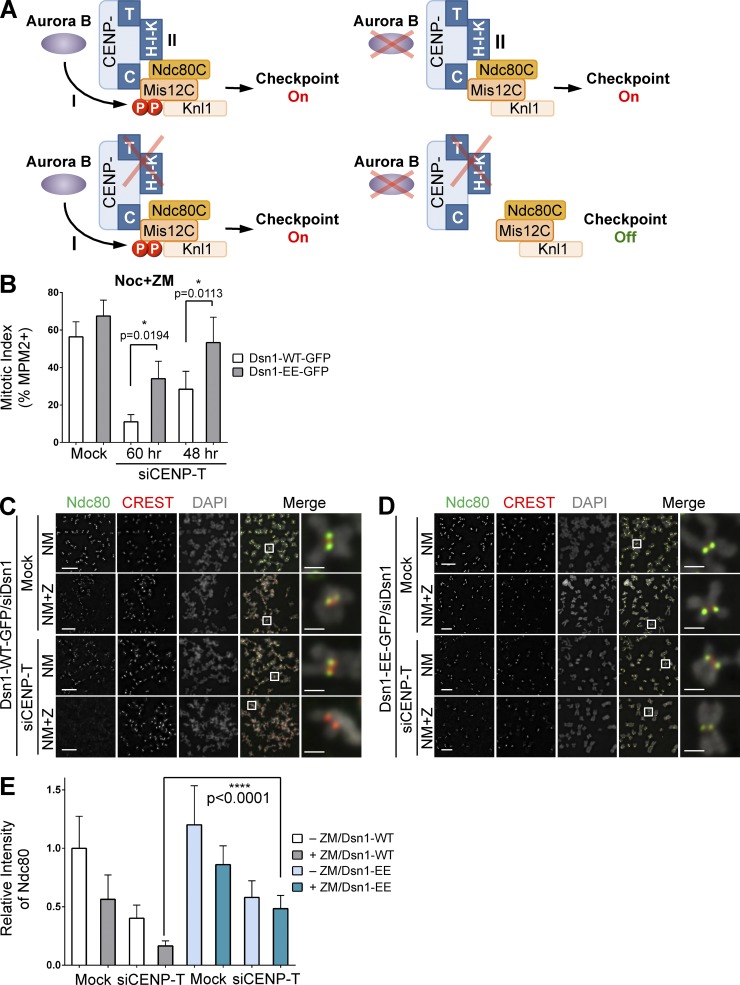
Figure 7.
Two semi-redundant mechanisms attach KMN to mitotic kinetochores. (A) Two proposed mechanisms of KMN assembly at kinetochores. In cells with depolymerized microtubules, inactivation of both is required to reduce the KMN level at kinetochores below that needed for checkpoint signaling. (B) Quantification of mitotic indices of Dsn1-WT/EE-GFP-expressing HeLa cells transfected with siCENP-T and treated with nocodazole and ZM (mean ± SD [error bars], n = 3). (C and D) Dsn1-WT/EE-GFP-expressing HeLa cells were transfected with siDsn1 and siCENP-T, arrested in mitosis by nocodazole, and further incubated with MG132 (NM) or with both MG132 and ZM (NM+Z). Mitotic cells were stained with DAPI and the indicated antibodies. The boxed regions of merged images are magnified and shown in the rightmost column. Bars, 5 µm (1 µm for magnified images). (E) Quantification of the relative kinetochore intensity of Ndc80 in C and D (mean ± SD [error bars], n = 400).
Two mechanisms contribute to KMN assembly at mitotic kinetochores
Our results so far suggest that at least two mechanisms attach KMN to kinetochores in mitosis (Fig. 7 A). In mechanism I, Aurora B phosphorylates Dsn1 and promotes the CENP-C–Mis12C interaction. In mechanism II, CENP-T anchors CENP-H-I-K at kinetochores, which interact with Ndc80C to promote KMN kinetochore targeting. It is well established that Aurora B inhibition alone is insufficient to abrogate the nocodazole-triggered mitotic arrest (Santaguida et al., 2011; Saurin et al., 2011). Thus, without the proper function of mechanism I, mechanism II is sufficient to maintain kinetochore KMN at a level above the threshold needed for checkpoint signaling. We wondered whether mechanism I could also transduce checkpoint signals when mechanism II was compromised. Expression of the phospho-mimicking Dsn1-EE, but not Dsn1-WT, partially restored nocodazole-induced mitotic arrest (Fig. 7 B) and the Ndc80 kinetochore signals (Fig. 7, C–E) in siCENP-T cells treated with ZM. Therefore, the two mechanisms of KMN attachment to kinetochores are not strictly dependent on each other for checkpoint signaling. When microtubules are depolymerized, only inactivation of both mechanisms can reduce the kinetochore level of KMN to below that required for checkpoint signaling.
Discussion
KMN as a checkpoint-signaling platform at mitotic kinetochores
Inactivation of certain KMN components causes spindle checkpoint defects in various organisms (McCleland et al., 2003; Kiyomitsu et al., 2007). Its components recruit key spindle checkpoint proteins, including Bub1–Bub3 and BubR1–Bub3, to kinetochores. In human cells, however, Ndc80C and Mis12C have not been definitively shown to be required for the mitotic arrest when all kinetochores are unattached (i.e., in the presence of high concentrations of nocodazole). A major reason for this stems from the high sensitivity of the spindle checkpoint. A few unattached kinetochores within a mitotic cell can engage the spindle checkpoint and cause prolonged mitotic arrest. Because a human cell contains 92 kinetochores (>100 for the aneuploid HeLa cell) during mitosis, the function of the kinetochore sensor has to be reduced to below a few percent of the wild-type level to reveal a strong checkpoint defect. This level of inactivation is difficult to attain experimentally, as KMN is a well-established kinetochore receptor for spindle microtubules, and is essential for cell viability. Insufficient inactivation of KMN often causes chromosome misalignment and checkpoint-dependent mitotic delay in human cells, complicating the analysis.
Through simultaneously targeting multiple components of two KMN subcomplexes—Mis12C or Ndc80C—we have suppressed both the kinetochore targeting and function of KMN in human cells below the thresholds needed to sustain a prolonged mitotic arrest, even when all kinetochores are unattached due to nocodazole-induced microtubule depolymerization. This mitotic arrest deficiency can be rescued by the ectopic expression of RNAi-resistant Mis12C or Ndc80C transgenes, ruling out dominant RNAi off-target effects. Our results confirm and extend numerous earlier findings, and clearly demonstrate a requirement for KMN in spindle checkpoint signaling in human cells. Being the kinetochore receptor for both microtubules and spindle checkpoint proteins, KMN is ideally suited to coordinate the generation and extinction of checkpoint signals.
The strategy of depleting multiple subunits of a given protein complex by RNAi to achieve more complete inactivation should be generally applicable. This strategy remains especially useful for protein complexes that are essential for cell viability, as simple deletion of their coding genes by new techniques, such as CRISPR (Mali et al., 2013), is not feasible.
KMN installation mechanisms at human mitotic kinetochores
Our study further reveals two semi-redundant mechanisms that install KMN at kinetochores during mitosis in human cells: the Aurora B–dependent CENP-C–Mis12C mechanism and the CENP-T–dependent Ndc80C mechanism (Fig. 7 A). Either mechanism is sufficient to install enough KMN at kinetochores to sustain prolonged mitotic arrest when microtubules are depolymerized. Inactivation of both mechanisms through Aurora B inhibition and partial depletion of CCAN or KMN components reduces KMN levels at kinetochores to below the threshold needed to sustain checkpoint signaling, lending further support for KMN being a critical sensor of the spindle checkpoint. Our model is consistent with an earlier study that demonstrated the synergistic effects of depleting both Knl1 and CENP-H-I-K in abolishing kinetochore functions in human cells (Cheeseman et al., 2008), although that study did not explicitly examine the spindle checkpoint.
We have further shown that Aurora B–dependent phosphorylation of Dsn1 is an important regulatory mechanism for mitosis-specific attachment of KMN at kinetochores. Strikingly, expression of the phospho-mimicking Dsn1 mutant, in conjunction with forced nuclear targeting of Ndc80, suffices to install the intact KMN at interphase kinetochores. In contrast, expression of phospho-mimicking CENP-T and forced nuclear targeting of Ndc80 only installs Ndc80C, but not Knl1 or Mis12C, at interphase kinetochores (Gascoigne and Cheeseman, 2013), which is consistent with the notion that CENP-T and Mis12C bind competitively to Ndc80C. Interestingly, forced nuclear targeting of Ndc80 alone (in the absence of phospho-mimicking Dsn1) is insufficient to install Ndc80C at interphase kinetochores, which indicates that the CENP-H-I-K–dependent mechanism on its own cannot install Ndc80C on interphase kinetochores. Because the CENP-H-I-K–dependent mechanism by itself can maintain a KMN pool at mitotic kinetochores in the absence of Aurora B activity, this result suggests the existence of additional, unidentified factors or modifications that promote this mechanism during mitosis. Further defining the CENP-H-I-K–Ndc80C interaction and its mitotic regulation remain important challenges for the future.
The mechanism by which CENP-T promotes CENP-H-I-K kinetochore localization is likely direct, as CENP-T-W binds to the CENP-H-I-K-M complex in vitro (Basilico et al., 2014). These two subcomplexes appear to be mutually dependent on each other for their kinetochore localizations, as disruption of the CENP-I-M interaction prevents the recruitment of CENP-T-W to kinetochores (Basilico et al., 2014).
We emphasize that our studies do not exclude the existence of additional KMN recruitment mechanisms. For example, in chicken cells, tethering of the N-terminal region of CENP-T to a noncentromeric locus is sufficient to establish an ectopic kinetochore that recruits KMN and supports chromosome segregation (Hori et al., 2013). Interestingly, the artificial kinetochore built with the CENP-T N-terminal region does not enrich CENP-A, -C, -H, -I, or -K, which suggests that this region of CENP-T on its own might be sufficient to recruit KMN.
Our results show that the small, conserved NBM in the N-terminal region of CENP-T (not the entire N-terminal region of CENP-T) contributes to the spindle checkpoint only when Aurora B is inhibited. Ndc80C bound to CENP-T NBM is, however, critical for chromosome segregation (Schleiffer et al., 2012; Malvezzi et al., 2013), and might contribute to kinetochore remodeling in anaphase (Bock et al., 2012).
Roles of Aurora B in mitotic regulation
Our results clearly demonstrate a role of Aurora B in mitosis-specific attachment of KMN to kinetochores through strengthening the CENP-C–Mis12C connection. However, this is not the only function of Aurora B in the spindle checkpoint, as expression of the phospho-mimicking Dsn1 mutant in HeLa cells treated with taxol fails to rescue the mitotic arrest deficiency caused by Aurora B inhibition (unpublished data). Aurora B phosphorylates other substrates at outer kinetochores, including Ndc80 and Knl1 (Welburn et al., 2010). These phosphorylation events likely contribute to the spindle checkpoint directly or indirectly. In addition, Aurora B is required for the kinetochore localization and activation of the checkpoint kinase Mps1, although the key substrates of Aurora B in that process have not been identified (Saurin et al., 2011; Nijenhuis et al., 2013).
Conclusion
The maturation of the outer kinetochore during mitosis in human cells is one of the most fascinating events in cell biology. Shortly after mitotic entry, tens of outer kinetochore proteins, including the KMN network, assemble onto the inner kinetochore in a hierarchical fashion. KMN then serves as the kinetochore receptor for both microtubules and spindle checkpoint proteins, and coordinates spindle checkpoint signaling. In this study, we have demonstrated a requirement for KMN in mediating mitotic arrest of human cells when all kinetochores are unattached. We have further delineated two mechanisms that anchor KMN to mitotic kinetochores. Our findings highlight a role of Aurora B in mitosis-specific KMN attachment to human kinetochores, and implicate a role of the CENP-H-I-K complex in mediating the interaction between CCAN and KMN.
Materials and methods
Cell culture and transfection
HeLa Tet-On (Takara Bio Inc.) cells with the reverse tetracycline-controlled transactivator (rtTA) integrated were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 6 mM l-glutamine, and 100 µg/ml penicillin and streptomycin. HeLa Tet-On cells stably expressing Dsn1-EGFP or mCherry-CENP-T were maintained with the growth medium containing 150 µg/ml hygromycin (Takara Bio Inc.). For G1/S arrest, cells were incubated in the growth medium containing 2 mM thymidine (Sigma-Aldrich) for 14–16 h. For prometaphase arrest, cells were arrested in G1/S with thymidine, washed twice with PBS, and incubated with the growth medium containing 500 nM or 5 µM nocodazole (Sigma-Aldrich) for 11–12 h. ZM447439 (Tocris Bioscience) was added to 4 µM to inhibit Aurora B.
For RNAi experiments, cells were transfected with Lipofectamine RNAiMAX (Invitrogen) and analyzed at 24–72 h after transfection. In the case of Dsn1 and Nsl1 RNAi, a second round of siRNA transfection was performed at 3 d after the initial transfection and a subsequent passage. The predesigned set of four siRNA oligonucleotides was purchased from Thermo Fisher Scientific and tested by Western blotting or immunostaining to identify the most effective siRNAs. The sequences (from 5′ to 3′) of siRNAs used in this study are: siControl (UCAUUCCGGAUACUGCGAUUU), siNdc80-4 (GAGUAGAACUAGAAUGUGA), siSpc24 (GGACACGACAGUCACAAUC), siSpc25 (CUACAAGGAUUCCAUCAAA), siDsn1 (GUCUAUCAGUGUCGAUUUA), siNsl1 (CAUGAGCUCUUUCUGUUUA), siCENP-C-6 (GAACAGAAUCCAUCACAAA), siCENP-C-9 (CGAAGUUGAUAGAGGAUGA), siCENP-T-1 (GAAGAACAUCCUACUAACU), siCENP-T-18 (AAGUAGAGCCCUUACACGA), siCENP-H (AGAUUGAUUUGGACAGUAU), siCENP-N (AUACACCGCUUCUGGGUCA), siCdc20 (CGAAAUGACUAUUACCUGA), siCENP-I (GAAGGUGUGUGACAUAUAU), siCENP-L (CCUCAAAGUCUGGACAUUU), siCENP-U-7 (GAAAGCCAUCUGCGAAAUA), siCENP-U-8 (GAAAAUAAGUACACAACGU), siCENP-U-9 (GGGAAGAUAUCUCAUGACA), siCENP-Q-17 (GAGUUAAUGACUGGGAAUA), siCENP-Q-19 (ACAAAGCACACUAACCUAA), and siCENP-Q-20 (UGUCAGAGAAUAAGGUUAG).
Site-directed mutagenesis was performed using the QuikChange kit (Agilent Technologies) to mutate the Aurora B phosphorylation sites (S100 and S109) in Dsn1 and to introduce silent mutations at the siRNA-targeting sites of Dsn1, Ndc80, and CENP-T transgenes. For artificial nuclear targeting of Ndc80, a plasmid encoding human Ndc80 fused at its C terminus to the SV40 NLS (with the amino acid sequence of PKKKRKV) was constructed by PCR. The CENP-T mutant with its Ndc80C-binding motif (residues 85–99) deleted (ΔNBM) and Dsn1 Δ92–113 were made with two-step PCR. A condon-optimized and RNAi-resistant CENP-C (Genscript) cDNA was used as a template to construct CENP-C1–71 or CENP-C Δ400 fragments by PCR. For transient or stable protein expression, the relevant cDNAs were cloned into pCS2 vectors with the CMV promoter or pTRE2hyg vectors with the tetracycline-responsive promoter (Takara Bio Inc.), respectively. All constructs were verified by DNA sequencing.
Plasmid transfection was performed with the Effectene reagent (QIAGEN) when cells reached a confluency of ∼60% according to the manufacturer’s instructions. For stable cell line generation, HeLa Tet-On cells were transfected with pTRE2 vectors encoding RNAi-resistant Dsn1 WT, S100E/S109E (EE), S100A/S109A (AA), and Δ92–113 tagged at the C terminus with EGFP, or RNAi-resistant CENP-T WT or ΔNBM tagged at the N terminus with mCherry. Cells were selected with 150 µg/ml hygromycin (Takara Bio Inc.). The surviving clones were screened for expression of the desired proteins in the presence of 1 µg/ml doxycycline (Sigma-Aldrich). For all the following experiments, 1 µg/ml doxycycline was added to induce expression unless otherwise indicated.
Protein purification
The pST39 Mis12C plasmids encoding Dsn1-WT or EE, Nsl11–258, Mis12, and Nnf1 were constructed from the original pST39 Mis12C plasmid (a gift from A. Desai, University of California, San Diego, San Diego, CA). For expression of Mis12C containing Dsn1-WT or -EE, the full-length (FL) Nsl1, Mis12, and Nnf1, a PreScission protease cleavage site and two tandem copies of the Strep tag were fused to the C terminus of Nsl1. These plasmids were transformed into BL21(DE3)pLysS cells (Invitrogen), and protein expression was induced with 0.1 mM IPTG at 16°C for overnight. The Mis12C complexes were purified essentially as described previously (Petrovic et al., 2010). In brief, cell pellets were resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% [vol/vol] glycerol, and 2 mM 2-mercaptoethanol) supplemented with a protease inhibitor cocktail (Roche) and TurboNuclease (Accelagen), and lysed with a cell disrupter. The cleared lysate was applied to Ni2+-NTA agarose beads (QIAGEN) or Strep-Tactin Superflow agarose beads (QIAGEN) preequilibrated in buffer A and incubated for 2 h. Beads were washed with 30 volumes of buffer A, and, in the case of Mis12C with Nsl1 FL, cleaved with the PreScission protease to remove the Strep tag. The proteins were further purified by anion exchange chromatography with a Resource Q column (GE Healthcare) followed by size exclusion chromatography with a Superdex 200 10/300 column (GE Healthcare).
Ndc80C was expressed in insect cells. Cell pellets were resuspended in buffer B (50 mM Tris-HCl, pH 8.0, 150 mM KCl, 0.1% Triton X-100, 5% [vol/vol] glycerol, 10 mM imidazole, and 5 mM 2-mercaptoethanol) supplemented with a protease inhibitor cocktail (Roche) and TurboNuclease (Accelagen), and lysed by a cell disrupter. The cleared lysate was applied to Ni2+-NTA agarose beads (QIAGEN) preequilibrated in buffer B and incubated for 2 h. Beads were washed with 40 volumes of buffer B, and eluted serially with buffer B containing 50 mM, 100 mM, and 250 mM imidazole. The protein was further purified by size exclusion chromatography with a Superdex 200 column (GE Healthcare).
A pGEX-duet vector encoding GST-tagged human Spc2492–197 and untagged Spc2570–224 was transformed into Rosetta (DE3)pLysS cells (EMD Millipore). Protein expression was induced with 0.2 mM IPTG at 16°C for overnight. The cells were lysed by sonication in the lysis buffer (20 mM Tris-HCl, pH 7.7, 300 mM NaCl, 2 mM DTT, and 1 mM EDTA). The GST-Spc24–Spc25 complex was bound to a Glutathione Sepharose 4B (GE Healthcare) column, and was incubated with HRV 3C protease at 4°C for overnight to remove the GST tag. The cleaved complex was further purified with Resource Q and Superdex 200 columns (GE Healthcare).
Antibodies, immunoblotting, and immunoprecipitation
For the generation of antibodies against human Knl1, CENP-C, and EGFP, GST- or His6-tagged fragments of Knl1 (residues 1,531–1,808), CENP-C (residues 1–165), and the full-length EGFP were expressed in bacteria and purified with the Glutathione Sepharose 4B (GE Healthcare) or Ni2+-NTA agarose (QIAGEN) resins. The purified fusion proteins were cleaved with HRV 3C or TEV proteases to remove the GST or His6 tags. For the generation of antibodies against human Mis12 complex (Mis12C), Mis12C containing Dsn1, Nsl1, Mis12, and Nnf1 was expressed in bacteria and purified as described in the Protein purification section. For the generation of the α-Spc24/25 antibody, an Spc24–Spc25 heterodimer containing residues 90–197 of human Spc24 and residues 70–224 of human Spc25 was expressed in bacteria and purified as described in the Protein purification section. The proteins were used to immunize rabbits at Yenzym Antibodies, LLC. Antibodies were affinity purified with the corresponding antigens coupled to aldehyde-activated agarose beads (Thermo Fisher Scientific) according to the manufacturer’s protocols. Production of rabbit polyclonal antibodies against human Mad1, Mad2, APC2, Bub1, and Ndc80 has been described previously (Bharadwaj et al., 2004; Kang et al., 2008; Kim et al., 2012). The α-Dsn1-pS100 antibody was produced at an in-house facility by immunizing rabbits with a mixture of the pS100 peptide (SWRRApSMKETNC) and the pS109 peptide (TNRRKpSLHPIHC) coupled to hemocyanin (Sigma-Aldrich). The α-Dsn1-pS100 antibody was purified with the pS100 peptide coupled to iodoacetyl-activated agarose beads (Thermo Fisher Scientific) according to the manufacturer’s protocols. The commercial antibodies used in this study were: rat anti–CENP-T (D286-3; MBL), rat anti–CENP-N (D285-3; MBL), rabbit anti–CENP-H (PD031; MBL), mouse anti–CENP-C (ab50974; Abcam), mouse anti-GFP (11814460001; Roche), mouse anti-mCherry (632543; Takara Bio Inc.), mouse anti-actin (MA1-37018; Thermo Fisher Scientific), mouse anti–α-tubulin (T9026; Sigma-Aldrich), mouse anti–β-tubulin (T4026; Sigma-Aldrich), human CREST autoimmune sera (HCT-0100; ImmunoVision), and mouse α-MPM2 (05-368; EMD Millipore).
For immunoblotting, antibodies were used at a concentration of 1 µg/ml for purified and monoclonal antibodies or at 1:1,000 dilution for crude sera. For immunoprecipitation, anti-Mis12C antibodies were affinity-purified and coupled to Affi-Prep Protein A beads (Bio-Rad Laboratories) at a concentration of 1 mg/ml. HeLa Tet-On cells were washed once with PBS and resuspended with the lysis buffer (25 mM Tris-HCl, pH 8.0, 75 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.1% NP-40, 10% [vol/vol] glycerol, 1 mM DTT, 10 mM NaF, 5 mM β-glycerophosphate, 0.5 µM okadaic acid, and protease inhibitor cocktail [Roche]) supplemented with 50 U/ml Turbo-nuclease (Accelagen). Cells were lysed with three cycles of freezing and thawing, and kept on ice for 1 h followed by a 10-min incubation at 37°C. The lysate was cleared by centrifugation for 20 min at 4°C at 20,817 g. The antibody beads were incubated with the supernatant for 2 h at 4°C with gentle rotation, and then washed four times with the lysis buffer. The proteins bound to beads were dissolved in SDS sample buffer, boiled, separated by SDS-PAGE, and blotted with the desired antibodies. The beads coupled with rabbit IgG (I5006; Sigma-Aldrich) were used as a negative control.
Live-cell imaging
Cells were grown and transfected in 12-well plates, passaged onto chambered coverslips (Lab-Tek), incubated with 2 mM thymidine (Sigma-Aldrich) for 14–16 h, and released into fresh media for 4–6 h before taking images. GFP or mCherry fluorescence images were acquired at 5-min intervals with a deconvolution fluorescence microscope (DeltaVision Core; Applied Precision) equipped with an environmental chamber (37°C and 5% CO2) and a CoolSNAP HQ2 camera (Roper Scientific). For each time point, five z-sections at 2.5-µm intervals were acquired by using a 100× 1.40 NA UPLS Apochromat objective lens (Olympus) with 2 × 2 binning. GFP or mCherry fluorescence were observed with the appropriate filters. The images were deconvolved with the Deconvolution tool in SoftWoRx (Applied Precision) that used the iterative-constrained algorithm. For 2D image presentation, z-series optical sections were projected by the Max Intensity method in SoftWoRx. Images were further processed with ImageJ and Photoshop (Adobe).
Immunofluorescence and chromosome spreads
For the staining of kinetochore proteins, mitotic cells were harvested by shake-off, washed once with PBS, resuspended in a prewarmed hypotonic solution (55 mM KCl), and incubated for 15 min at 37°C. Swollen cells were spun onto slides with a Shandon Cytospin 4 centrifuge (Thermo Fisher Scientific), extracted with PBS containing 0.2% Triton X-100, and fixed in 4% paraformaldehyde at room temperature for 5 min. After washing three times with PBS, the fixed cells were incubated with the desired primary antibodies and CREST in PBS containing 0.1% Triton X-100 and 3% BSA for overnight at 4°C. After washing three times with PBS containing 0.1% Triton X-100, cells were further incubated with 1 µg/ml fluorescent secondary antibodies (Invitrogen) in PBS containing 0.1% Triton X-100 and 3% BSA for 1 h at room temperature. Cells were again washed three times with PBS containing 0.1% Triton X-100 and stained with 1 µg/ml DAPI for 5 min. After the final washes, the slides were mounted and analyzed with the deconvolution fluorescence microscope (DeltaVision Core; Applied Precision). Images were acquired with 100× 1.40 NA UPLS Apochromat or 60× 1.42 NA Plan-Apochromat N objective lenses (Olympus) at 37°C through glycerol-based Aqua-Poly/Mount media (PolySciences). Alexa Fluor 488, 568, or 647, or DAPI florescence was observed with the appropriate filter sets. A series of z-stack images were captured at 0.5-µm intervals for kinetochore images. All images in each experiment were taken with the same light intensity and exposure time. Images were deconvolved as described in the Live-cell imaging section, and projected by the Sum Intensity method in SoftWoRx (Applied Precision). Quantification of the relative intensity of kinetochore signals was done with ImageJ. In brief, a circle that enclosed CREST signals from a pair of kinetochores was drawn and set as the region of interest (ROI). The integrated density of the gray value for the selected ROI was measured from each channel. The value of object intensity was then divided by the corresponding value of CREST intensity. 20 ROIs per cell chosen at random were measured. The graphs and statistics were generated with Prism (GraphPad Software). For presentation, images were further processed with Photoshop (Adobe).
Flow cytometry
Cells were collected by trypsinization, washed once with PBS, and fixed with cold 70% ethanol overnight at −20°C. Cells were washed once with PBS, permeabilized with PBS containing 0.25% Triton X-100 for 5 min, and then incubated with the mouse α-MPM2 antibody diluted in PBS containing 1% BSA for 3 h. After being washed once with PBS containing 1% BSA, cells were incubated with Alexa Fluor 488 fluorescent secondary antibodies against mouse IgG (A-21202; Invitrogen) diluted in PBS plus 1% BSA for 30 min. After one more wash with PBS, cells were stained with propidium iodide (Sigma-Aldrich) at a final concentration of 20 µg/ml, and simultaneously treated with 200 µg/ml RNase A (QIAGEN). The samples were analyzed with FACSCalibur (BD) and the FlowJo software (Tree Star).
Kinase and protein-binding assays
About 0.1 µg of recombinant Aurora B–INCENP was incubated with 2 µg of Mis12C or MBP substrates for 30 min at 30°C in 25 µl of the kinase buffer (50 mM Tris-HCl, pH 7.5, 0.2 M NaCl, 1 mM DTT, 10 mM NaF, 5 mM β-glycerophosphate, 0.5 µM okadaic acid, protease inhibitor cocktail [Roche], 0.1 mM ATP, and 0.1 µCi/µl γ-[32P]ATP). Reaction mixtures were quenched with the SDS sample buffer, separated on SDS-PAGE, and analyzed using a phosphorimager (Fuji). For testing the specificity of the α-Dsn1-pS100 antibody, kinase assays on Mis12C-WT and Mis12C-EE were performed with cold ATP, and samples were analyzed by immunoblotting.
For protein-binding assays, purified Mis12C were immobilized on Ni2+-NTA beads (QIAGEN) preequilibrated in the binding buffer (25 mM Tris-HCl, pH 8.0, 75 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.1% NP-40, 10% [vol/vol] glycerol, 1 mM DTT, 10 mM NaF, 5 mM β-glycerophosphate, and protease inhibitor cocktail [Roche]), and incubated with the blocking solution (the binding buffer containing 5% nonfat milk). The [35S]methionine-labeled proteins were produced with coupled in vitro transcription and translation of the pCS2-myc-CENP-C1–71 or pCS2-CENP-H, -I, or -K vectors in reticulocyte lysate using the TNT SP6 kit (Promega), and incubated with the Mis12C-bound beads in the blocking solution for 1 h at room temperature. The beads were washed three times with the binding buffer, and the bound proteins were separated by SDS-PAGE and analyzed with a phosphorimager (Fuji). The Myc-CENP-C full-length and Δ400 proteins were produced with in vitro translation in the presence of unlabelled methionine and detected by immunoblotting.
For competition assays, 0.1 µM of His6-tagged Mis12C was incubated with 0.05 µM of purified Spc24–Spc25 complex in the binding buffer (25 mM Tris-HCl, pH 8.0, 75 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.1% NP-40, 10% [vol/vol] glycerol, 1 mM DTT, 10 mM NaF, 5 mM β-glycerophosphate, and protease inhibitor cocktail [Roche]), in the presence of 0, 0.1, 1.0, and 10 µM of the synthetic phospho-T85 CENP-T peptide containing residues 80–104. The reaction mixtures were then incubated with Ni2+-NTA beads (QIAGEN) preequilibrated in the binding buffer, and washed three times with the binding buffer. The bound proteins were separated by SDS-PAGE and analyzed by immunoblotting.
Microscale thermophoresis (MST) measurements were performed with a Monolith NT.115 instrument (NanoTemper) at 24°C. The fluorescein-labeled CENP-C1–28 peptide (5FAM-MAASGLDHLKNGYRRRFCRPSRARDINT) at a fixed concentration (200 nM) was mixed with increasing concentrations of Mis12C-WT or -EE in PBS containing 0.1% NP-40. The samples were loaded into hydrophilic capillaries (NanoTemper) after a 90-min incubation at room temperature. Measurements were performed with 20% LED power and 40% IR-laser power according to the manufacturer’s instructions. Data analyses were performed using the PALMIST software.
Online supplemental material
Fig. S1 shows the interdependency between Mis12C and Ndc80C at kinetochores. Fig. S2 shows that Dsn1 phosphorylation contributes to spindle checkpoint signaling. Fig. S3 provides additional data showing that phospho-mimicking and Δ92–113 mutants of Dsn1 localize to interphase kinetochores and that Dsn1 phosphorylation enhances the Mis12C–CENP-C interaction. Fig. S4 contains data showing that depletion of CENP-T and Aurora B inhibition compromise KMN kinetochore localization and that depletion of several other CENP proteins does not synergize with Aurora B inhibition to produce checkpoint defects. Fig. S5 shows that CENP-T contributes to CENP-H kinetochore localization independently of its NBM. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201407074/DC1.
Supplementary Material
Acknowledgments
We thank Arshad Desai for providing the pST39-Mis12C plasmid, Haydn Ball for peptide synthesis, Chad Brautigam for assistance with microscale thermophoresis, and the UT Southwestern Proteomics Core Facility (supported by Cancer Prevention and Research Institute of Texas grant RP120613) for mass spectrometry analysis. H. Yu is an investigator with the Howard Hughes Medical Institute.
This work is supported by the Cancer Prevention and Research Institute of Texas (RP110465-P3 and RP120717-P2) and the Welch Foundation (I-1441).
The authors declare no further competing financial interests.
Footnotes
Abbreviations used in this paper:
- CCAN
- constitutive centromere-associated network
- IP
- coimmunoprecipitation
- NBM
- Ndc80-binding motif
- ZM
- ZM447439
References
- Akiyoshi B., Nelson C.R., and Biggins S.. 2013. The Aurora B kinase promotes inner and outer kinetochore interactions in budding yeast. Genetics. 194:785–789 10.1534/genetics.113.150839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico F., Maffini S., Weir J.R., Prumbaum D., Rojas A.M., Zimniak T., De Antoni A., Jeganathan S., Voss B., van Gerwen S., et al. 2014. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 3:e02978 10.7554/eLife.02978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj R., Qi W., and Yu H.. 2004. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 279:13076–13085 10.1074/jbc.M310224200 [DOI] [PubMed] [Google Scholar]
- Black B.E., and Cleveland D.W.. 2011. Epigenetic centromere propagation and the nature of CENP-a nucleosomes. Cell. 144:471–479 10.1016/j.cell.2011.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock L.J., Pagliuca C., Kobayashi N., Grove R.A., Oku Y., Shrestha K., Alfieri C., Golfieri C., Oldani A., Dal Maschio M., et al. 2012. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 14:614–624 10.1038/ncb2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., and Desai A.. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., and Desai A.. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., and Sullivan K.F.. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Collin P., Nashchekina O., Walker R., and Pines J.. 2013. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat. Cell Biol. 15:1378–1385 10.1038/ncb2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick A.E., and Gerlich D.W.. 2013. Kinetic framework of spindle assembly checkpoint signalling. Nat. Cell Biol. 15:1370–1377 10.1038/ncb2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S., and Stukenberg P.T.. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., and Kapoor T.M.. 2013. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 14:25–37 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D.R., Jansen L.E., Black B.E., Bailey A.O., Yates J.R. III, and Cleveland D.W.. 2006. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8:458–469 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- Gascoigne K.E., and Cheeseman I.M.. 2013. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol. 201:23–32 10.1083/jcb.201301006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne K.E., Takeuchi K., Suzuki A., Hori T., Fukagawa T., and Cheeseman I.M.. 2011. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 145:410–422 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S., Geissen E.M., Kamenz J., Trautmann S., Widmer C., Drewe P., Knop M., Radde N., Hasenauer J., and Hauf S.. 2013. Determinants of robustness in spindle assembly checkpoint signalling. Nat. Cell Biol. 15:1328–1339 10.1038/ncb2864 [DOI] [PubMed] [Google Scholar]
- Hori T., Amano M., Suzuki A., Backer C.B., Welburn J.P., Dong Y., McEwen B.F., Shang W.H., Suzuki E., Okawa K., et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 135:1039–1052 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- Hori T., Shang W.H., Takeuchi K., and Fukagawa T.. 2013. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J. Cell Biol. 200:45–60 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L., Kim S., and Yu H.. 2013. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 38:302–311 10.1016/j.tibs.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Kang J., Yang M., Li B., Qi W., Zhang C., Shokat K.M., Tomchick D.R., Machius M., and Yu H.. 2008. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol. Cell. 32:394–405 10.1016/j.molcel.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Sun H., Tomchick D.R., Yu H., and Luo X.. 2012. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl. Acad. Sci. USA. 109:6549–6554 10.1073/pnas.1118210109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., and Yanagida M.. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Krenn V., Overlack K., Primorac I., van Gerwen S., and Musacchio A.. 2014. KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr. Biol. 24:29–39 10.1016/j.cub.2013.11.046 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., and Cheeseman I.M.. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P., Westhorpe F.G., and Taylor S.S.. 2012. The spindle assembly checkpoint. Curr. Biol. 22:R966–R980 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., and Biggins S.. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Esvelt K.M., and Church G.M.. 2013. Cas9 as a versatile tool for engineering biology. Nat. Methods. 10:957–963 10.1038/nmeth.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi F., Litos G., Schleiffer A., Heuck A., Mechtler K., Clausen T., and Westermann S.. 2013. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J. 32:409–423 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V.M., and Nigg E.A.. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- Matson D.R., and Stukenberg P.T.. 2014. CENP-I and Aurora B act as a molecular switch that ties RZZ/Mad1 recruitment to kinetochore attachment status. J. Cell Biol. 205:541–554 10.1083/jcb.201307137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson D.R., Demirel P.B., Stukenberg P.T., and Burke D.J.. 2012. A conserved role for COMA/CENP-H/I/N kinetochore proteins in the spindle checkpoint. Genes Dev. 26:542–547 10.1101/gad.184184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., Gardner R.D., Kallio M.J., Daum J.R., Gorbsky G.J., Burke D.J., and Stukenberg P.T.. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114 10.1101/gad.1040903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami Y., Hori T., Kimura H., and Fukagawa T.. 2005. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol. Cell. Biol. 25:1958–1970 10.1128/MCB.25.5.1958-1970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijenhuis W., von Castelmur E., Littler D., De Marco V., Tromer E., Vleugel M., van Osch M.H., Snel B., Perrakis A., and Kops G.J.. 2013. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201:217–231 10.1083/jcb.201210033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Rago F., Hori T., Tomii K., Cheeseman I.M., and Fukagawa T.. 2013. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J. 32:424–436 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I.M., Hori T., Okawa K., McLeod I.X., Yates J.R. III, Desai A., and Fukagawa T.. 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8:446–457 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- Petrovic A., Pasqualato S., Dube P., Krenn V., Santaguida S., Cittaro D., Monzani S., Massimiliano L., Keller J., Tarricone A., et al. 2010. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190:835–852 10.1083/jcb.201002070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A., Mosalaganti S., Keller J., Mattiuzzo M., Overlack K., Krenn V., De Antoni A., Wohlgemuth S., Cecatiello V., Pasqualato S., et al. 2014. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol. Cell. 53:591–605 10.1016/j.molcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Primorac I., Weir J.R., Chiroli E., Gross F., Hoffmann I., van Gerwen S., Ciliberto A., and Musacchio A.. 2013. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2:e01030 10.7554/eLife.01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., and Sluder G.. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., and Earnshaw W.C.. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Vernieri C., Villa F., Ciliberto A., and Musacchio A.. 2011. Evidence that Aurora B is implicated in spindle checkpoint signalling independently of error correction. EMBO J. 30:1508–1519 10.1038/emboj.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.T., van der Waal M.S., Medema R.H., Lens S.M., and Kops G.J.. 2011. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2:316 10.1038/ncomms1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiffer A., Maier M., Litos G., Lampert F., Hornung P., Mechtler K., and Westermann S.. 2012. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 14:604–613 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- Screpanti E., De Antoni A., Alushin G.M., Petrovic A., Melis T., Nogales E., and Musacchio A.. 2011. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 21:391–398 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., and Millar J.B.. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke V.M., Baumann C., and Nigg E.A.. 2004. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 113:1–15 10.1007/s00412-004-0288-2 [DOI] [PubMed] [Google Scholar]
- Subramanian R., and Kapoor T.M.. 2013. Slipping past the spindle assembly checkpoint. Nat. Cell Biol. 15:1261–1263 10.1038/ncb2876 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., and Fukagawa T.. 2012. Molecular architecture of vertebrate kinetochores. Exp. Cell Res. 318:1367–1374 10.1016/j.yexcr.2012.02.016 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., and Nasmyth K.. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Vleugel M., Tromer E., Omerzu M., Groenewold V., Nijenhuis W., Snel B., and Kops G.J.. 2013. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J. Cell Biol. 203:943–955 10.1083/jcb.201307016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R. III, Lampson M.A., Fukagawa T., and Cheeseman I.M.. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., and Watanabe Y.. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., Yan F., Wu Q., Wang Z., McGlothen T., Peng W., You T., Sun M., et al. 2008. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 283:26726–26736 10.1074/jbc.M804207200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.